Abstract
Although levels of minimal residual disease (MRD) decrease below the detection limit in most adult patients with standard-risk acute lymphoblastic leukemia (ALL) after consolidation treatment, about 30% of these patients will ultimately relapse. To evaluate the power of MRD monitoring as an indicator of impending relapse, we prospectively analyzed postconsolidation samples of 105 patients enrolled in the German Multicenter ALL (GMALL) trial by real-time quantitative polymerase chain reaction (PCR) of clonal immune gene rearrangements. All patients were in hematologic remission, had completed first-year polychemotherapy, and tested MRD negative prior to study entry. Twenty-eight of 105 patients (27%) converted to MRD positivity thereafter, and 17 of 28 (61%) relapsed so far. Median time from molecular (MRD-positive) to clinical relapse was 9.5 months. In 15 of these patients, MRD within the quantitative range of PCR was measured in hematologic remission, and 13 of these patients (89%) relapsed after a median interval of 4.1 months. Of the 77 continuously MRD-negative patients, only 5 (6%) have relapsed. We conclude that conversion to MRD positivity during the early postconsolidation phase in adult standard-risk ALL patients is highly predictive of subsequent hematologic relapse. As a result of the study, as of spring 2006, salvage treatment in the ongoing GMALL trial is intended to be started at the time of recurrence of quantifiable MRD.
Introduction
Treatment of adult patients with acute lymphoblastic leukemia (ALL) is becoming increasingly complex as diverse treatment protocols are introduced for different subtypes of the disease, reflecting the intention to optimally tailor therapy to specific risk-adapted disease entities. Recent improvements have been achieved by introducing new therapeutic principles, such as the early addition of the tyrosine kinase inhibitor imatinib in Ph-positive ALL1 or the use of the anti-CD20 antibody rituximab in CD20+ cases of B-lineage ALL. Diagnostic improvements were achieved by assessing the level of minimal residual disease (MRD) either by molecular genetic methods or by flow cytometry, which has been shown to be predictive for outcome in a number of studies in children2-6 and adults.7-10
Survival rates with modern treatment protocols for adult ALL patients have reached a plateau where the potential benefit of more aggressive chemotherapeutic regimens is often offset by an excess mortality due to complications, thus making efforts to individualize treatment even more important. Whereas standard-risk patients without conventional risk factors, who have a greater than 50% chance of long-term survival with chemotherapy alone,11 are potentially put at unnecessary risk by intensified and prolonged therapy, outcome in patients with relapsed ALL is extremely poor, even if a second remission is achieved. Therefore, starting with the German Multicenter ALL (GMALL) 06/99 trial and continuing in the 07/03 trial, we prospectively monitored MRD in standard-risk patients during and after completion of therapy. Molecular detection of an impending relapse may help to initiate salvage therapy prior to hematologic relapse, with a respective lower tumor burden and possibly improved treatment results. Whereas there are data in acute promyelocytic leukemia (APL) suggesting that administration of salvage therapy early in molecular relapse may improve prognosis,12 conclusive similar data are lacking in ALL. This may partly be due to the fact that, apart from the Ph-positive cases, until now no study has shown conclusively in a predefined cohort that the timely anticipation of an impending relapse in ALL is possible.
MRD monitoring during the first year of intensive chemotherapy led to an MRD-based risk stratification that has been published recently.10 This classification allows the identification of an MRD low-risk group consisting of about 10% of patients with a minimal chance of relapse at 3 years, an MRD high-risk group of about 25% of patients with an almost 100% risk of relapse, and an MRD intermediate-risk group. In the latter group, about 30% of patients will eventually relapse despite becoming MRD negative or reaching MRD levels below 10−4 at the end of the first year of therapy.
Therefore, the aim of this study was to evaluate whether additional MRD monitoring during the early postconsolidation phase in MRD-negative patients detects the recurrence of disease prior to hematologic relapse. Based on our findings, molecular relapse is now defined as conversion to molecularly quantifiable MRD that leads to an anticipation of salvage therapy prior to hematologic relapse in the ongoing GMALL trial.
Patients, materials, and methods
Patients
All investigated patients were enrolled into the 06/99 and 07/03 trials of the GMALL Study Group and thus were confirmed to have ALL by reference morphology and immunophenotyping. In these studies, patients without clinical (high leukocyte count at presentation, no complete remission [CR] after induction phase I), immunologic (pro-B, pre-T, and mature T-ALL), or cytogenetic (detection of the translocations t(4;11) and t(9;22)) risk factors were stratified into the standard-risk group and were prospectively monitored for MRD during the first year of treatment, as published recently.10 Data of 56 patients of that cohort are included in this study (9 MRD low-, 39 MRD intermediate-, and 8 MRD high-risk patients based on the criteria developed in that study). The complementary data presented here were obtained by further monitoring MRD-negative patients after the first year of intensive chemotherapy to test whether reconversion to MRD positivity precedes hematologic relapse. After the end of the first year of therapy, prospective MRD monitoring was recommended for MRD-negative patients in 3-month intervals during maintenance therapy and follow-up. Consecutive patients participating in the GMALL 06/99 and 07/03 trials were eligible if their diagnostic sample was available to us before the end of 2003 and if they fulfilled the following criteria: (1) patients belonged to the standard-risk group, (2) patients had become MRD negative during the first year of therapy as determined by at least one molecular marker with a quantitative range of at least 10−4 (ie, the patient was reproducibly MRD negative at that level), (3) after the first year of therapy (week 52) patients were either MRD negative or MRD positive below a quantitative range of 10−4, and (4) at least one follow-up material in hematologic remission was available after the first year of treatment.
Treatment
Treatment for the standard-risk patients consists of an induction, reinduction, and 6 cycles of consolidation therapy lasting 1 year based on the drugs dexamethasone, cyclophosphamide, vincristine, daunorubicin, methotrexate, asparaginase, cytosine arabinoside, 6-MP, VP16, and VM26 (for details, refer to the study protocol available in short form from the website of the European Leukemia Net13 and in complete form from the study coordination center in Frankfurt, Germany, the address of which is also available on the website13 ). The recommended maintenance therapy consists of alternating drug combinations similar to the consolidation cycles with intermediary application of 6-mercaptopurine plus methotrexate for 52 weeks.
The study was approved by the institutional committees of the study coordination center in Frankfurt, Germany, and of the participating centers. All patients had given signed informed consent as to the scientific use of their data.
MRD assessment
Samples obtained at the time of diagnosis were screened for clonal immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangements of the IgH, Igκ-Kde, TCRβ, γ, and δ locus using published primer sets.14-17 Clonal immune gene rearrangements were identified by GeneScanning (using ABI Prism 310 and 377 genetic analyzers from Applied Biosystems, Darmstadt, Germany) and/or heteroduplex gel analysis and sequenced.15 MRD then was determined in cryopreserved Ficoll-separated cells from bone marrow samples by quantitative real-time polymerase chain reaction (PCR) using clone-specific primers for the leukemia-specific Ig/TCR gene rearrangements and a set of different germ line TaqMan probes (13 TCRβ-Jβ, 8 TCRγ-Vγ, 1 TCRδ-Jδ1, 1 TCRδ-Dδ2, 3 TCRδVδ, 4 IgH-JH, and 1 Igκ-KDE) and germ line primers.18-21 MRD levels were stated as the proportion of leukemic cells to all nucleated cells in the sample. The aim was to obtain 2 molecular markers per patient with a sensitivity of at least 10−4. If MRD levels differed between assays, the higher MRD level was assumed to be more accurate. PCR results were interpreted according to the common criteria of the laboratories organized in the European Study Group on MRD-Detection in ALL (ESG MRD-ALL; coordinator, J. J.M. van Dongen, Rotterdam, The Netherlands). These criteria define the sensitivity of the assay as the lowest dilution step giving specific amplification, with at least 1 well positive within 20 cycles from the undiluted sample and a threshold cycle (Ct) of at least 1.0 Ct value lower than lowest Ct value of background. The quantitative range is defined as the lowest dilution giving specific amplification where the ΔCt replicates differ by not more than 1.5 Ct values and all Ct values are at least 3.0 Ct values lower than lowest Ct value of background. Furthermore, the mean Ct value of each 10-fold dilution step has to be 2.6 to 4.0 higher than the previous 10-fold dilution with a standard curve of at least 2 previous dilutions with a slope between −3.1 and −3.9 and a correlation coefficient of at least 0.98. All MRD results were qualitatively interpreted as MRD negative, MRD below quantitative range (QR), and MRD positive within QR with quantitative MRD values being stated only in the latter case. The terms were defined according to the sensitivity and QR of PCR as follows: MRD negativity means the absence of a specific amplification or amplification within 1.0 Ct of the background or amplification with a distance of more than 20 Cts (ΔCt) from the undiluted diagnostic sample. MRD positivity was stated if the follow-up sample showed a specific amplification product of more than 1 Ct lower than the background and separated less than 20 Cts (ΔCt) from the undiluted diagnostic sample. An exact MRD value was given if the result was within the QR of the PCR and the ΔCt between replicates was less than 1.5. For additional technical details, see Brüggemann et al19 and van der Velden et al.20
Statistical analysis
Survival curves were estimated according to the method of Kaplan and Meier22 ; survival was compared using the Anderson-Gill reformulation of the Cox proportional hazards model as a counting process23 with the median times to relapse after conversion to MRD positivity or MRD within quantitative range used as the cutting intervals (Figure 3). Briefly, every patient's observation period is split into multiple, independently treated intervals, each with the length of the cutting interval or the remaining time to event, whichever is shorter. MRD positivity and MRD within quantitative range of PCR were treated as binary time-dependent covariates (BTDCs) reflecting a nonreversible change of group membership (ie, the conversion from MRD negativity to MRD positivity or the first time the MRD was measured inside the quantitative range of the PCR changed the status of the patient for all further observations). The proportional hazards assumption of the Cox model was confirmed by the method of Grambsch and Therneau24 using the correlation coefficient between survival time and the scaled Schoenfeld residuals.25 The influence of age at diagnosis and length of initial therapy were analyzed by univariate Cox regression. All calculations were done using the open source R implementation of the S statistical language26 (V 2.2.1; packages used: base, survival, MASS, graphics, lattice).
Results
Patients and samples
A total of 105 patients fulfilled the criteria outlined in “Patients, materials, and methods,” from whom we analyzed a total of 519 samples with a median time of active monitoring of 16.1 months (range, 0.7-41.1 months) and a median number of 3 samples (range, 1-12 samples) per patient after the end of the first year of therapy. Summary data of the patients including samples analyzed and molecular markers used for MRD monitoring as well as the sensitivities and quantitative ranges of the assays are given in Table 1. There was a 2.5:1 male-female ratio and a 1.8:1 ratio of B-lineage ALL to T-lineage ALL. The characteristics of the patients from which this subset is derived are statistically not different from those of the total study population of standard-risk patients.
Characteristics of patients, samples, and assays used
| . | Data . |
|---|---|
| Patients | |
| No. of patients | 105 |
| Median age at diagnosis, y (range) | 28 (15-61) |
| Median duration of therapy, mo (range) | 14.4 (8.2-20.4) |
| ALL lineage | |
| Precursor B | 67 |
| Thymic T | 38 |
| Sex | |
| Female | 30 |
| Male | 75 |
| Samples | |
| Total, no. (no. after wk 52) | 519 (414) |
| Median no. samples per patient (range) | 3 (1-12) |
| Assays | |
| No. of patients investigated with only 1 molecular marker | 22 |
| No. of patients investigated with 2 molecular markers | 83 |
| Median duration of monitoring after end of first y of therapy, mo (range) | 16.1 (0.7-41.1) |
| Usage of molecular markers | |
| IgH, incomplete | 5 |
| IgH, complete | 49 |
| KDE | 21 |
| TCRβ, incomplete | 12 |
| TCRβ, complete | 41 |
| TCRγ | 29 |
| TCRδ, incomplete | 19 |
| TCRδ, complete | 12 |
| Quality of assays | |
| Sensitivities reached | |
| 1 × 10−5 | 26 |
| 5 × 10−5 | 77 |
| 1 × 10−4 | 51 |
| 5 × 10−4 | 21 |
| 1 × 10−3 | 10 |
| 1 × 10−2 | 3 |
| Quantitative ranges | |
| 1 × 10−4 | 108 |
| 5 × 10−4 | 37 |
| 1 × 10−3 | 32 |
| 1 × 10−2 | 10 |
| 1 × 10−1 | 1 |
| . | Data . |
|---|---|
| Patients | |
| No. of patients | 105 |
| Median age at diagnosis, y (range) | 28 (15-61) |
| Median duration of therapy, mo (range) | 14.4 (8.2-20.4) |
| ALL lineage | |
| Precursor B | 67 |
| Thymic T | 38 |
| Sex | |
| Female | 30 |
| Male | 75 |
| Samples | |
| Total, no. (no. after wk 52) | 519 (414) |
| Median no. samples per patient (range) | 3 (1-12) |
| Assays | |
| No. of patients investigated with only 1 molecular marker | 22 |
| No. of patients investigated with 2 molecular markers | 83 |
| Median duration of monitoring after end of first y of therapy, mo (range) | 16.1 (0.7-41.1) |
| Usage of molecular markers | |
| IgH, incomplete | 5 |
| IgH, complete | 49 |
| KDE | 21 |
| TCRβ, incomplete | 12 |
| TCRβ, complete | 41 |
| TCRγ | 29 |
| TCRδ, incomplete | 19 |
| TCRδ, complete | 12 |
| Quality of assays | |
| Sensitivities reached | |
| 1 × 10−5 | 26 |
| 5 × 10−5 | 77 |
| 1 × 10−4 | 51 |
| 5 × 10−4 | 21 |
| 1 × 10−3 | 10 |
| 1 × 10−2 | 3 |
| Quantitative ranges | |
| 1 × 10−4 | 108 |
| 5 × 10−4 | 37 |
| 1 × 10−3 | 32 |
| 1 × 10−2 | 10 |
| 1 × 10−1 | 1 |
Relapse after the first year of therapy
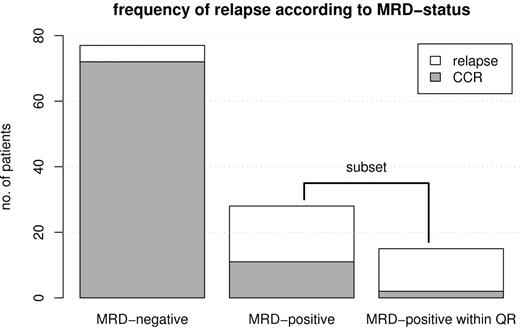
Of the 105 patients included in the analysis, 22 relapsed after the first year of therapy (21%). In 17 of these, MRD was detected before relapse while in 5 patients MRD was not detected before relapse (Figure 1). Seventeen of the relapsed patients (77% of relapses) had precursor B-ALL, and 5 had thymic T-ALL (23% of relapses).
Frequency of relapse. Bar plot showing the frequency of relapse according to MRD status for MRD-negative patients (n = 77, 5 relapses), MRD-positive patients (n = 28, 17 relapses), and MRD-positive patients within the quantitative range of the PCR (n = 15, 13 relapses, being a subset of the MRD-positive group). The 2 patients who did not relapse in the last group were censored at the time of stem cell transplantation.
Frequency of relapse. Bar plot showing the frequency of relapse according to MRD status for MRD-negative patients (n = 77, 5 relapses), MRD-positive patients (n = 28, 17 relapses), and MRD-positive patients within the quantitative range of the PCR (n = 15, 13 relapses, being a subset of the MRD-positive group). The 2 patients who did not relapse in the last group were censored at the time of stem cell transplantation.
Influence of MRD on relapse-free survival (RFS).
Using the MRD-based risk stratification based on MRD monitoring during the first year as recently published,10 8 patients belonged to the MRD high-risk group, 44 belonged to the MRD intermediate-risk group, 11 belonged to the MRD low-risk group, and 42 were not classifiable because critical time points during the first year were lacking. Six of the 8 patients in the MRD high-risk group relapsed during the observation period through the end of 2005 (75%), 10 of 44 (23%) relapsed in the MRD intermediate-risk group, and there were no relapses in the MRD low-risk group and 6 of 42 (14%) in the unclassifiable group.
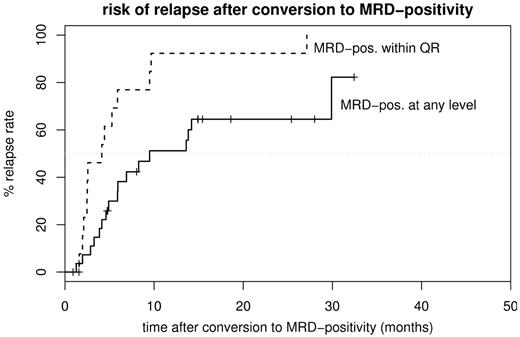
As shown in Figure 2, conversion to MRD positivity was associated with a significantly shorter relapse-free survival (RFS). Accordingly, in the time-dependent proportional hazards model MRD positivities within or below the quantitative range of the PCR were both associated with a higher risk of relapse (MRD detectable at any level: P < .001; hazard ratio, 11.4; 95% confidence interval [CI], 4.2-31.2; MRD within quantitative range: P < .001; hazard ratio, 18.7; 95% CI, 7.8-44.8). The assumption of proportional hazards in the model is in both cases statistically confirmed by the Grambsch-Therneau test (P = .25 and .93, respectively). To exclude a significant dependence of the results on the presence of patients already classified to be MRD high risk based on the MRD results during the first year of therapy, these data were reanalyzed excluding the MRD high-risk patients. The reanalysis confirmed the results (MRD detectable at any level: P = .001, hazard ratio, 7.9; 95% CI, 2.7-23.0; MRD within quantitative range: P < .001, hazard ratio, 20.8; 95% CI, 7.2-59.6). Seventeen of 28 patients who became MRD positive at any level relapsed during the observation period (61%). Of the remaining 11 patients, 7 became MRD negative again later and, as of the end of 2005, have remained so up to 32.4 months after conversion. Of the patients with MRD detectable within the quantitative range of the PCR assay, 13 of 15 (87%) relapsed, with the 2 nonrelapsed patients having been censored due to allogeneic stem cell transplantation (SCT), which was performed as an individual therapeutic decision while still in hematologic remission (Figure 3). Median RFS of the patients who eventually became MRD positive during follow-up was 17.7 months after the end of initial therapy; median RFS for the patients who are still MRD negative is not reached after a median follow-up of 23.2 months after the first year of therapy. As of the end of 2005, conversion to MRD positivity at any level resulted in a median RFS of 9.5 months; when MRD was detectable within the quantitative range of the PCR, the median RFS was shortened to 4.1 months (Figure 3). Of the 77 patients in the MRD negative group, only 5 (6%) relapsed. Three of them had their last available MRD samples obtained 6, 6, and 8 months before relapse. One patient had an extramedullary relapse (in the cheek) with MRD in the bone marrow detectable below quantitative range at that time point, and one patient had an ongoing rearrangement of his Ig/Tcr genes that led to a loss of both molecular markers used to follow up this patient.
Relapse-free survival. RFS is shown after initial therapy for the total cohort (n = 105; 5 events; median survival, 39.8 months) and the 2 subgroups of patients continuously MRD negative (MRD-neg.; n = 77; 22 events; median survival not reached) or converting to MRD positivity (MRD-pos.; n = 28; 17 events; median, 16.7 months).
Relapse-free survival. RFS is shown after initial therapy for the total cohort (n = 105; 5 events; median survival, 39.8 months) and the 2 subgroups of patients continuously MRD negative (MRD-neg.; n = 77; 22 events; median survival not reached) or converting to MRD positivity (MRD-pos.; n = 28; 17 events; median, 16.7 months).
Risk of relapse. Cumulative risk of relapse is shown after conversion to MRD positivity at any level (MRD-pos. at any level; n = 28; median time to relapse, 9.5 months) or after the first time MRD was measurable within the quantitative range of the PCR (MRD-pos. within QR; n = 15; median time to relapse, 4.1 months).
Risk of relapse. Cumulative risk of relapse is shown after conversion to MRD positivity at any level (MRD-pos. at any level; n = 28; median time to relapse, 9.5 months) or after the first time MRD was measurable within the quantitative range of the PCR (MRD-pos. within QR; n = 15; median time to relapse, 4.1 months).
Influence of age at diagnosis and duration of initial therapy.
In a univariate Cox regression, neither age at diagnosis (P = .11; hazard ratio, 0.97; 95% CI, 0.92-1.01) nor duration of initial therapy (P = .68; hazard ratio, 0.96; 95% CI, 0.78-1.18) was significantly associated with relapse.
Discussion
The significance of MRD determination for risk stratification in patients treated for ALL has been well established for pediatric2-6 and adult7-9 patients. In line with these studies, in our recently reported10 large cohort of prospectively monitored standard-risk adult ALL patients we could clearly identify 2 groups: a good-risk group with a rapid decline in MRD and a poor-risk group with persisting high levels of MRD. The complementary data presented here were obtained to assess the possibility to predict relapse by molecular methods during maintenance and follow-up in MRD-negative patients after intensive therapy. As shown in Figure 2, the relapse rate of these patients, who by definition successfully completed induction and consolidation therapy, is still above 20%. Using a risk stratification based on MRD in the first year of therapy10 we would have predicted 6 of the 16 relapses observed in the group of patients classifiable according to that stratification, suggesting that a sizable proportion of patients at risk are not identified at this time. Therefore, for those patients continuous monitoring during and after maintenance therapy provides extremely relevant additional information in terms of relapse prediction.
To our knowledge, the study presented here is the first that systematically analyzes the clinical significance of conversion to MRD positivity in Ph-negative ALL after the completion of initial therapy in a completely prospective manner. Although there are other studies that include late follow-up time points (up to 120 weeks after start of therapy) in their analyses by either molecular genetic methods6,8,27 or by flow cytometry,3 these have either not systematically followed up a predefined cohort or used very few predefined time points late in the disease (eg, 24 months27 ), not allowing any insight into the dynamics of recurring disease. Therefore, a direct comparison of these studies with the results presented here regarding accuracy and predictive value of MRD late in the course of the disease is not possible. Conversely, for Ph-positive ALL there are data showing that after allogeneic SCT an increase of MRD as measured by the level of BCR/ABL transcripts precedes relapse.28
During the observation period, which ended by the end of 2005, 28 of the 105 patients became MRD positive, 17 of whom have relapsed so far, with 4 additional patients being converted to MRD positivity rather recently, not yet having reached the median time from MRD positivity to relapse of 9.5 months of the total cohort (Figure 2). Once the MRD level was measurable within the quantitative range of the PCR, only 2 of 15 patients have not yet relapsed, both of them censored at the time of allogeneic SCT, which was in both cases performed because of an individual treatment decision while the patients were still in hematologic remission. The observation that not all patients with MRD detectable at very low levels even at later time points might relapse is in line with other studies.8,29 Due to the rather ample definition of sensitivity of the PCR assays with a ΔCt of only 1.0 to the background in order not to miss any true positives, this may in some cases well be due to false positive results of the PCR. In addition, a longer follow-up might show relapse in some of the cases with a short observation period after MRD conversion. The alternative hypothesis of late elimination of the leukemic clone at very low levels as proposed by Roberts et al30 and supported by the data of Marshall et al27 also exists. These data, however, have to be interpreted cautiously because the authors have used extremely sensitive PCR techniques and therefore reported a very high percentage of MRD-positive patients very late in the course of the disease with relatively low predictive values for the individual patient. Nevertheless, these data are supported by some clinical observations: because maintenance chemotherapy does not introduce a new therapeutic principle to an already intensive pharmacological treatment, the reason for this elimination seems to be a combination of chemotherapeutic suppression of the leukemic clone together with immunologic containment and killing of the remaining leukemic blasts. This is in keeping with the results of clinical studies where maintenance chemotherapy has been shown to improve survival rates in ALL, partly but not exclusively dependent on the dose intensity administered.31-33 The 7 patients in this study who became MRD positive at very low levels and then declined again to MRD negativity may provide indirect evidence for such a mechanism.
Five of 23 patients relapsed without MRD being detectable previously; in 3 MRD possibly could have been detected if they had their MRD samples taken at the 3-month intervals proposed by the study protocol. One further patient had an initially extramedullary relapse that was, not unexpectedly, not detected in advance by MRD analysis of the bone marrow. In only one patient was MRD not detected due to an evolution of the leukemic clone. Frequency of false MRD negativity is lower than expected from the literature,34-36 possibly reflecting different populations (adult versus pediatric) and/or selection criteria (systematic continuous follow-up of patients versus comparison of the diagnostic sample and sample at the time of the established relapse).
The dynamics of relapse after converting to MRD positivity as shown in Figure 3 illustrate that relying exclusively on detecting MRD within the quantitative range of the PCR and thereby improving the accuracy of the method to predict relapse will, in a clinical setting, detect a significant proportion of patients too late for timely treatment intervention due to the strict definition of the quantitative range in order to reach a high specificity. With a median time of 4.1 months between MRD positivity within quantitative range of the PCR and hematologic relapse, using a 2-step approach seems to be justified: Accordingly, patients who convert to MRD positivity should be tested again after 2 to 4 weeks. In parallel, necessary preparations for the planned salvage therapy (eg, donor search) should be performed. If MRD becomes detectable within the quantitative range of the PCR, the strategy of an immediate allogeneic SCT while still in CR should be pursued. Because recurrence of measurable MRD within the quantitative range of the PCR in MRD-negative patients is such a powerful predictor for subsequent hematologic relapse, we propose to define this as a molecular relapse. Following this approach, 7 of 28 patients would have been subjected to possibly unnecessary control bone marrow aspirations. Given the consequences of a full-blown ALL relapse, this relatively minor diagnostic procedure seems to be acceptable. None of the temporarily MRD-positive patients would have received salvage chemotherapy because none of them had MRD measurable within the quantitative range of the PCR at any time after the first year of therapy. As to the required length of MRD monitoring, a follow-up of 12 months after the end of the first year of therapy would have detected 14 of the 17 patients who later relapsed. Thus, this monitoring interval seems to be a good trade-off between cost and benefit.
Whether MRD-based anticipation of salvage treatment at the time of molecular relapse will improve the outcome of relapsed ALL, as seems to be the case in APL,12 remains to be investigated. However, the data provided in this study support the concept that hematologic relapse can be predicted by monitoring the course of the disease using quantitative PCR, thus making individual treatment escalation and deescalation for adult ALL patients belonging to the MRD-defined intermediate-risk groups feasible. Although MRD monitoring is time-consuming and costly by any method currently available, avoiding overtreatment of a substantial number of patients, while at the same time offering the security of reliable detection of an impending relapse, seems to be worthwhile.
Authorship
Contribution: T.R., M.B., and M.K. were responsible for analysis of the data and preparation of the manuscript; S.L., S.I., M.R., and S.B. provided MRD data; H.-A.H. is a reference morphologist; and D.H., N.G., and R.R. are principal investigator, coordinator, and data manager of the GMALL 06/99 and 07/03 studies, respectively.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Participating centers of the GMALL study group from which patients were included into this study are listed as a data supplement to the online version of this article.
Correspondence: Thorsten Raff, 2nd Medical Department, University Hospital Schleswig-Holstein, Campus Kiel, Chemnitzstraβe 33, 24116 Kiel, Germany; e-mail: t.raff@med2.uni-kiel.de.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Wilhelm-Sander-Stiftung (grant 2001.074.2); Bundesministerium für Bildung und Forschung (BMBF), Competence Network “Acute and Chronic Leukemias” (grant 01GI 9971); and Deutsche Krebshilfe (grant 70-2657-Ho2). The authors thank Bettina Brix, Frauke Hemken, Lena Lorenzen, and Heidi Seppelt for excellent technical assistance and the members of the GMALL Study Group for their support.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal