Abstract
Genes that are strongly repressed after B-cell activation are candidates for being inactivated, mutated, or repressed in B-cell malignancies. Krüppel-like factor 4 (Klf4), a gene down-regulated in activated murine B cells, is expressed at low levels in several types of human B-cell lineage lymphomas and leukemias. The human KLF4 gene has been identified as a tumor suppressor gene in colon and gastric cancer; in concordance with this, overexpression of KLF4 can suppress proliferation in several epithelial cell types. Here we investigate the effects of KLF4 on pro/pre–B-cell transformation by v-Abl and BCR-ABL, oncogenes that cause leukemia in mice and humans. We show that overexpression of KLF4 induces arrest and apoptosis in the G1 phase of the cell cycle. KLF4-mediated death, but not cell-cycle arrest, can be rescued by Bcl-XL overexpression. Transformed pro/pre-B cells expressing KLF4 display increased expression of p21CIP and decreased expression of c-Myc and cyclin D2. Tetracycline-inducible expression of KLF4 in B-cell progenitors of transgenic mice blocks transformation by BCR-ABL and depletes leukemic pre-B cells in vivo. Collectively, our work identifies KLF4 as a putative tumor suppressor in B-cell malignancies.
Introduction
The Krüppel-like factor (KLF) family of transcription factors is related to the protein Krüppel in Drosophila melanogaster.1,2 KLFs regulate cell-fate decisions, including development and differentiation of specific tissues. Each contains 3 conserved zinc finger DNA-binding motifs in the c-terminal domain and bind GC-rich regions with a consensus sequence of “CACCC.” The functional specificity of distinct KLF family members is maintained through the transactivation and/or repression domains in the N-terminus of the protein and through differential tissue expression. KLF4 is highly expressed in epithelial tissues, and mouse genetic models have established a role for Klf4 in terminal epidermal differentiation.3,4 KLF4 is also highly expressed in resting lymphocytes but strongly down-regulated in activated cells.5,6 During human B-cell development, KLF4 is differentially expressed, with greater levels of mRNA in the small pre B-I and pre B-II stages compared with the large pre–B-cell stage.7 Thus, KLF4 expression in the B lineage correlates with stages with low proliferation, implying a role in B-cell development similar to its function in terminal skin differentiation.
Many KLFs have been implicated in suppression of proliferation. For example, KLF6 is a tumor suppressor in human prostate cancer, and disruption of KLF3 (BKLF) function induces a myeloproliferative disorder in mice.8,9 KLF2, the isoform most closely related to KLF4, has been established as a quiescence factor in T lymphocytes.10,11 KLF4 itself has been identified as a tumor suppressor gene in colon and gastric cancer based on studies that show loss of heterozygosity, and is also down-regulated in bladder cancer.12-14 KLF4 promotes cell-cycle arrest as shown by serum withdrawal and overexpression studies in multiple cell lines including in NIH 3T3 fibroblasts and RKO cells (a colonic epithelial cancer cell line).15 KLF4 is up-regulated upon DNA damage in a p53-dependent fashion and can prevent both G1-to-S transition and G2-to-M transitions in colon cancer cell lines.15-17 KLF4 promotes expression of the cell-cycle inhibitor protein p21CIP but in situations where p21CIP is inactivated, KLF4 can act as a tumor promoter by suppressing p53.18,19
In a fraction of patients with B-cell acute lymphoblastic leukemia (B-ALL), tumor cells carry the Philadelphia (Ph) chromosome (9;22). The translocation encodes a fusion protein combining the breakpoint cluster region (BCR) protein with the Abelson tyrosine kinase (ABL).20 Although the ABL kinase inhibitor imatinib has been effective with patients with Ph+ chronic myelogenous leukemia, appearance of resistance to this compound has mitigated the prospect of a cure for this leukemia. Furthermore, acute lymphoblastic leukemia caused by BCR-ABL is more refractory to treatment.21 BCR-ABL, and the related murine oncoprotein v-Abl, activate many mitogenic signaling enzymes and transcription factors that have been extensively studied.22,23 Identifying factors that must be silenced or inactivated to maintain the high proliferative state in ABL-transformed leukemias and other B-cell malignancies provides another avenue with therapeutic potential. In this paper, we test the hypothesis that KLF4 acts as a tumor suppressor in B cells, using ABL oncogenes and murine pro/pre-B cells as a primary model system. We find that KLF4 expression can oppose proliferation and survival of several B-lymphoid cell lines, and can block pre–B-cell transformation and leukemogenesis by BCR-ABL.
Materials and methods
Animals
Mouse procedures were approved by the Institutional Animal Care and Use Committee of the University of California-Irvine. Balb/c and FVB/N mice were purchased from Charles River Laboratories (Wilmington, MA). Transgenic TRE-mycKLF4 (FVB/N background)3 and MMTV-tTA24,52,53 (FVB/N) mice were bred together and given water containing doxycycline (DOX; Sigma-Aldrich, St Louis, MO) at 200 μg/mL throughout gestation and nursing unless otherwise noted. Progeny were genotyped for TRE-mycKLF4 as described3 and for MMTV-tTA by using the following primers: 5′-GCTAGGTGTAGAGCCGCATAC-3′ and 5′-GGCGGCATACTATCAGTAGTA-3′.
Antibodies and inhibitors
Primary antibodies used for immunoblot were: anti–β-actin (Sigma-Aldrich), anti-KLF4 (described previously25 ), anti-KLF4 and anti–c-Myc, (Santa Cruz Biotechnology, Santa Cruz, CA), anti-p21CIP (BD Biosciences, Mountain View, CA), and anti–Bcl-XL (BD Transduction Labs, Lexington, KY). Reagents used for flow cytometry were annexin V–phycoerythrin (PE) (Caltag, Burlingame, CA), anti–B220-fluorescein isothiocynate (FITC) and Streptavidin-Cychrome (BD Biosciences), anti–Thy1.1-Biotin, anti–BP-1-PE, CD43-PE, anti–human CD4-allophycocyanin (APC) (eBiosciences, San Diego, CA), and anti-caspase3 (BD Biosciences). The Abl kinase inhibitor imatinib mesylate was obtained from Calbiochem (San Diego, CA), zVAD-fmk (100 μM) from Alexis Biochemicals (San Diego, CA), and etoposide from Sigma-Aldrich.
Retroviral vectors and generation of virus stocks
cDNAs for KLF4,26 KLF4 (ΔTA), and KLF227 (gift of Kathleen Anderson, University of Cincinnati) were subcloned into the retroviral vectors MSCV-IRES-GFP (pMIG)28 and MSCV-IRES-hCD4 (pMIC)29 (gift from Jason Cyster, University of California-San Francisco). KLF4 (ΔTA) contains an N-terminal deletion of the transcriptional activation domain and is missing the first 109 amino acids. Myc-tagged KLF4 was subcloned into the retroviral vector MSCV-IRES-Thy1.1 (pMIT),28 (gift from Philippa Marrack, National Jewish Medical Center). KLF (ΔDBD), which lacks the zinc finger DNA-binding domain (amino acids 350-483), was subcloned into pMIC. pMIG-Bcl-XL30 was obtained from Craig Walsh (University of California-Irvine). A cDNA for c-Myc in pCDNA3 (gift from Christopher Hughes, University of California-Irvine) was subcloned into pSP72 and then into pMIC. Further cloning details are available on request. Helper-free retrovirus stocks were prepared and titered as described.28
RNA isolation and quantitative real-time polymerase chain reaction (QPCR)
RNA was isolated using TRIZOL (Invitrogen, Carlsbad, CA) and RNAqueous Micro Kit containing DNAse (Ambion, Austin, TX) from primary murine B cells that were purified using magnetic CD43 beads (Miltenyi Biotech, Auburn, CA) as described,31 from B220-positive/IgM-negative bone marrow cells sorted by flow cytometry (MoFlo; Cytomation, Fort Collins, CO), and from BCR-ABL–transformed cells. cDNA was synthesized using the iScript cDNA synthesis kit (Biorad, Hercules, CA). cDNA was then used for quantitative real-time polymerase chain reaction (QPCR; Biorad, iCycler) using SYBR green (Molecular Probes, Eugene, OR) as previously described,5 using primers that span intron boundries (sequences available upon request).
Leukemia and lymphoma cell lines and colony transformation assays
Bone marrow cells were isolated from femurs and tibias of Balb/c mice, double transgenic (TRE-mycKLF4/MMTV-tTA), or nontransgenic littermates. To generate transformed pro-B and pre-B cells, bone marrow was infected with AbMuLV stock (gift of Naomi Rosenberg, Tufts University) or MIG-p190-BCR-ABL (gift of Owen Witte, University of California-Los Angeles) as described.28 Lines were expanded initially in pre–B-cell medium (RPMI 1640, 20% heat-inactivated fetal calf serum [FCS], penicillin (100 U/mL), streptomycin (100 μg/mL), 2 mM l-glutamine, 50 μg/mL gentamicin, 50 μM β-mercaptoethanol). Established lines were later switched to B-cell medium, whose composition is identical except for containing 10% FCS. DOX (1 μg/mL) was included in cultures of double transgenic cells, in order to suppress KLF4 expression. Bulk cell lines established with v-Abl or BCR-ABL were B220+IgM− and were mixed populations of cells expressing markers for either pro-B (CD43intBP-1−) or pre-B cells (CD43lowBP-1+).32 For colony assays, aliquots of 1×105 infected cells were added to methylcellulose METHOCULT M3630 with rhIl-7 (StemCell Technologies, Vancouver, BC, Canada) in duplicate dishes, in the absence or presence of DOX (1 μg/mL). Colony assays were scored 7 days after transduction by adding the total counts from both dishes. Clonal cell lines were established by transferring colonies into liquid culture; these were predominantly pro-B based on surface phenotype (CD43intCD24+CD25−) although some contained cells expressing lower levels of CD43 along with the large pre–B-cell marker CD25.33
Other murine B-lineage cell lines used in this study were Baf/3 cells stably expressing p210-BCR-ABL (Baf/3p210, gift of Tiong Ong, University of California-Irvine); 70Z/334 and A2035 (Ig− pre-B leukemia and mature IgG+ lymphoma, respectively; gift of Owen Witte, University of California-Los Angeles) and M1235 (mature IgG+ lymphoma; gift of Michael Gold, University of British Columbia). These were grown in B-cell medium.
Retroviral transductions of transformed cell lines
We used the MSCV-based retroviral vectors described in “Retroviral vectors and generation of virus stocks” to infect 70Z/3 cells and to superinfect v-Abl cells as well as cell lines established with MIG-p190-BCR-ABL. Cells (2 × 106) in 1 mL pre–B-cell medium were mixed with 1 mL virus and 8 μg/mL polybrene and centrifuged in 24-well plates at 33°C for 45 minutes at 450g. Transduction efficiencies after 24 hours were typically 5% to 20% for KLF2 and KLF4, and 30% to 50% for empty vector and KLF4 mutants. Bcl-XL–transduced cells were used after 1 week of culture at which time approximately 80% to 90% were marker-positive. For A20 and M12 cells, plasmid DNA was electroporated as described previously.36 After incubation for 20 to 24 hours at 37°C, cells were either analyzed by FACS or expanded into 3 mL fresh medium. To prepare an enriched population from cells infected with MIC, MIC-KLF2, or MIC-KLF4, we used anti–human CD4 micro beads (Miltenyi Biotech) 24 hours after infection. Sorted cells were tested for purity by FACS (FACSCalibur; BD Biosciences) and then used for either injection or immunoblot analysis.
FACS analysis and BrdU
v-Abl–transformed cells at 42 hours after infection with MIC or MIC-KLF4 were pulsed with BrdU for 6 hours prior to surface staining for human CD4, then fixed and stained with anti–BrdU-FITC according to the manufacturer's instructions (BD BioSciences). For apoptotic assays, 32-hour postinfection cells were untreated or treated with zVAD or etoposide and then stained at 48 hours with the following reagents: 7-aminoactinomycin D (7-AAD; Molecular Probes) to assess plasma-membrane integrity; annexin V–PE to detect flipped phosphatidylserine; and/or 3,3′-dihexyloxacarbocyanine iodide (DiOC6; Molecular Probes) to detect loss of mitochondrial-membrane potential. Cells were also stained with antibodies to human CD4 or Thy1.1 to distinguish the infected cell population, and then analyzed by flow cytometry. For cell-cycle analysis, cells were fixed in ethanol and stained with propidium iodide or stained with 7-AAD as described.31,37 The percentages of cells in different phases of the cell cycle were calculated with the flow cytometry analysis software FlowJo (Treestar, Ashland, OR), using the Watson pragmatic model.
In vivo leukemogenesis assay
Bone marrow cells (1 × 106) from double transgenic (TRE-mycKLF4/MMTV-tTA) or nontransgenic littermates (all in FVB/N background) were infected with MIG-p190-BCR-ABL and injected intravenously into syngeneic (FVB/N) mice that had been lethally irradiated (9 Gy from 60Co source, in 2 doses separated by 4 hours). Animals were monitored daily for symptoms of leukemia as defined by partial paralysis and poor grooming. Sick animals were humanely killed, and bone marrow and spleen were harvested for analysis of disease phenotype as described.28
Immunoprecipitation/immunoblotting
Cells (cell numbers shown in the figure legends) were washed in 1x phosphate buffered saline (PBS), then lysed in 40 μL lysis buffer (50 mM Tris, pH 7.6, 150 mM NaCl, 1% Triton X-100, 10% glycerol) for direct immunoblot analysis. Lysates corresponding to equal cell numbers were electrophoresed (7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis [SDS-PAGE]) and transferred to nitrocellulose (GE Osmonics, Minnetonka, MN). Immunoblotting was done as described.31
Oncomine analysis
The Oncomine DNA microarray database contains compiled mean expression values from the following number of samples for each of the 4 separate studies. The first study contained 6 human normal germinal center B-cell samples, 11 large B-cell lymphoma, and 11 follicular lymphoma samples (Figure 1C, Figure 5D).38 The second study contained 3 normal skin and 3 normal tonsil tissue with 8 cutaneous diffuse large B-cell and 5 marginal zone lymphoma patient samples (Figure 1C, Figure 5C).39 The third study contained 3 normal resting CD19+ B cells, 3 activated CD19+ B cells, and 4 germinal center B cells, 3 chronic lymphocytic leukemia, and 274 diffuse large B-cell lymphoma samples (Figure 1D).40 Expression of KLF1, KLF2, and KLF3 were not tested in the third study. The last study contained 31 normal lymphoid, 9 follicular lymphoma, and 12 chronic lymphocytic leukemia samples (Figure 5C-D).41 For further information on the statistical tools used for the Oncomine analysis refer to previously published work.42
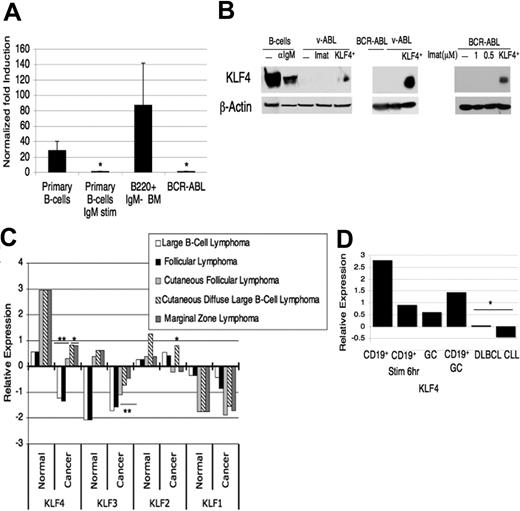
KLF4 is expressed at low levels in B cells transformed by p190 BCR/ABL compared with untransformed B cells. (A) cDNA was prepared from purified resting splenic B cells, splenic B cells activated by anti-IgM, B220+IgM− precursor cells from the bone marrow of wild-type mice, and BCR-ABL–transformed pre-B cells. Real-time quantitative PCR was performed, expression was normalized to β-actin, the value for BCR-ABL–transformed cells was set to 1, and fold-increase was calculated. Data are representative of at least 4 experiments. *P < .02 relative to the BCR-ABL–transformed cells. Error bars indicate standard deviation (SD). (B) Immunoblots were prepared with lysates from indicated cell preparations. The left blot was probed with rabbit anti-KLF4,24 and the right blots with rabbit anti-KLF4 from Santa Cruz Biotechnology. β-Actin was used as a loading control. v-Abl and BCR-ABL indicate transformed pre-B-cell lines; Imat, 4 hours treatment with imatinib (10 μM), the ABL kinase inhibitor; KLF4+, v-Abl cells infected with MIC-KLF4 and sorted for hCD4 24 hours later. The blot on the far right shows cells treated with subtoxic concentrations of imatinib for 24 hours, with sorted KLF4+ infected cells used as a positive control. (C) RNA-expression data from the Oncomine microarray database was compiled for different B-cell malignancies and the normal tissue control for that study, either germinal center B cells or tonsil tissue. Numbers on the y-axis represent the number of standard deviations from the median expression value from thousands of genes tested in each microarray experiment and the P values indicate the significance between the normal and cancer tissue for the respective study (*P < .05, **P < .001). (D) Oncomine data derived from a study41 comparing peripheral blood CD19+ B cells, either resting or activated for 6 hours, total germinal center (GC) cells, a large panel of diffuse large B-cell lymphomas, and chronic lymphocytic leukemias. CD19+GC refers to the average of all nontransformed B-cell samples in the study.
KLF4 is expressed at low levels in B cells transformed by p190 BCR/ABL compared with untransformed B cells. (A) cDNA was prepared from purified resting splenic B cells, splenic B cells activated by anti-IgM, B220+IgM− precursor cells from the bone marrow of wild-type mice, and BCR-ABL–transformed pre-B cells. Real-time quantitative PCR was performed, expression was normalized to β-actin, the value for BCR-ABL–transformed cells was set to 1, and fold-increase was calculated. Data are representative of at least 4 experiments. *P < .02 relative to the BCR-ABL–transformed cells. Error bars indicate standard deviation (SD). (B) Immunoblots were prepared with lysates from indicated cell preparations. The left blot was probed with rabbit anti-KLF4,24 and the right blots with rabbit anti-KLF4 from Santa Cruz Biotechnology. β-Actin was used as a loading control. v-Abl and BCR-ABL indicate transformed pre-B-cell lines; Imat, 4 hours treatment with imatinib (10 μM), the ABL kinase inhibitor; KLF4+, v-Abl cells infected with MIC-KLF4 and sorted for hCD4 24 hours later. The blot on the far right shows cells treated with subtoxic concentrations of imatinib for 24 hours, with sorted KLF4+ infected cells used as a positive control. (C) RNA-expression data from the Oncomine microarray database was compiled for different B-cell malignancies and the normal tissue control for that study, either germinal center B cells or tonsil tissue. Numbers on the y-axis represent the number of standard deviations from the median expression value from thousands of genes tested in each microarray experiment and the P values indicate the significance between the normal and cancer tissue for the respective study (*P < .05, **P < .001). (D) Oncomine data derived from a study41 comparing peripheral blood CD19+ B cells, either resting or activated for 6 hours, total germinal center (GC) cells, a large panel of diffuse large B-cell lymphomas, and chronic lymphocytic leukemias. CD19+GC refers to the average of all nontransformed B-cell samples in the study.
Results
B-cell lineage malignancies and stimulated primary B cells express low levels of KLF4
To identify possible B-cell tumor suppressors, we searched for genes whose expression is diminished in activated primary B cells compared with resting cells. Analysis of microarray datasets revealed that KLF4 was one of the transcription factors down-regulated in B cells activated by antigen receptor crosslinking (I.Y., M.G.K., Jing Chen, Autumn Maruniak, Pratibha Sareen, Mary Tomayko, Mark Schlomchik, V.W.Y., Klaus Kaestner, and D.A.F.; “KLF4 is a FOXO target gene that suppresses B-cell proliferation,” manuscript submitted July 2006).5,43 We confirmed this observation by QPCR (Figure 1A). In sorted B220+/IgM− murine bone marrow cells (containing pro-B and pre-B cells), KLF4 RNA was expressed to a similar degree as in unstimulated splenic B cells (Figure 1A). In contrast, murine pro-B/pre-B cell lines transformed with BCR-ABL or v-Abl expressed amounts of KLF4 mRNA and protein that were comparable to or lower than stimulated splenic B cells (Figure 1A-B). To investigate further the correlation between B-cell malignancy and KLF4 down-regulation, we examined the Oncomine database42 for KLF4 expression. This analysis showed KLF4 to be consistently expressed at low levels in several types of B-lineage lymphomas and leukemias relative to normal tissue (Figure 1C). This pattern of down-regulation was specific in that it was not seen in the closely related Krüppel-like factors KLF2 or KLF1. KLF3 expression was low in some but not all B-lymphoid malignancies examined. In concordance with KLF4-expression data in murine B cells (I.Y., M.G.K., Jing Chen, Autumn Maruniak, Pratibha Sareen, Mary Tomayko, Mark Schlomchik, V.W.Y., Klaus Kaestner, and D.A.F.; “KLF4 is a FOXO target gene that suppresses B-cell proliferation,” manuscript submitted July 2006)5,43 (Figure 1A), the Oncomine data indicated that normal human blood CD19+ B cells down-regulated KLF4 after stimulation and had higher expression compared with leukemia and lymphoma samples (Figure 1D).
Overexpression of KLF4 inhibits propagation of B-cell lines
In several cell types, including fibroblasts and colon cancer cell lines, overexpression of KLF4 induces cell-cycle arrest.15-17 We tested if forced expression of KLF4 can affect proliferation or survival in transformed murine B-lineage cell lines. Primary pre-B cells transformed by v-Abl or p190-BCR-ABL, BaF/3 cells stably expressing p210-BCR-ABL (BaF/3p210), mature lymphoma cell lines A20 and M12, and pre–B-lymphoma 70Z/3 cells were either transfected or transduced with retroviruses to express KLF4, KLF2, and KLF4 mutants. In these constructs, the murine stem-cell virus long terminal repeat (MSCV-LTR) drives expression of the gene of interest and an internal ribosome entry site (IRES)–linked marker gene to identify transduced cells by fluorescence-activated cell sorting (FACS). To examine the effect of KLF4 on cell proliferation and survival, we measured the maintenance of transduced cells in the population over time. We determined the percentage of marker-positive cells by FACS at 24, 48, and 84 hours after infection, and normalized to the percentage at 24 hours (Figure 2A). In each B-lymphoid cell line, KLF4-expressing cells were depleted rapidly whereas cells infected with control virus were maintained or increased. The overexpression of KLF4 was verified by sorting transduced v-Abl cells or BCR-ABL cells at 24 hours and immunoblotting lysates for KLF4 protein (Figure 1B). Expression of a mutant KLF4 lacking the transactivation domain (KLF4(ΔTA)) had variable effects among cell lines, but was generally less effective than wild-type KLF4 at depleting cells. Expression of this mutant and the other KLF constructs described in Figure 3 was verified in transduced cells by immunoblotting (data not shown).
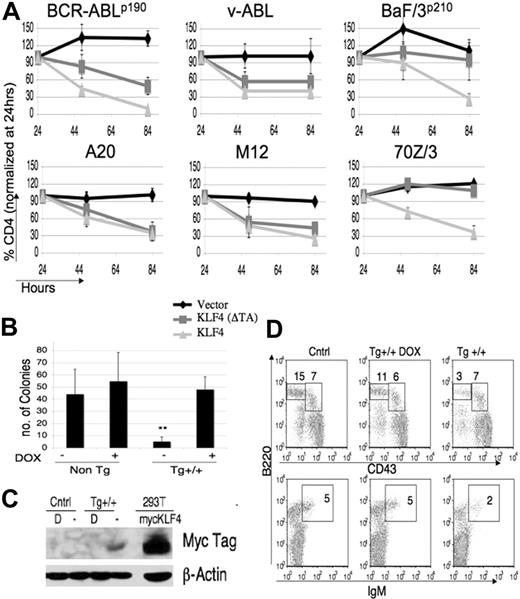
KLF4 overexpression inhibits propagation of B-lymphoid cell lines and blocks transformation of pre-B cells by BCR-ABL. (A) BCR-ABL–transformed pre-B cells, v-Abl–transformed pre-B cells, BaF/3p210 cells, A20, M12, or 70Z/3 cells were transfected or transduced with retroviral vectors including empty vector (MIT or MIC), KLF4(ΔTA) (mutant lacking transactivation domain), KLF4(ΔDBD) (lacking DNA binding domain), or KLF2, a closely related transcription factor. The percentage of marker-positive cells was determined 24, 48, and 84 hours after infection. Data were normalized to the percent positive at 24 hours, which was set at 100% for each virus. All graphs depict the mean plus or minus the standard deviation (SD) of 3 to 5 experiments. (B) A colony formation assay using MMTV-tTA/TRE-KLF4 double transgenic or control bone marrow, transformed with BCR-ABL and incubated with or without DOX. Colonies were scored after 7 days (**P < .01). Data are representative of 5 experiments. (C) Bone marrow from indicated mice was grown in the presence of IL-7 for 5 days treated with or without DOX (“D”), and 1 × 106 cells were immunblotted for myc-tagged KLF4 and β-actin as a loading control. 293T cells transfected with myc-tagged KLF4 were used as a positive control. (D) Bone marrow from indicated mice maintained with or without DOX was stained to determine percentages of progenitor IgM− B cells (pre-B cells: IgM−, B220+, CD43low; pro-B cells: IgM−, B220+, CD43int) and then analyzed by FACS. Cntrl indicates nontransgenic littermates. Results are representative of 5 sets of mice.
KLF4 overexpression inhibits propagation of B-lymphoid cell lines and blocks transformation of pre-B cells by BCR-ABL. (A) BCR-ABL–transformed pre-B cells, v-Abl–transformed pre-B cells, BaF/3p210 cells, A20, M12, or 70Z/3 cells were transfected or transduced with retroviral vectors including empty vector (MIT or MIC), KLF4(ΔTA) (mutant lacking transactivation domain), KLF4(ΔDBD) (lacking DNA binding domain), or KLF2, a closely related transcription factor. The percentage of marker-positive cells was determined 24, 48, and 84 hours after infection. Data were normalized to the percent positive at 24 hours, which was set at 100% for each virus. All graphs depict the mean plus or minus the standard deviation (SD) of 3 to 5 experiments. (B) A colony formation assay using MMTV-tTA/TRE-KLF4 double transgenic or control bone marrow, transformed with BCR-ABL and incubated with or without DOX. Colonies were scored after 7 days (**P < .01). Data are representative of 5 experiments. (C) Bone marrow from indicated mice was grown in the presence of IL-7 for 5 days treated with or without DOX (“D”), and 1 × 106 cells were immunblotted for myc-tagged KLF4 and β-actin as a loading control. 293T cells transfected with myc-tagged KLF4 were used as a positive control. (D) Bone marrow from indicated mice maintained with or without DOX was stained to determine percentages of progenitor IgM− B cells (pre-B cells: IgM−, B220+, CD43low; pro-B cells: IgM−, B220+, CD43int) and then analyzed by FACS. Cntrl indicates nontransgenic littermates. Results are representative of 5 sets of mice.
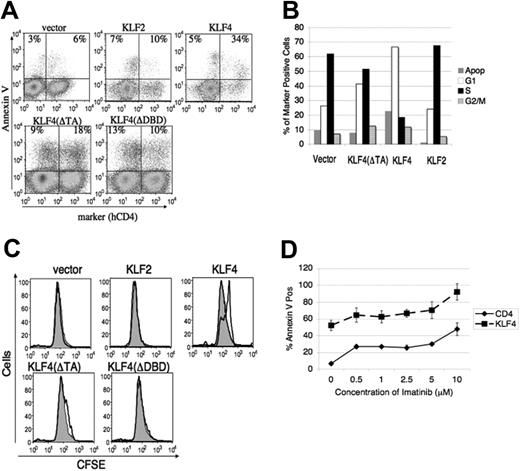
KLF4 induces cell-cycle arrest/delay and death that is nonredundant with imatinib. (A) v-Abl–transformed cells were harvested at 48 hours after infection and stained for hCD4 and annexin V for FACS analysis. The percentages of death in marker-negative and -positive cells are displayed in the top left and right quadrants, respectively. Data are representative of 3 to 5 experiments. (B) v-Abl–transformed cells were infected and harvested as in panel A and then stained for marker and propidium iodide to measure cell cycle. Percentages of G1, S, and G2/M were calculated from a live cell gate, with apoptosis calculated as the fraction of total cells with subdiploid DNA content. Data are representative of at least 5 experiments. (C) v-Abl–transformed cells were stained with CFSE, infected with indicated retroviruses, and analyzed by FACS 24 hours after transduction. The gray shaded trace represents the population that is negative for the marker gene while the darker line represents cells that have been infected and express the marker. Data are representative of 3 experiments. (D) v-Abl cells were incubated with the indicated concentration range of imatinib 24 hours after transduction with MIC or MIC-KLF4 and then stained with annexin V 24 hours later for analysis by FACS. Representative plot from 2 experiments is shown and error bars represent SD.
KLF4 induces cell-cycle arrest/delay and death that is nonredundant with imatinib. (A) v-Abl–transformed cells were harvested at 48 hours after infection and stained for hCD4 and annexin V for FACS analysis. The percentages of death in marker-negative and -positive cells are displayed in the top left and right quadrants, respectively. Data are representative of 3 to 5 experiments. (B) v-Abl–transformed cells were infected and harvested as in panel A and then stained for marker and propidium iodide to measure cell cycle. Percentages of G1, S, and G2/M were calculated from a live cell gate, with apoptosis calculated as the fraction of total cells with subdiploid DNA content. Data are representative of at least 5 experiments. (C) v-Abl–transformed cells were stained with CFSE, infected with indicated retroviruses, and analyzed by FACS 24 hours after transduction. The gray shaded trace represents the population that is negative for the marker gene while the darker line represents cells that have been infected and express the marker. Data are representative of 3 experiments. (D) v-Abl cells were incubated with the indicated concentration range of imatinib 24 hours after transduction with MIC or MIC-KLF4 and then stained with annexin V 24 hours later for analysis by FACS. Representative plot from 2 experiments is shown and error bars represent SD.
Inducible expression of KLF4 blocks transformation
We next tested if KLF4 could inhibit the initiation of transformation. Due to the low efficiency of double retroviral transductions on primary murine bone marrow, we used an inducible “Tet-off” transgenic system in which overexpression of KLF4 can be induced in target tissues by withdrawal of tetracycline or its analog doxycyline (DOX). For this approach, we crossed transgenic mice carrying a KLF4 cDNA driven by tetracycline-responsive elements (TRE-KLF4) with transgenic mice expressing the tTA transactivator protein from the MMTV promoter (MMTV-tTA). TRE-KLF4 transgenics have been used previously to explore the role of KLF4 in skin development and dysplasia, by crossing with transgenic mice in which expression of tTA was driven by keratinocyte-specific promoters.3,44 In MMTV-tTA mice, transactivator is expressed in several tissues including B-cell progenitors in the bone marrow.24 We bred the 2 transgenic lines in the presence of DOX to assure normal development and to create double transgenic mice that could induce KLF4 expression in B-cell progenitors upon withdrawal of DOX. We transformed bone marrow from double transgenic mice or nontransgenic mice with p190-BCR-ABL and performed colony assays using methylcellulose supplemented with or without DOX. There was a dramatic reduction in colony formation when DOX was withheld from the double transgenic cells to induce KLF4 expression (Figure 2B). Colony formation in double transgenic cells plated in the presence of DOX was equivalent to colony numbers in wild-type samples plated in the absence or presence of DOX. We were not able to detect myc-KLF4 in transgenic cell lines or colonies after DOX withdrawal (see “Discussion”). However, we verified the inducible expression of KLF4 by growing IL-7–supplemented bone marrow pro/pre–B-cell cultures in the presence or absence of DOX and immunoblotting for the myc tag (Figure 2C). We conclude that this system allows inducible expression of KLF4 in developing B cells, and blocks B-lymphoid transformation by BCR-ABL in vitro.
Interestingly, when DOX was withheld from double transgenic mice during gestation and early postnatal life, the percentage of bone marrow pre-B cells was consistently reduced (Figure 2D; (–DOX) = 6% ± 3% vs (+DOX) = 21% ± 6%, P < .001) even though there was no statistical difference in the absolute numbers of bone marrow cells (data not shown). This suggests that KLF4 expression can also suppress proliferation or survival of normal pre-B cells in vivo.
KLF4 induces cell death and cell-cycle arrest/delay
We then investigated whether the effects of KLF4 were the result of reduced proliferation, increased death, or both. For most of these assays we used v-Abl–transformed cells, which lacked green fluorescent protein (GFP) expression thus facilitating FACS-based assays that use the first fluorescence channel. KLF4-transduced cells displayed markedly greater death as determined by annexin V staining (Figure 3A) and DNA content analysis (Figure 3B), compared with cells transduced with empty virus. KLF4 also induced cell-cycle arrest or delay, as shown by an increased percentage in G1 phase and a decrease in S phase (Figure 3B). To further examine proliferation we stained the cells with the cell division tracker dye carboxy-fluorescein diacetate, succinimidyl ester (CFSE). Labeled cells were infected with retroviruses and analyzed for CFSE fluorescence 48 hours after infection (Figure 3C). KLF4-transduced cells proliferated at a slower rate of about 1 division behind the uninfected cells in the same culture.
To determine if these effects were specific to KLF4, we overexpressed the most closely related KLF family member, KLF2. In addition, we examined the role of the KLF4 transactivation domain using the KLF4(ΔTA) mutant, and the KLF4 DNA-binding domain using the KLF4(ΔDBD) mutant. KLF2 had no appreciable effect on cell cycle, proliferation, or survival of v-Abl–transformed cells (Figure 3A-C). The maximal KLF4-mediated cell death and cycle arrest required both DNA binding activity and the transactivation domains (Figure 3A-B). However, KLF4(ΔTA) did mediate a modest but reproducible increase in G1 phase and death, consistent with decreased maintenance seen in Figure 2A. Collectively, these data provide evidence that KLF4 has a potent and specific ability to suppress proliferation and survival in transformed pre-B cells.
The effects of ABL kinase inhibition and KLF4 are additive
In order to distinguish if imatinib-induced death and KLF4-mediated death occur via shared or separate pathways, we first assessed whether ABL kinase inhibition led to increased KLF4 expression. When we treated BCR-ABL–transformed pro/pre-B cells with varying concentrations of imatinib (0.5 μM-50 μM) for various times (4-24 hours), we found in about 70% of cell lines (9/13) that KLF4 protein was maintained at low levels (Figure 1B and data not shown). These data indicated that BCR-ABL tyrosine kinase activity does not suppress KLF4 expression in established cell lines. To determine if KLF4 expression and ABL kinase inhibition have redundant effects on cell death, we infected cells with MIC-KLF4 or empty virus and measured cell death over a range of imatinib concentrations (Figure 3D). The effect of KLF4 on cell death was additive with the effect of imatinib at all concentrations. These data support the conclusion that the death pathways occur in parallel and are nonredundant.
KLF4-mediated death occurs in the G1 phase
To elucidate the specific stage of the cell cycle during which KLF4 overexpression causes cell death, we measured BrdU incorporation and DNA content (Figure 4A). The fraction of KLF4-expressing cells entering S-phase during a 6-hour BrdU pulse was substantially reduced compared with cells expressing empty vector (50% ± 10% vs 84% ± 12%). The majority of remaining cells appeared to be in G1 phase (KLF4: 28% ± 0.6% vs vector: 7% ± 0%), and most had not degraded their DNA as judged by subdiploid DNA content (KLF4: 8% ± 2% vs vector: 0.7% ± 0.5%). Nevertheless, most cells in the G1 gate were dead as judged by forward and side scatter parameters (KLF4: 74% ± 11% vs vector: 10% ± 10%, Figure 4A, bottom panels). The cell cycle, CFSE, and BrdU experiments provide strong evidence that cells overexpressing KLF4 are delayed in exiting G1, and are susceptible to death in this cell-cycle phase.
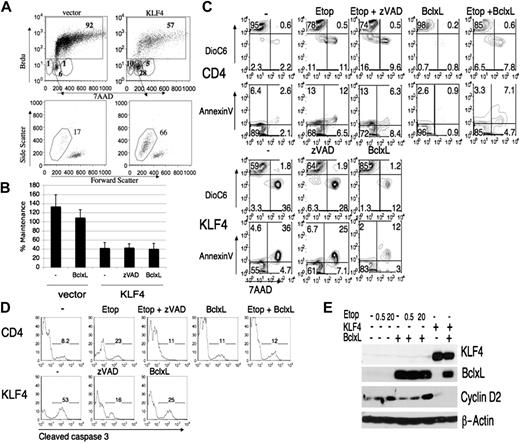
KLF4 infected cells arrest in G1 and die by apoptosis. (A) v-Abl cells were infected with MIC or MIC-KLF4 and then pulsed with BrdU for 6 hours before harvesting at 48 hours. Cells were stained with anti-BrdU antibody and 7-AAD to assess cell-cycle status. Top panel includes the percentages of each phase of the cell cycle, with BrdU-positive cells boxed. Bottom circular gates represent, from left to right, subdiploid, G1, combined S/G2 that have not divided during the BrdU pulse. The bottom panel shows the forward–side scatter dot plot of the G1 cells that have not entered S phase during the BrdU pulse. Many cells within this gate were no longer live cells based on laser scatter parameters and percent death is displayed above the gate. (B) BCR-ABL cells, before or after infection with MIG-Bcl-XL, were superinfected with MIC-KLF4 or empty MIC virus. Some KLF4-transduced samples were treated with zVAD-fmk (100 μM), which was not toxic under these conditions (data not shown). Cells were stained for hCD4 at 24 hours or 48 hours and then analyzed by FACS. The percentage of hCD4+ cells remaining from 24 hours to 48 hours (maintenance) was calculated as in Figure 2A. Error bars represent standard deviations of 3 independent experiments. (C) BCR-ABL cells were treated with etoposide (20 μM) or infected with MIC-KLF4, in the absence or presence of Bcl-XL overexpression, as indicated. Staining with 7-AAD, annexin V, and DioC6 was used to assess plasma-membrane integrity, phosphatidylserine exposure, and mitochondrial-membrane potential. (D) BCR-ABL cells infected with empty MIC virus (CD4) or MIC-KLF4 were treated as in panel C then fixed, permeabilized, and stained for cleaved caspase-3. Data are representative of at least 3 experiments. (E) Cells were infected with MIC-KLF4 and/or MIG-Bcl-XL, then cultured in the absence or presence of etoposide (concentrations in μM) for 24 hours before sorting for hCD4 marker gene expression. Equivalent numbers of sorted cells were then immunoblotted with indicated antibodies, and β-actin was used as a loading control.
KLF4 infected cells arrest in G1 and die by apoptosis. (A) v-Abl cells were infected with MIC or MIC-KLF4 and then pulsed with BrdU for 6 hours before harvesting at 48 hours. Cells were stained with anti-BrdU antibody and 7-AAD to assess cell-cycle status. Top panel includes the percentages of each phase of the cell cycle, with BrdU-positive cells boxed. Bottom circular gates represent, from left to right, subdiploid, G1, combined S/G2 that have not divided during the BrdU pulse. The bottom panel shows the forward–side scatter dot plot of the G1 cells that have not entered S phase during the BrdU pulse. Many cells within this gate were no longer live cells based on laser scatter parameters and percent death is displayed above the gate. (B) BCR-ABL cells, before or after infection with MIG-Bcl-XL, were superinfected with MIC-KLF4 or empty MIC virus. Some KLF4-transduced samples were treated with zVAD-fmk (100 μM), which was not toxic under these conditions (data not shown). Cells were stained for hCD4 at 24 hours or 48 hours and then analyzed by FACS. The percentage of hCD4+ cells remaining from 24 hours to 48 hours (maintenance) was calculated as in Figure 2A. Error bars represent standard deviations of 3 independent experiments. (C) BCR-ABL cells were treated with etoposide (20 μM) or infected with MIC-KLF4, in the absence or presence of Bcl-XL overexpression, as indicated. Staining with 7-AAD, annexin V, and DioC6 was used to assess plasma-membrane integrity, phosphatidylserine exposure, and mitochondrial-membrane potential. (D) BCR-ABL cells infected with empty MIC virus (CD4) or MIC-KLF4 were treated as in panel C then fixed, permeabilized, and stained for cleaved caspase-3. Data are representative of at least 3 experiments. (E) Cells were infected with MIC-KLF4 and/or MIG-Bcl-XL, then cultured in the absence or presence of etoposide (concentrations in μM) for 24 hours before sorting for hCD4 marker gene expression. Equivalent numbers of sorted cells were then immunoblotted with indicated antibodies, and β-actin was used as a loading control.
Caspases are not required for KLF4-mediated death
Although we observed a portion of cells that had subdiploid amounts of DNA, indicative of apoptosis, a majority of the dead cells had not degraded their DNA (Figure 4A). This prompted us to investigate further the mechanism of this form of cell death. Apoptotic cells undergo an ordered series of changes including flipping of phosphatidylserine in the membrane, loss of mitochondrial-membrane potential, caspase activation, and DNA degradation. An alternative form of cell death is necrosis, in which DNA is not degraded and there is a usually rapid loss of membrane integrity.
As a benchmark for apoptosis we treated cells with etoposide, a topoisomerase inhibitor that induces apoptosis in various cell lines. To inhibit caspases we used zVAD-fmk. We transduced BCR-ABL–transformed cells with KLF4 or empty vector, then cultured the cells in the presence or absence of etoposide and/or zVAD-fmk. At various time points thereafter we measured maintenance of marker-positive cells, mitochondrial-membrane potential (decrease in DioC6 staining), and membrane integrity (annexin V or 7-AAD) (Figure 4B-C). In cells expressing KLF4 or treated with etoposide there was a loss of mitochondrial-membrane potential and an increase in annexin V/7-AAD staining. Inhibition of caspases by zVAD-fmk in the KLF4-transduced cells did not prevent the loss of maintenance and mitochondria-membrane potential (Figure 4B-C). zVAD-fmk also had little effect on cell death induced by etoposide. In control experiments we observed that both KLF4 expression and etoposide treatment augmented caspase-3 cleavage that was mostly blocked by zVAD treatment (Figure 4D). Our results indicate that caspase activation occurs but is not required for induction of cell death in response to either KLF4 expression or etoposide treatment in this cell system.
Bcl-XL uncouples cell death and cell-cycle arrest
The observation that the cells lost mitochondrial-membrane potential prompted us to determine whether expression of proteins that preserve mitochondrial integrity could protect from KLF4-induced cell depletion. Bcl-XL is a pro-survival member of the Bcl-2 family that opposes apoptosis through the mitochondrial pathway. We infected BCR-ABL–transformed cells with a virus expressing Bcl-XL and then superinfected these cells with viruses containing KLF4, or with empty vector retroviruses followed by treatment with etoposide. The Bcl-XL–expressing cells were partially rescued from the effects of etoposide as judged by measurement of mitochondrial-membrane potential and annexin V (Figure 4C). In contrast, KLF4-transduced Bcl-XL–expressing cells showed a decrease in maintenance even though there was an increase in cells with intact mitochondrial-membrane potential and plasma-membrane integrity and a decrease in caspase-3 activation compared with KLF4-transduced cells lacking Bcl-XL (Figure 4B-D).
These results could be explained by continued cell-cycle arrest by KLF4 even in cells with antiapoptotic Bcl-XL expression. To explore this possibility, we sorted KLF4/Bcl-XL and KLF4-expressing cells 24 hours after infection and immunoblotted for KLF4, Bcl-XL, and cyclin D2, an important regulator of cell-cycle progression in B-lineage cells.45 This time point was chosen on the assumption that depletion of the transduced cells at later time points (Figure 2A) is the result of transcriptional changes likely to be initiated by 24 hours. We verified the overexpression of Bcl-XL and KLF4 and found that uninfected cells express cyclin D2 that is down-regulated in KLF4-transduced cells regardless of Bcl-XL expression (Figure 4E). The cells that were treated with varying concentrations of etoposide maintained their cyclin D2 expression even in the presence of Bcl-XL, indicating that etoposide was mainly inducing apoptosis and not affecting cell cycle. These data indicate that Bcl-XL overexpression uncouples apoptosis from cell-cycle arrest in KLF4-transduced cells. Overall, these analyses show that KLF4 overexpression induces cell-cycle arrest, and an intrinsic form of cell death that is blocked by Bcl-XL, similar to the death caused by the known apoptotic inducer etoposide.
KLF4 modulates c-Myc and p21CIP expression
To investigate further the molecular mechanisms by which KLF4 promotes cell-cycle arrest and apoptosis, we compared the expression of possible KLF target genes in transduced and nontransduced cells. We chose to examine the expression of p21CIP, a known target gene of KLF4 in fibroblasts,46 and c-Myc, a gene down-regulated by KLF2 in T cells.10 We observed that KLF4 expression decreased c-Myc and increased p21CIP expression (Figure 5A).
KLF4 regulates expression of candidate target genes. (A) Forced KLF4 expression reduces c-Myc and augments p21CIP expression. BCR-ABL–transformed cells were infected with MIC-KLF4 and magnetically sorted after 24 hours. Parental cells were left untreated or treated with imatinib for 4 hours. Lysates were immunoblotted with indicated antibodies. Representative blot from 3 experiments is shown. (B) Restoration of c-Myc does not rescue depletion by KLF4. v-Abl cells were double infected as in Figure 4B and maintenance of cells expressing both markers was measured from 24 to 48 hours. (+) indicates empty vector. (C, D) Compiled Oncomine expression data for p21CIP and cyclin D2 with normal lymphoid tissue compared with the indicated B-cell malignancies (*P < .05, **P < .01).
KLF4 regulates expression of candidate target genes. (A) Forced KLF4 expression reduces c-Myc and augments p21CIP expression. BCR-ABL–transformed cells were infected with MIC-KLF4 and magnetically sorted after 24 hours. Parental cells were left untreated or treated with imatinib for 4 hours. Lysates were immunoblotted with indicated antibodies. Representative blot from 3 experiments is shown. (B) Restoration of c-Myc does not rescue depletion by KLF4. v-Abl cells were double infected as in Figure 4B and maintenance of cells expressing both markers was measured from 24 to 48 hours. (+) indicates empty vector. (C, D) Compiled Oncomine expression data for p21CIP and cyclin D2 with normal lymphoid tissue compared with the indicated B-cell malignancies (*P < .05, **P < .01).
The marked decrease in c-Myc expression in KLF4-expressing cells suggested a possible role for c-Myc repression in the mechanism of death or cell-cycle arrest. We used combinations of viruses to assess if reintroduction of c-Myc could rescue KLF4-mediated depletion. The percentage of cells infected with both MIC-KLF4 and MIG-c-Myc was determined at 24 and 48 hours and the percent maintenance was calculated as in Figure 2. This analysis showed that forced expression of c-Myc could not prevent KLF4-mediated depletion (Figure 5B). We then re-examined the Oncomine database to determine if expression of these potential KLF4 targets (p21CIP, c-Myc, and cyclin D2) correlated with KLF4 expression in human leukemias and lymphomas. This analysis showed that p21CIP was down-regulated and cyclin D2 was expressed at a higher degree in leukemias and lymphomas compared with normal lymphoid tissue (Figure 5C-D), similar to the pattern of KLF4 (Figure 1C). The expression of c-Myc did not correlate as closely although it was up-regulated in some transformed B-lineage malignancies (data not shown).
Inducible expression of KLF4 in vivo depletes pre–B-cell leukemia
The ability of KLF4 overexpression to block transformation by BCR-ABL in vitro prompted us to assess whether leukemia could be delayed by KLF4. In initial experiments we infected v-ABL–transformed pro/pre-B cell lines with MIC-KLF4 or MIC alone, sorted for hCD4-positive cells after 24 hours, and injected the cells into sublethally irradiated hosts. All mice died of leukemia with equivalent latency of approximately of 17 to 22 days (data not shown). However, when we examined the cells by FACS, we could not detect the hCD4 marker gene in cells obtained from animals that had received bone marrow cells transduced with MIC-KLF4. In contrast, hCD4 was present on nearly all leukemic cells from mice injected with empty vector-transduced cells. These findings indicated that there was selection against the cells that overexpressed KLF4. Presumably, a small population of uninfected cells (or cells silencing the retrovirus) was able to expand in vivo to produce leukemia.
To assess leukemogenesis in a system with regulatable KLF4 expression, we used the double transgenic system described in Figure 2B. We transduced nontransgenic or MMTV-tTA/TRE-KLF4 bone marrow cells with p190-BCR-ABL, and then injected the cells into lethally irradiated syngeneic recipients. The recipient mice were given water with or without DOX, then monitored for disease progression. We found that mice died of leukemia with equivalent latency regardless of treatment or genotype (data not shown). We used flow cytometry to determine if there were any differences in cell lineage among the leukemias. The results showed that leukemias arising from nontransgenic cells contained a mixture of pro-B and pre-B cells whereas leukemias arising from double transgenic cells in the absence of DOX contained almost exclusively pro-B cells (Figure 6B-C). We were not able to detect myc-tagged KLF4 protein in double transgenic pro-B-cell leukemias derived from DOX-withdrawn mice (data not shown), as we observed for cell lines established in vitro (see “Discussion”). Double transgenic leukemias arising in the presence of DOX showed an intermediate effect on the pro-B/pre-B ratio (Figure 6), perhaps due to leakiness of the Tet-Off system in vivo. These data indicate that targeted inducible expression of KLF4 in vivo can deplete a specific lineage of leukemic cells.
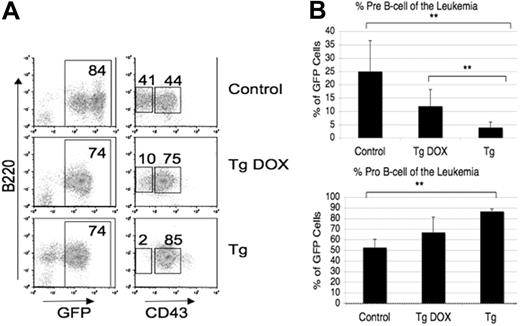
DOX-regulated KLF4 expression depletes BCR-ABL–transformed pre-B cells in vivo. Bone marrow from FVB/N mice that were either nontransgenic or double transgenic for MMTV-tTA and TRE-KLF4 was transduced with BCR-ABL and injected in lethally irradiated FVB/N mice. Recipients were given water with or without DOX. (A, B) Leukemic mouse bone marrow was analyzed for progenitor B-cell markers as in Figure 2D. Percentages are displayed on a representative plot (A). Mean percentage of pre-B and pro-B cells was calculated and graphed (B). Error bars indicate SD. **P < .01. Of the 5 mice receiving nontransgenic cells, 3 were treated with DOX and 2 without, and there was no appreciable difference in disease latency or phenotype.
DOX-regulated KLF4 expression depletes BCR-ABL–transformed pre-B cells in vivo. Bone marrow from FVB/N mice that were either nontransgenic or double transgenic for MMTV-tTA and TRE-KLF4 was transduced with BCR-ABL and injected in lethally irradiated FVB/N mice. Recipients were given water with or without DOX. (A, B) Leukemic mouse bone marrow was analyzed for progenitor B-cell markers as in Figure 2D. Percentages are displayed on a representative plot (A). Mean percentage of pre-B and pro-B cells was calculated and graphed (B). Error bars indicate SD. **P < .01. Of the 5 mice receiving nontransgenic cells, 3 were treated with DOX and 2 without, and there was no appreciable difference in disease latency or phenotype.
Discussion
As B cells progress through the many stages of development, they undergo ordered changes in gene expression and several cycles of proliferation and growth arrest.7,47,48 Interestingly, there are common gene-expression signatures (ie, gadd45 and jun) among stages that maintain a block in cell-cycle progression, such as small pre-B cells and naive mature B cells.48 In addition, global gene-expression patterns in some B-cell malignancies are similar to patterns in normal activated B cells.41 These commonalities support the idea that a similar set of transcriptional regulators must be inactivated for proliferation to occur at various stages of B-cell development and activation, and for transformation to occur. To identify transcription factors that could act as B-cell tumor suppressors, we searched microarray datasets for genes down-regulated after BCR crosslinking and in B-cell malignancies. Klf4 displayed a dramatic reduction in expression after activation, and human KLF4 was expressed at low levels in several B lymphomas and activated B cells compared with normal resting B cells.
We observed that Klf4 expression was lower in transformed murine pro-B/pre-B cell lines versus normal B-lineage precursors or mature resting B cells. Based on experiments with the ABL kinase inhibitor imatinib, we determined that the BCR-ABL oncogene does not actively suppress KLF4 expression in most established cell lines. However, it is possible that BCR-ABL induces stable silencing during transformation, or selectively transforms a B-lineage developmental stage at which KLF4 expression is low. It has been shown that the KLF4 promoter region is hypermethylated in human T-cell leukemia virus–infected T cells and forced expression of KLF4 induces death.49 It will be interesting to determine whether the KLF4 gene is hypermethylated in B-cell malignancies.
We determined that forced expression of KLF4 blocks proliferation of BCR-ABL–expressing pro/pre-B cells and induces cell death or cycle arrest in multiple murine B-cell lines. These data coincide with observations in other cell types that KLF4 possesses antiproliferative properties. When we expressed the closely related KLF2, we found that it did not induce cell death or cell-cycle arrest. This suggests that KLF4 has unique functions in B cells, not shared by the closely related KLF2, and that the effects of KLF4 are not simply due to competition with other factors that bind similar DNA elements.
There have been conflicting reports about how KLF4 mediates its growth inhibitory effects in fibroblasts and colon cancer cell lines. Some reports indicate a clear apoptotic mechanism with DNA fragmentation, whereas other studies show mainly cell-cycle arrest.15,26,49,50 We compared cell death in etoposide-treated cells versus KLF4-transduced cells. We found that KLF4 expression induced death similar to etoposide treatment, in that caspases were activated but not required and this death was attenuated by Bcl-XL overexpression. These data are in line with other studies showing that caspases are not required for etoposide-mediated apoptosis.51 Expression of Bcl-3 or constitutively active Akt also failed to rescue cells from KLF4-mediated depletion (data not shown). Even when KLF4-mediated death was blocked by Bcl-XL expression, cells expressing KLF4 were not maintained in culture relative to nontransduced cells in the same population. This is probably the result of cell-cycle arrest, as suggested by continued down-regulation of cyclin D2. In summary, KLF4 overexpression induces a coordinated change in gene expression that depletes transformed pre-B cells by both cell-cycle arrest in G1 phase and apoptosis.
Similar to the finding that KLF2 suppresses c-Myc expression in T cells,10 we found that KLF4 expression leads to reduced c-Myc expression in transformed pro/pre-B cells and activated mature B cells (I.Y., M.G.K., Jing Chen, Autumn Maruniak, Pratibha Sareen, Mary Tomayko, Mark Schlomchik, V.W.Y., Klaus Kaestner, and D.A.F.; “KLF4 is a FOXO target gene that suppresses B-cell proliferation,” manuscript submitted July 2006). However, restoration of c-Myc expression was not sufficient to overcome the effects of KLF4 on survival and proliferation in v-Abl–transformed cells. The Oncomine database also failed to show a strong correlation between KLF4 down-regulation and increased c-Myc RNA expression in human B-lineage malignancies. The expression of 2 other KLF targets identified in this paper, p21CIP and cyclin D2, showed a better correlation in these microarray datasets. Further experiments are necessary to determine if p21CIP up-regulation and/or cyclin D2 down-regulation are required for KLF4-induced effects in transformed B-lineage cells.
We found that inducible KLF4 expression could block transformation in vitro, even though leukemic disease latency was not increased in vivo. Furthermore, expression of myc-KLF4 protein was not detectable by immunoblot in lysates of DOX-withdrawn pro-B-cell lines established in vitro or in vivo. These results could be explained by low levels of myc-KLF4 expression in pro-B cells, which is sufficient to prevent transformation in vitro but not in vivo due to the presence of additional growth factors and survival signals. Importantly, KLF4 induction did greatly reduce the percentage of leukemic cells with a pre–B-cell phenotype (B220+CD43low), perhaps owing to greater expression of myc-KLF4. In support of this model, we could detect myc-KLF4 in DOX-withdrawn IL-7 cultures from transgenic bone marrow, conditions in which both pre-B and pro-B cells are present. When the MMTV-tTA strain was used to drive expression of a TRE-linked BCR-ABL cDNA, the leukemias that emerged were of pre–B-cell phenotype rather than pro-B,24 providing further indirect evidence for greater transgene expression in the pre–B-cell stage.
Our laboratory has shown that mice in which the Klf4 gene is disrupted, specifically in B cells, do not exhibit spontaneous B-cell activation or malignancies (I.Y., M.G.K., Jing Chen, Autumn Maruniak, Pratibha Sareen, Mary Tomayko, Mark Schlomchik, V.W.Y., Klaus Kaestner, and D.A.F.; “KLF4 is a FOXO target gene that suppresses B-cell proliferation,” manuscript submitted July 2006). Other KLFs, including KLF2 and KLF3, are expressed in the B lineage and it is reasonable to propose that these or other transcription factors compensate to prevent transformation. There might be less redundancy among KLFs in other cell types, as single deletion of KLF3 or KLF2 results in myeloproliferation and spontaneous T-cell activation, respectively.9,11 It would be interesting to determine if combined deletion of multiple KLFs in the B lineage promotes leukemic transformation, and whether loss of one or more KLFs enhances the effects of B-cell oncogenes.
In summary, we have shown that KLF4 is expressed at low levels in several B-cell malignancies and in cells transformed by ABL oncogenes. Additionally, forced expression of KLF4 blocks transformation, inhibits proliferation, and induces cell death. These data support the idea that KLF4 expression opposes B-cell malignancy and identifies KLF4 as a candidate tumor suppressor gene in B cells. Further experimentation is required to determine whether the loss of multiple KLFs in B cells can drive cancer progression and how expression of these factors can be harnessed for therapeutic benefit.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: M.G.K. performed and designed research and wrote the paper; I.Y. and V.M.S. performed research; V.W.Y., J.A.S., and C.S.H. contributed vital reagents; D.A.F. designed research and wrote the paper.
Acknowledgments
We thank Craig Walsh for helpful suggestions and critical reading of the manuscript, Keith Luhrs for assistance in the apoptotic assays, and members of the Walsh laboratory for Bcl-XL virus. We thank Kathleen Anderson, Jason Cyster, Philippa Marrack, Christopher Hughes, Naomi Rosenberg, Owen Witte, Michael Gold, and Tiong Ong for valuable reagents. We thank Travis Moore for help with bone marrow reconstitution and Jing Chen for assisting with QPCR.
This work was supported by National Institutes of Health grants (AI50831 to D.A.F., T32 CA9054 to M.G.K.), a Research Scholar Grant from the American Cancer Society (D.A.F.), and a grant from the University of California Cancer Research Coordinating Committee (D.A.F.).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal