Abstract
This study demonstrates a unique and crucial role of plasmacytoid dendritic cells (pDCs) and pDC-derived type I interferons (IFNs) in the pathogenesis of mouse coronavirus infection. pDCs controlled the fast replicating mouse hepatitis virus (MHV) through the immediate production of type I IFNs. Recognition of MHV by pDCs was mediated via TLR7 ensuring a swift IFN-α production following encounter with this cytopathic RNA virus. Furthermore, the particular type I IFN response pattern was not restricted to the murine coronavirus, but was also found in infection with the highly cytopathic human severe acute respiratory syndrome (SARS) coronavirus. Taken together, our results suggest that rapid production of type I IFNs by pDCs is essential for the control of potentially lethal coronavirus infections.
Introduction
Type I IFNs (IFNα/β) play a decisive role in shaping antiviral immune responses.1 Signaling through the type I IFN receptor leads to the activation of a particular set of genes, including protein kinase R, and Mx proteins,2 which exert potent direct antiviral effects. Other type I IFN–stimulated gene products, such as IFN-γ, activate downstream elements of the innate immune system that further promote rapid clearance of the viral pathogen.3 Although almost all hematopoietic and nonhematopoietic cells are able to produce IFN-α/β after viral infections, plasmacytoid dendritic cells (pDCs) are the major source of IFN-α, both in humans and mice.4-7 One key feature of pDCs is the expression of receptors for single-stranded RNA or DNA such as Toll-like receptor 7 (TLR7)8,9 and TLR9,10,11 respectively, which are essential to sense the viral pathogen and to trigger the innate immune response. In mouse cytomegalovirus (MCMV) infection, pDCs respond quickly and generate the first wave of IFN-α.12,13 These previous studies have clearly established an important role of pDCs for the rapid production of type I IFNs in antiviral immune responses.
Coronaviruses are positive-stranded RNA viruses that are able to infect a broad range of vertebrates and are associated mainly with respiratory, enteric, and, sometimes, systemic diseases.14 Human coronaviruses are known to generally cause mild upper-respiratory tract disease, including common cold, and occasional enteric infections. The emergence of a novel coronavirus (COV) causing severe acute respiratory syndrome (SARS) highlighted the potential of coronaviruses to seriously impact on human health.14 Phylogenetic analyses revealed a close relationship of SARS-CoV to group II coronaviruses,15 which is prototyped by the mouse hepatitis virus (MHV). MHV causes enteritis, pneumonia, hepatitis, and demyelinating encephalomyelitis in mice and is one of the most extensively studied coronaviruses in vitro and in vivo.16 Both SARS-CoV and MHV have a similar genome organization and share common mechanisms and enzymes involved in genome expression.15,17 Because of these similarities, it is conceivable that the pathogenesis of and the immune response against systemic MHV infection in mice recapitulates some of the essential features of SARS-CoV infection.
The immune response against MHV is characterized by a strong cytotoxic T-cell (CTL) response that mediates initial clearance of the virus,18,19 whereas neutralizing antibodies appear to be required to prevent re-emergence of the persistent infection.20,21 In contrast to the well-characterized defense mechanisms of the adaptive immune response against MHV, the innate immune response has not been sufficiently characterized. Similarly, the importance of innate immune mechanisms triggered by SARS-CoV has remained enigmatic. Several studies have shown that peripheral-blood mononuclear cells (PBMCs) from SARS-CoV–infected individuals produce high amounts of inflammatory cytokines and chemokines, but not type I IFNs.22-24 Furthermore, in vitro studies have shown that neither macrophages nor monocyte-derived DCs respond to SARS-CoV infection with significant IFN-α production.25-28 Nonetheless, there is clear evidence that treatment with recombinant IFN-β or IFN-α can inhibit SARS-CoV replication in vitro,29-31 and, most importantly, diminish the severity of SARS-CoV infection in vivo.32,33 Thus, although the antiviral activity of type I IFNs in SARS-CoV infections has been clearly demonstrated, it remains to be established whether a significant production of type I IFNs can be achieved upon coronavirus infection and how this may impact on virus replication and disease. The present study addressed this issue and identified pDCs as the major source of type I IFNs during cytopathic coronaviral replication. We show furthermore that pDCs are crucial during the initial phase of MHV infection since widespread replication in various nonlymphoid organs, and the associated organ damage, is efficiently kept in check through pDC-derived type I IFNs.
Materials and methods
Mice and viruses
C57BL/6 (B6) mice were obtained from Charles River Laboratories (Sulzfeld, Germany). TLR3−/−,34 TLR7−/−,35 TLR3−/−/TLR7−/−, and MyD88−/−36 mice were bred in the Institut für Labortierkunde (University of Zurich, Switzerland). 129Sv and type I IFN receptor–deficient (IFNAR−/−) mice37 were obtained from the Institut für Labortierkunde (University of Zurich) and bred in our facilities. All mice were maintained in individually ventilated cages and were used between 6 and 9 weeks of age. All animal experiments were performed in accordance with the Swiss federal legislation on animal protection.
MHV A59 was generated from a molecularly cloned cDNA38 based on the Albany strain of MHV A59 and propagated on L929 cells. SARS-CoV (isolate Frankfurt-1) was kindly provided by Stephan Becker (University of Marburg, Germany) and propagated on Vero cells. The Newcastle disease virus (NDV; strain H53) stock was grown on 10-day-old embryonated chicken eggs.
Isolation of dendritic cells and flow cytometry
Murine conventional DCs (cDCs) and pDCs were obtained from spleens of 129Sv or IFNAR−/− mice following digestion with collagenase type II (Invitrogen, Basel, Switzerland) for 20 minutes at 37°C. Cells were resuspended in PBS supplemented with 2% FCS and 2 mM EDTA and overlaid on 20% Optiprep density gradient medium (Sigma-Aldrich, Basel, Switzerland). After centrifugation at 700g for 15 minutes, low-density cells were depleted of CD3+ and CD19+ cells using DYNAL magnetic beads according to the instructions of the manufacturer (Invitrogen). The DC-enriched preparations were stained with α–PDCA-1, α-CD11b, and α-CD11c, and the distinct pDC and cDC populations were sorted using a fluorescence-activated cell sorter (FACS) ARIA (BD Biosciences, Allschwil, Switzerland). Purity of both cell preparations was always more than 98%.
Murine bone marrow–derived cDCs or pDCs were generated by 6 to 7 days of culture with either granulocyte-monocyte colony-stimulating factor (GM-CSF)–containing supernatant from the cell line X63-GM-CSF (kindly provided by Dr Antonius Rolink, University of Basel) or Flt3-L (R&D systems, Oxford, United Kingdom) at 20 ng/mL, respectively. Bone marrow–derived cDCs were further purified using Optiprep density gradient centrifugation. Bone marrow–derived pDCs were purified using the mouse pDC isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) adapted for the isolation of in vitro–derived pDCs by adding CD11b-biotin to the negative selection cocktail.
Human circulating conventional dendritic cells and plasmacytoid dendritic cells were isolated from healthy donors (obtained from the Blood Bank of the University Hospital Freiburg, Germany). PBMCs were obtained by Ficoll-Hypaque density gradient centrifugation. Human pDCs or cDCs were further purified by magnetic sorting with human plasmacytoid dendritic-cell isolation kit (Miltenyi Biotec) or human BDCA-1 dendritic-cell isolation kit (Miltenyi Biotec), respectively. Purity of both cell preparation was for all donors more than 90% (with B cells being the major contaminating cell type).
Cells were analyzed with a FACSCalibur flow cytometer using CellQuest software (BD Biosciences). The antibodies CD123 biotin, CD11c PE, B220 APC, and CD11b FITC were purchased from Biolegend (San Diego, CA); and CD303 (BDCA-2)–PE, mPDCA-1-FITC, mPDCA-1-PE, and CD11c-APC, from Miltenyi Biotec.
Virus infections, determination of virus titers, and liver enzyme values
Human pDCs and cDCs were exposed to SARS-CoV or NDV for 1 hour at 37°C, washed, and plated at 1.5 × 105/mL. Murine pDCs or cDCs were infected with MHV A59 for 1 hour at 37°C, washed, and plated at 1 to 2 × 105/mL. CpG ODN 2216 was used as a positive control for IFN-α production as described previously.39
Mice were injected intraperitoneally with 5 pfu MHV A59 and killed at the indicated time points. For depletion of pDCs, mice were injected intraperitoneally with 0.5 mg α–mPDCA-1 (Miltenyi Biotec) or 0.5 mg rat IgG2b isotype control antibody (Biolegend) 12 hours prior to infection. Organs were stored at −70°C until further analysis. Blood was incubated at room temperature to coagulate and was then centrifuged, and serum was used for alanine 2-oxoglutarate-aminotransferase (ALT) measurements using a Hitachi 747 autoanalyzer (Tokyo, Japan). Virus titers in organs were determined from frozen organs after weighing and homogenization. Viral titers were determined by standard plaque assay using L929 cells.
Histology, IFN-α ELISA, and reverse-transcription–polymerase chain reaction (RT-PCR)
Organs were fixed in 4% formalin and embedded in paraffin. Sections were stained with hematoxylin and eosin. Human IFN-α and mouse IFN-α concentration in cell-culture supernatants or serum was measured by enzyme-linked immunosorbent assay (ELISA; PBL Biomedical Laboratories, Piscataway, NJ) according to the manufacturer's instructions. To detect cellular and viral mRNAs in pDCs and cDCs, total cellular RNA was prepared using the RNeasy Kit for murine DCs (Qiagen, Basel, Switzerland) or Trifast (PEQLAB, Erlangen, Germany) for human DCs. Reverse transcription was done with 100 ng total RNA using the SuperScript II First-Strand Synthesis System (Invitrogen). PCR was performed with Red Taq Polymerase (Sigma-Aldrich) using standard protocols. The following primers were used for amplification: mIFN-β forward, 5′-CATCAACTATAAGCAGCTCCA-3′; mIFN-β reverse, 5′TTCAAGTGGAGAGCAGTTGAG3′; mIFN-α4 forward, 5′CTGGTCAGCCTGTTCTCTAGGATGT3′; mIFN-α4 reverse, 5′TCAGAGGAGGTTCCTGCATCAC3′; mIFN-α total forward, 5′ATGGCTAGRCTC TGTGCTTTCCT3′; mIFN-α total reverse, 5′AGGGCTCTCCAGAYTTCTGCTCTG3′; mGAPDH forward, 5′CATCAAGAAGGTGGTGAAGC3′; mGAPDH reverse, 5′CCTGTTGCTGTAGCCGTATT3′; MHV-N forward, 5′TCCTGGTTTTCTGGCATTACCCAG3′; and MHV-N reverse, 5′CTGAGGCAATACCGTGCCGGGCGC3′. Primer sequences for amplifying human IFN-β were 5′CATACCCACGGAGAAAGGACATT3′ and 5′TGATAGACATTAGCCAGGAGGTTC3′; for ISG56, 5′AAGTGGACCCTGAAAACCCTGAAT3′ and 5′TGCCCTTTTGTAGCCTCCTTGAT3′; for MxA, 5′GTTGTTTCCGAAGTGGACATCGCAAAA3′ and 5′CGGGCATCTGGTCACGAT3′; and for γ-actin, 5′GCCGGTCGCAATGGAAGAAGA3′ and 5′CATGGCCGGGGTGTTGAAGGTC3′. SARS-CoV transcription was detected by using primer 5′TGTCTAGCAGCAATAGCGCGAGGGC3′ for reverse transcription and primers 5′GGAAAAGCCAACCAACCTCGATCTCT3′ and 5′AAGTTGTAGCACGGTGGCAGC3′ for PCR.
Results
Rapid type I IFN production in pDCs following MHV infection
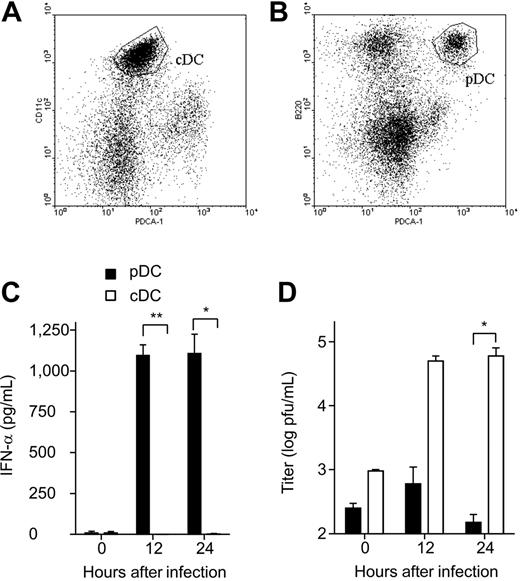
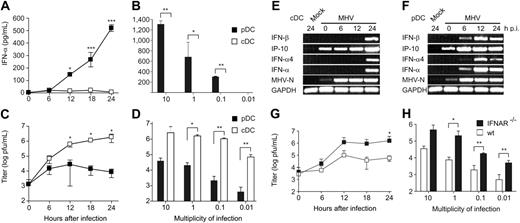
In a first set of experiments, the type I IFN response of pDCs and cDCs following encounter with MHV was determined. To this end, CD11clowB220+PDCA-1+ pDCs and CD11c+B220− cDCs were sorted from spleen-cell suspensions (Figure 1A-B) and infected with MHV. Primary pDCs but not cDCs responded with rapid and significant production of IFN-α (Figure 1C). The high IFN-α production in pDCs correlated well with the control of the virus infection (Figure 1D). Bone marrow–derived pDCs and cDCs that were differentiated with the growth factors Flt-3L or GM-CSF, respectively, responded in a similar pattern: rapid and high production of IFN-α in pDCs but not in cDCs (Figure 2A-B) and a good containment of the viral replication by pDCs (Figure 2C-D). A time-course RT-PCR analysis confirmed that the type I IFN response is considerably slower in cDCs (Figure 2E) compared with pDCs (Figure 2F). Furthermore, pDCs lacking the type I IFN receptor were more susceptible to MHV infection than wild-type pDCs (Figure 2G-H). It appears therefore that pDCs are well equipped to respond efficiently against MHV with a strong type I IFN production, and that this early reaction exerts a potent protective effect against this cytopathic virus.
Type I IFN production and viral replication in MHV-infected splenic cDCs and pDCs. (A) Flow cytometric analysis of splenic cDCs (CD11c+PDCA-1−) and (B) splenic pDCs CD11clow B220+ PDCA-1− before FACS sorting. Gates for sorting are indicated. (C) Primary FACS-purified murine splenic cDCs or pDCs were infected with MHV at a multiplicity of infection (moi) of 1. IFN-α secretion to culture supernatants was determined by ELISA at the indicated time points. (D) Virus titers in culture supernatants were determined by plaque assay. (C-D) Data represent mean values ± SD pooled from 2 experiments. Statistical analysis was performed using Student t test (*P < .05; **P < .01).
Type I IFN production and viral replication in MHV-infected splenic cDCs and pDCs. (A) Flow cytometric analysis of splenic cDCs (CD11c+PDCA-1−) and (B) splenic pDCs CD11clow B220+ PDCA-1− before FACS sorting. Gates for sorting are indicated. (C) Primary FACS-purified murine splenic cDCs or pDCs were infected with MHV at a multiplicity of infection (moi) of 1. IFN-α secretion to culture supernatants was determined by ELISA at the indicated time points. (D) Virus titers in culture supernatants were determined by plaque assay. (C-D) Data represent mean values ± SD pooled from 2 experiments. Statistical analysis was performed using Student t test (*P < .05; **P < .01).
Type I IFN production and viral replication in MHV-infected in vitro–derived cDCs and pDCs. Infection of bone marrow–derived pDCs and cDCs with MHV. (A,C) IFN-α and virus titers in tissue culture supernatants were determined at the indicated times after MHV infection (moi = 1), or (B,D) at 24 hours after MHV infection with different moi as indicated. Values in panels A-D represent means ± SD from triplicate measurements. Experiments in panels A-D were repeated 3 times with comparable results. Expression of IFN-β, IP-10, IFN-α4, IFN-α, GAPDH, or MHV nucleoprotein (MHV-N) mRNAs was determined by RT-PCR using total RNA from bone marrow–derived (E) cDCs or (F) pDCs infected with MHV (moi = 1) or treated with PBS (mock). (G-H) Viral replication in MHV-infected wt or IFNAR−/− pDCs. Cells were infected with MHV A59 at (G) an moi of 1 or (H) at different moi as indicated. Virus titers in culture supernatants were determined by plaque assay at (G) the indicated times after MHV A59 infection (moi = 1) or (H) 24 hours after MHV infection. (A-D,G-H) Data represent mean values ± SD pooled from 2 experiments. Statistical analysis was performed using Student t test (*P < .05; **P < .01, ***P < .001).
Type I IFN production and viral replication in MHV-infected in vitro–derived cDCs and pDCs. Infection of bone marrow–derived pDCs and cDCs with MHV. (A,C) IFN-α and virus titers in tissue culture supernatants were determined at the indicated times after MHV infection (moi = 1), or (B,D) at 24 hours after MHV infection with different moi as indicated. Values in panels A-D represent means ± SD from triplicate measurements. Experiments in panels A-D were repeated 3 times with comparable results. Expression of IFN-β, IP-10, IFN-α4, IFN-α, GAPDH, or MHV nucleoprotein (MHV-N) mRNAs was determined by RT-PCR using total RNA from bone marrow–derived (E) cDCs or (F) pDCs infected with MHV (moi = 1) or treated with PBS (mock). (G-H) Viral replication in MHV-infected wt or IFNAR−/− pDCs. Cells were infected with MHV A59 at (G) an moi of 1 or (H) at different moi as indicated. Virus titers in culture supernatants were determined by plaque assay at (G) the indicated times after MHV A59 infection (moi = 1) or (H) 24 hours after MHV infection. (A-D,G-H) Data represent mean values ± SD pooled from 2 experiments. Statistical analysis was performed using Student t test (*P < .05; **P < .01, ***P < .001).
Type I IFN signaling is essential for the control of MHV infection
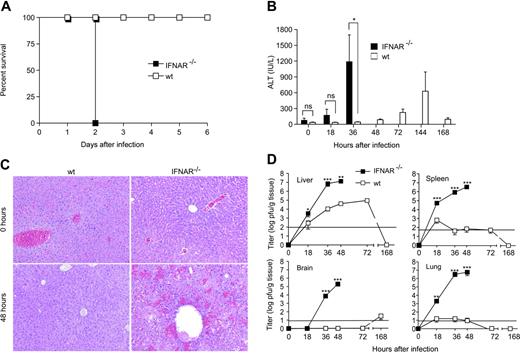
Signaling through the type I IFN receptor (IFNAR) is essential for the control of several viral infections.40 To assess the importance of type I IFN signaling in the course of an MHV infection, IFNAR-deficient (IFNAR−/−) and 129Sv wild-type (wt) mice were infected with 5 pfu MHV. MHV infection in IFNAR−/− mice was lethal within only 48 hours, while wt mice survived without showing signs of MHV infection–associated clinical disease (Figure 3A). Furthermore, IFNAR−/− but not wt mice showed rapidly rising liver enzyme values in serum (Figure 3B) and an acute hemorrhagic liver disease with massive hepatocyte necrosis (Figure 3C). The detailed time-course analysis of viral spread in both IFNAR−/− and wt mice indicated that a functional type I IFN system is essential to restrict the initial viral replication to the spleen and to prevent spread to nonhematopoietic organs such as the lung and the central nervous system (Figure 3D). Notably, the replication of the strongly hepatotropic MHV in the liver was efficiently reduced in the presence of a functional type I IFN system with a reduction of viral titers of 3 to 4 orders of magnitude (Figure 3D). It is thus most likely that the rapidly lethal disease in IFNAR−/− mice following MHV infection is a consequence of an insufficient initial control of the cytopathic virus in the spleen and the subsequent high-level replication in various organs, eventually causing an acute multiorgan failure.
Impact of type I IFN signaling during MHV infection. IFNAR−/− or wt mice were injected intraperitoneally with 5 pfu MHV A59. (A) Health status of IFNAR−/− and wt mice was monitored twice daily after infection (n = 6). (B) ALT values were measured at the indicated time points after infection. (C) Liver pathology in IFNAR−/− and wt mice before or 48 hours after MHV A59 infection. Hematoxylin-eosin staining of 4% formaldehyde-fixed sections. Images were acquired using a Leica DMRA microscope (Leica, Heerbrugg, Switzerland) with a 25×/0.65 NA objective (total magnification, ×162). Images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA). (D) Viral titers in liver, spleen, brain, and lung of MHV A59–infected IFNAR−/− or wt mice were determined at different time points after infection. Results represent the mean of 6 individual mice per time point. Solid horizontal lines in panel D represent limit of detection in the plaque assay. Data in panels B and D represent means ± SD from 2 experiments with a total of 3 or 6 mice evaluated per time point. Statistical analysis was performed using Student t test (ns, P > .05; *P < .05; **P < .01; ***P < .001).
Impact of type I IFN signaling during MHV infection. IFNAR−/− or wt mice were injected intraperitoneally with 5 pfu MHV A59. (A) Health status of IFNAR−/− and wt mice was monitored twice daily after infection (n = 6). (B) ALT values were measured at the indicated time points after infection. (C) Liver pathology in IFNAR−/− and wt mice before or 48 hours after MHV A59 infection. Hematoxylin-eosin staining of 4% formaldehyde-fixed sections. Images were acquired using a Leica DMRA microscope (Leica, Heerbrugg, Switzerland) with a 25×/0.65 NA objective (total magnification, ×162). Images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA). (D) Viral titers in liver, spleen, brain, and lung of MHV A59–infected IFNAR−/− or wt mice were determined at different time points after infection. Results represent the mean of 6 individual mice per time point. Solid horizontal lines in panel D represent limit of detection in the plaque assay. Data in panels B and D represent means ± SD from 2 experiments with a total of 3 or 6 mice evaluated per time point. Statistical analysis was performed using Student t test (ns, P > .05; *P < .05; **P < .01; ***P < .001).
Early control of MHV infection through pDC-derived type I IFN
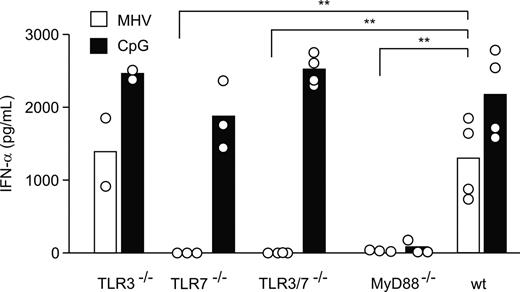
The above results suggested that the initial control of MHV requires an efficient type I IFN response that might be generated by pDCs. It has been shown that pDCs use the TLR pathway rather than the RNA helicase RIG-I for recognition of RNA viruses and to produce type I IFN.41 Therefore, to investigate how pDCs recognize MHV, bone marrow–derived pDCs from TLR3-deficient (TLR3−/−), TLR3 and TLR7 double knock-out (TLR3−/−/TLR7−/−), TLR7-deficient (TLR7−/−), and MyD88-deficient (MyD88−/−) mice were infected with a low dose of MHV (moi = 1), and the production of IFN-α was determined after 24 hours. Comparable amounts of IFN-α were found in the supernatants of TLR3−/− and wt control pDC cultures (Figure 4). In contrast, pDCs from neither TLR7−/−, TLR3−/−/TLR7−/−, nor MyD88−/− mice responded with a significant IFN-α production to MHV infection (Figure 4), clearly indicating that MHV recognition by pDCs is triggered exclusively via the TLR7/MyD88 pathway.
pDCs sense MHV via TLR7. Bone marrow–derived pDCs from TLR3−/−, TLR7−/−, TLR3−/−/TLR7−/−, MyD88−/−, or wt mice were infected with MHV A59 (moi = 1) or treated with CpG oligonucleotides. IFN-α in tissue culture supernatants was determined 24 hours after infection by ELISA. Bars represent means with values from individual mice shown as open circles. Statistical analysis was performed using Student t test (**P < .01).
pDCs sense MHV via TLR7. Bone marrow–derived pDCs from TLR3−/−, TLR7−/−, TLR3−/−/TLR7−/−, MyD88−/−, or wt mice were infected with MHV A59 (moi = 1) or treated with CpG oligonucleotides. IFN-α in tissue culture supernatants was determined 24 hours after infection by ELISA. Bars represent means with values from individual mice shown as open circles. Statistical analysis was performed using Student t test (**P < .01).
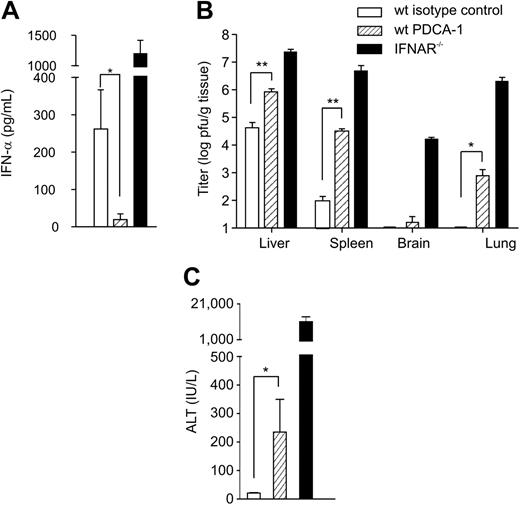
To assess the importance of pDC-derived IFN-α during MHV infection in vivo, pDCs were ablated by the depleting antibody PDCA-1. As described for MCMV by Krug et al,11 pDC depletion was accompanied by severely diminished IFN-α levels in serum following MHV infection (Figure 5A). The treatment with PDCA-1 resulted in an 80% depletion of splenic pDCs for roughly 48 hours (not shown). Although profound effects on viral titers could be observed (Figure 5B), transient pDC depletion did not lead to lethality following low-dose MHV infection. Nevertheless, initial viral titers in spleens were more than 1000-fold increased in pDC-depleted compared with isotype control antibody–treated mice, and virus was found in other organs such as lung or brain (Figure 5B). To exclude complement-mediated global changes in the status of the immune system, we evaluated the effect of natural killer (NK) cell depletion via anti–asialo GM1. Depletion of NK cells altered neither initial viral replication and distribution nor IFN-α levels in serum (not shown). Finally, ALT values in PDCA-1–depleted mice were elevated compared with control animals (Figure 5C), indicating significant liver damage. These data clearly show that pDCs are important for the early control of MHV infection and that the lack of pDCs not only leads to uncontrolled viral replication and spread to different organs but also impacts on the severity of viral disease.
Effect of antibody-mediated pDC depletion on MHV infection. 129Sv mice were treated with rat IgG2b (wt isotype control) or α–mPDCA-1 (wt PDCA-1) and infected intraperitoneally with 5 pfu MHV A59 (n = 6). IFNAR−/− mice (n = 3) were used to demonstrate uncontrolled MHV spread in the absence of a functional IFN system. (A) IFN-α concentration in serum (means ± SD), (B) viral titers (means ± SD) in liver, spleen, brain, and lung, and (C) serum ALT values (means ± SD) were assessed at 48 hours after infection. (A-C) Statistical analysis was performed using Student t test (*P < .05; **P < .01).
Effect of antibody-mediated pDC depletion on MHV infection. 129Sv mice were treated with rat IgG2b (wt isotype control) or α–mPDCA-1 (wt PDCA-1) and infected intraperitoneally with 5 pfu MHV A59 (n = 6). IFNAR−/− mice (n = 3) were used to demonstrate uncontrolled MHV spread in the absence of a functional IFN system. (A) IFN-α concentration in serum (means ± SD), (B) viral titers (means ± SD) in liver, spleen, brain, and lung, and (C) serum ALT values (means ± SD) were assessed at 48 hours after infection. (A-C) Statistical analysis was performed using Student t test (*P < .05; **P < .01).
Rapid induction of type I IFNs in pDCs following SARS-CoV infection
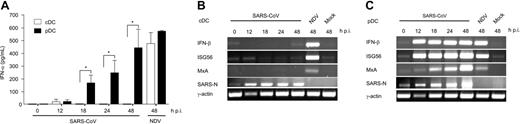
In order to relate the above findings to a pathogenic and potentially lethal human coronavirus infection, the ability of pDCs to produce IFN-α following encounter with SARS-CoV was assessed. Primary pDCs and cDCs were isolated from peripheral blood of healthy donors and infected with SARS-CoV. As described for monocyte-derived cDCs,28 primary cDCs from healthy donors were also not able to produce significant amounts of IFN-α (Figure 6A) and did not up-regulate transcripts of IFN-β and IFN-stimulated genes such as ISG56 and MxA, located downstream in the type I IFN signaling pathway (Figure 6B). In contrast and as expected from the MHV experiments, pDCs were able to produce IFN-α early after SARS-CoV infection (Figure 6A). Furthermore, IFN-β, MxA, or ISG56 mRNA expression was found in infected pDCs (Figure 6C). Based on this evidence and the unsuccessful efforts from previous studies to determine a cell type that produces IFN-α in response to SARS-CoV,25-28 we conclude that pDCs are most likely the major source of type I IFNs in SARS-CoV infection.
Infection of human pDCs and cDCs with SARS-CoV. Primary pDCs or cDCs were isolated from peripheral blood of healthy donors and infected with either SARS-CoV (moi = 1) or Newcastle disease virus (NDV) as positive control (moi = 5), or were left uninfected (mock). (A) IFN-α in culture supernatants was determined at the indicated time points. Results represent pooled data (means ± SD) using pDCs and cDCs from 4 healthy donors. Statistical analysis was performed using Student t test (*P < .05). (B-C) Expression of IFN-β, ISG56, MxA, SARS-CoV N protein, and γ-actin mRNAs in (B) cDCs and (C) pDCs was determined by RT-PCR. One representative result of 4 individual experiments is shown.
Infection of human pDCs and cDCs with SARS-CoV. Primary pDCs or cDCs were isolated from peripheral blood of healthy donors and infected with either SARS-CoV (moi = 1) or Newcastle disease virus (NDV) as positive control (moi = 5), or were left uninfected (mock). (A) IFN-α in culture supernatants was determined at the indicated time points. Results represent pooled data (means ± SD) using pDCs and cDCs from 4 healthy donors. Statistical analysis was performed using Student t test (*P < .05). (B-C) Expression of IFN-β, ISG56, MxA, SARS-CoV N protein, and γ-actin mRNAs in (B) cDCs and (C) pDCs was determined by RT-PCR. One representative result of 4 individual experiments is shown.
Discussion
In this study, we demonstrate a unique and essential function for type I IFN–producing pDCs: protection against the rapidly replicating and cytopathic murine coronavirus. Furthermore, we have identified pDCs as the source of type I IFN in response to human SARS-CoV, suggesting an important biologic role of pDC-derived type I IFNs for highly pathogenic coronavirus infections in humans.
The expression of TLRs that recognize viral products such as CpG oligonucleotides10,11 or ssRNA9 has indicated that pDCs represent a highly specialized cell type that provides an early response against a particular set of infectious agents. A further characteristic of pDCs is the constitutively high expression of IFN regulatory factor-7 (IRF-7),42,43 which directly stimulates IFN-α expression, independently of an IFN-β–mediated feedback loop.44 In MHV infection, this efficient direct IFN-α induction appears to be not only essential for the regulation of the magnitude of the type I IFN response, but also important to restrict replication of this cytopathic virus within pDCs. Furthermore, pDC-derived type I IFNs provide an efficient “bystander” protection because the initial replication of MHV in lymphoid organs such as the spleen was diminished in the presence of pDCs. Notably, in MHV infection this function of pDCs cannot be substituted by other cells as demonstrated, for example, in MCMV infection.11,13 It is possible that viruses that replicate rather slowly, such as MCMV, cannot reveal the full importance of pDCs in the control of cytopathic viruses that require a swift type I IFN response.
Within secondary lymphoid organs, macrophages are the major target cell of MHV,14 and recent studies indicate that cDCs can also be readily infected with MHV A5945 or MHV JHM.46 It is important to note that the uncontrolled infection of cDCs by MHV is detrimental for the initiation of the adaptive antiviral immune response.46 The rapid control of MHV through pDC-derived type I IFN in secondary lymphoid organs ensures therefore the subsequent induction of adaptive immune responses. Likewise, a recent study by Smit et al47 indicates that pDCs not only help to minimize respiratory syncytial virus infection–associated immunopathologic damage, but also facilitate establishment of antiviral T-cell responses in the lung.
Fatal SARS-CoV infection–associated clinical disease is characterized by respiratory insufficiency and, eventually, respiratory failure. One of the reasons for this outcome may be the down-regulation of angiotensin-converting enzyme 2 by the viral spike protein leading to an exacerbation of the pulmonary damage.48 Furthermore, it is well-documented that SARS-CoV infects macrophages and lymphocytes leading to a pronounced atrophy of lymphoid organs in those patients who succumbed to the infection.49 Extrapolating from the data obtained in the MHV model, it is likely that these patients may have suffered from an insufficient early control of the virus within lymphoid organs that eventually led to the unrestrained replication in the respiratory tract. Because SARS-CoV is sensitive to type I IFNs both in vitro30,31,50 and in vivo,32,33 an early control of SARS-CoV by type I IFNs might have been a decisive advantage for those patients who have survived the infection. It is noteworthy that neither macrophages, cDCs, fibroblasts, nor lung epithelial cells28 are able to mount a significant type I IFN response against SARS-CoV. The lack of a significant type I IFN response in PBMCs of SARS-CoV–infected patients23 might be due to a partial inhibition of type I IFN signaling not only in nonlymphoid cells,51,52 but also in pDCs. Therefore, the potential of the various SARS-CoV nonstructural proteins that might inhibit and/or modulate type I IFN responses in pDCs and other important target cells should be addressed in future studies.
Taken together, the results of this study provide insight into the immunopathogenesis of coronavirus-associated diseases by demonstrating an exclusive role of pDC-derived type I IFNs for initial viral control. Triggering of this pathway, for example via specific TLR agonists, might open new avenues for the treatment of coronavirus infections. Indeed, stimulation of TLR3 at the vaginal mucosa can protect mice against herpes simplex virus-2 challenge via the mucosal route.53 In a clinical setting, systemic administration of a TLR7 agonist elicited potent antiviral effects against hepatitis C virus with significant reduction of plasma viremia.54 Our data provide the rationale that such a treatment approach might help to reduce initial viral load and eventually favor a mild course of SARS.
Authorship
Contribution: B.L. and V.T. designed the study and wrote the paper; L.C.-B. performed research and wrote the paper; R.Z., F.W., and M.S. performed research; K.S.L. and S.A. provided mice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Burkhard Ludewig, Research Department, Kantonsspital St Gallen, 9007 St Gallen, Switzerland; e-mail: burkhard.ludewig@kssg.ch; Volker Thiel, Research Department, Kantonsspital St Gallen, 9007 St Gallen, Switzerland; e-mail: volker.thiel@kssg.ch.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Elke Scandella, Philippe Krebs, and Reinhard Maier for critical reading of the paper. We thank Simone Miller, Beat Ryf, and Valentina Wagner for excellent technical assistance.
This work was supported by the Gebert-Rüf-Stiftung, the UBS Optimus Stiftung, the Swiss National Science Foundation, the European Commission (SARS-DTV), the Deutsche Forschungsgemeinschaft (We 2616/4), and the Sino-German Center for Research promotion (GZ Nr 239 202/12).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal