Abstract
We recently cloned a novel human nuclear factor (designated THAP1) from postcapillary venule endothelial cells (ECs) that contains a DNA-binding THAP domain, shared with zebrafish E2F6 and several Caenorhabditis elegans proteins interacting genetically with retinoblastoma gene product (pRB). Here, we show that THAP1 is a physiologic regulator of EC proliferation and cell-cycle progression, 2 essential processes for angiogenesis. Retroviral-mediated gene transfer of THAP1 into primary human ECs inhibited proliferation, and large-scale expression profiling with microarrays revealed that THAP1-mediated growth inhibition is due to coordinated repression of pRB/E2F cell-cycle target genes. Silencing of endogenous THAP1 through RNA interference similarly inhibited EC proliferation and G1/S cell-cycle progression, and resulted in down-regulation of several pRB/E2F cell-cycle target genes, including RRM1, a gene required for S-phase DNA synthesis. Chromatin immunoprecipitation assays in proliferating ECs showed that endogenous THAP1 associates in vivo with a consensus THAP1-binding site found in the RRM1 promoter, indicating that RRM1 is a direct transcriptional target of THAP1. The similar phenotypes observed after THAP1 overexpression and silencing suggest that an optimal range of THAP1 expression is essential for EC proliferation. Together, these data provide the first links in mammals among THAP proteins, cell proliferation, and pRB/E2F cell-cycle pathways.
Introduction
In the adult, endothelial cells (ECs) normally are quiescent and play an important role in maintaining the integrity of blood vessels.1 However in several physiologic conditions (eg, ovarian and uterine vascular beds) and diverse pathologic conditions (eg, cancer, rheumatoid arthritis, diabetic retinopathy, and atherosclerosis) ECs have the ability to leave their normal quiescent state, re-enter the cell cycle, proliferate, and form neo-vessels in a process called neovascularization or angiogenesis.2-4 A better understanding of the molecular mechanisms and factors controlling EC proliferation and cell-cycle progression may therefore lead to novel therapeutic approaches for the control of angiogenesis-dependent diseases, including cancer and chronic inflammatory diseases.
Whereas many secreted angiogenic growth factors (including vascular endothelial growth factor [VEGF] and fibroblast growth factor [FGF] family members), angiogenesis inhibitors (eg, angiostatin, endostatin), and their corresponding receptors, which modulate EC growth and survival during angiogenesis, have been described,2-4 comparatively little is known about the nuclear transcription factors involved in the control of EC proliferation and cell-cycle progression.5 We recently identified a novel nuclear factor in a cDNA library from postcapillary high endothelial venule ECs (HEVECs), isolated from human tonsils.6 This factor, designated THAP1, is the prototype of a previously uncharacterized family of cellular factors, the THAP proteins (> 100 distinct members in the animal kingdom), defined by the presence at their amino-terminus of an evolutionarily conserved, approximately 90-residue protein motif, the THAP domain.7 We demonstrated that the THAP domain of THAP1 is an atypical zinc-dependent, sequence-specific, DNA-binding domain belonging to the zinc finger superfamily.8 We also showed that the THAP domain shares its metal-coordinating C2CH signature (CX2-4C-X35-53-CX2H) with the site-specific, DNA-binding domain of Drosophila P element transposase, indicating that the THAP domain constitutes a novel example of a DNA-binding domain shared between cellular proteins and transposases from mobile genomic parasites.7,8
Although orthologous relationships with the human THAP proteins were not obvious, analysis of THAP proteins in model animal organisms gave interesting clues to the functions of these proteins in vivo.8 First, in zebrafish and 2 other unrelated fish species, the orthologue of cell-cycle transcription factor E2F6 was found to contain a THAP domain at its amino-terminus which exhibits significant homologies with the THAP domain of human THAP1.8 Although E2F6 does not bind retinoblastoma gene product (pRB) family members, it functions as a repressor of E2F-dependent transcription during S phase that is critical to distinguish G1/S and G2/M transcription during the cell cycle.9 The identification of the THAP-E2F6 fusion gene in fish species provided the first link in vertebrates between the THAP proteins and the pRB/E2F pathway, and nicely complemented previous findings in Caenorhabditis elegans that have revealed the existence of genetic interactions between LIN-35/Rb, the sole C elegans retinoblastoma homologue, and 4 distinct C elegans THAP proteins (LIN-36, LIN-15B, LIN-15A, and HIM-17), initially characterized for their role in the specification of vulval cell fates (synthetic multivulva genes [synMuv]) or meiotic chromosome segregation.10-13 Among these, LIN-36 and LIN-15B have been found to function as inhibitors of the G1/S cell-cycle transition.12 LIN-36 behaved most similar to LIN-35/Rb and Efl-1/E2F,12,14 the orthologue of mammalian cell-cycle transcription factors E2F4/5,15 and was therefore proposed to act in a pathway or complex with LIN-35/Rb and Efl-1/E2F to repress G1/S control genes.12,16 Finally, a third C elegans THAP protein, GON-14, was recently shown to play a critical role in cell proliferation and development since gon-14 null mutants were found to exhibit larval growth arrest associated with cell division defects in intestine, gonad, and vulva.17 Together, these observations in zebrafish and C elegans suggested important roles for THAP proteins in cell proliferation and cell-cycle control.
Although several protein partners of THAP1, THAP0/DAP4, and THAP7 have been identified,6,18-20 the biologic roles of human THAP proteins remain largely unknown and potential functions in cell proliferation or cell-cycle regulation have not yet been described. In the present manuscript, we show that THAP1 is an endogenous physiologic regulator of EC proliferation and G1/S cell-cycle progression, which modulates expression of several pRb/E2F cell-cycle target genes. In addition, we identify RRM1, a G1/S-regulated gene required for S-phase DNA synthesis, as a direct transcriptional target of endogenous THAP1.
Materials and methods
Retroviral vector construction, virus packaging, and transduction of primary human ECs
A THAP1 retroviral expression vector (pMLV-THAP1) was generated using a pseudotyped vesicular stomatitis virus G protein (VSV-G)–Moloney murine leukemia virus (MuLV)–derived retrovirus vector, which allows very efficient transduction (> 90%) of proliferating human umbilical vein ECs (HUVECs). The multiple-cloning site of retroviral vector pBullet21 was modified by incorporation of synthetic oligonucleotides containing NaeI and MfeI restriction sites, between the NcoI and BamHI sites of the polylinker. The modified vector was called pMLV-MCS. The full-length coding region of human THAP16 was amplified by polymerase chain reaction (PCR) with primers THAP1-5′ (5′-ATGGTGCAGTCCTGCTCCGC-3′) and THAP1-MfeI-3′ (5′-GCCAATTGTTATGCTGGTACTTCAACTATTT-3′), and cloned into the pMLV-MCS vector cleaved with NaeI and MfeI restriction enzymes, to generate the pMLV-THAP1 retroviral expression vector. Retroviral vectors were produced by transient triple transfection of the packaging plasmid (pVPack-GP; Stratagene, La Jolla, CA), the envelope plasmid (pVPack-VSV-G; Stratagene), and the transducing vectors pMLV-MCS or pMLV-THAP1, in 293T cells (ATCC no. CRL11 268; ATCC, Rockville, MD), using a DNA–calcium phosphate procedure. Cell supernatants containing viral particles were harvested 48 hours after the transfection, clarified of cell debris using low-speed centrifugation, and filtered on 0.45-μm filters. A total of 106 primary HUVECs were transduced in a 75-cm2 plate with viral supernatant in the presence of 8 μg/mL polybren (Sigma, St Louis, MO) as previously described.22 After 4 hours, the supernatant was replaced by fresh endothelial cell medium consisting of MCDB131 (Invitrogen, Carlsbad, CA) supplemented with 20% heat-inactivated serum, endothelial cell growth supplement (ECGS; Sigma), and 5 U/mL sodium heparin. When applicable, a second transduction was performed using the same protocol a day after the first transduction. At 48 hours after infection, cells were trypsinized and pelleted for RNA preparation. Total RNA was isolated from 106 cells with the Absolutely RNA miniprep kit according to manufacturer's instructions (Stratagene).
Antibody production, Western blotting, and immunofluorescence microscopy
Affinity-purified rabbit polyclonal antibodies were raised against the peptide FQKEKDDVSERGYVI, corresponding to amino acids 186 to 200 of the human THAP1 sequence (Eurogentec, Seraing, Belgium). For Western blotting, lysates from retrovirally transduced HUVECs or siRNA-treated HUVECs (each corresponding to ∼105 cells) were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 10%). Detection was performed with rabbit antiserum to THAP1 (1:500) or mouse antibody to α-tubulin (1:20 000; Sigma), followed by horseradish peroxidase (HRP)–conjugated goat anti–rabbit or anti–mouse Ig (1:10 000; Promega), and finally an enhanced chemiluminescence kit (Amersham Bioscience, Freiburg, Germany). Indirect immunofluorescence staining was performed as previously described,6 with rabbit polyclonal anti-THAP1 (1:500) or mouse monoclonal anti–Ki-67 (1:250; clone B56; BD Pharmingen, San Diego, CA) antibodies. Microscopy was performed using a Nikon Eclipse TE300 fluorescence microscope equipped with a Nikon 20 ×/0.75 numerical aperture (NA) plan Apo objective (Mowiol mounting media) and a Nikon DXM1200 digital camera (Nikon, Tokyo, Japan). Nikon ACT-1 software version 2.10 was used for image acquisition.
Analysis of DNA synthesis by BrdU labeling, FACS, and TUNEL assays
DNA synthesis in retrovirally transduced or siRNA-treated HUVECs was analyzed by measuring bromodeoxyuridine (BrdU) incorporation as described previously.23 For cell-cycle analysis, 1 × 106 cells were collected, washed in phosphate-buffered saline (PBS), fixed with 70% cold ethanol for at least 30 minutes, washed with PBS, treated for 30 minutes at 37°C with RNase A (0.1 mg/mL), and stained with propidium iodide (69 μM; Sigma) in 38 mM sodium citrate. Cell-cycle analysis was carried out with a fluorescence-activated cell sorter (FACScan; Becton Dickinson, San Jose, CA). Analyses were performed 3 times on 40 000 cells with similar results. Transferase-mediated deoxyuridine triphosphate nick-end labeling (TUNEL) assays were performed as described previously.6
Colony assay
Human U2OS osteosarcoma cells were transfected with pcDNA3, pcDNA3-THAP1, or pcDNA3-p21 expression vectors, using a calcium-phosphate procedure. Transfected cells were selected in neomycin (750 μg/mL) and the ability to form colonies was assayed after 2 weeks by counting crystal violet–stained cells. Image acquisition was performed using Alpha Imager TM1200 software in a MultiImage Light Cabinet (Alpha Innotech, San Leandro, CA).
Microarray analysis
Total RNA quality control was performed by running 100 ng on an RNA 6000 Nano assay (Agilent, Wilmington, DE) using Bio-analyser 2100. Probe synthesis and labeling with Cy3- or Cy5-fluorescent CTP (Perkin-Elmer, Shelton, CT) were carried out with 500 ng total RNA using a low RNA input fluorescent linear amplification kit (Agilent) according to the manufacturer's instruction. One microgram of fragmented Cy3- and Cy5-labeled cRNAs were hybridized to Agilent human genome 1A oligonucleotide array (Agilent G4110A, 22 000 unique 60-nt oligonucleotide probes representing > 17 000 human genes) for 17 hours at 60°C, then washed with 0.6 × and 0.01 × sodium chloride–sodium citrate (SSC) buffers containing triton, and dried with a nitrogen gun. Microarrays were scanned using the Agilent DNA microarray scanner. Data analysis was performed with the Feature extraction software (Agilent) and Resolver software (Rosetta, Seattle, WA). Dye-swap experiments were performed to eliminate the effect of dye bias. The final ratio (ie, fold change) is the average of the 2 individual ratios from dye-swap experiments (combined experiments). Microarray data have been submitted to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/; accession no. E-TABM-24).
Quantitative real-time PCR (qPCR)
qPCR was performed using the ABI7700 Prism SDS Real-Time PCR Detection System (Applied Biosystems, Foster City, CA) with a SYBR Green PCR Master Mix kit (Applied Biosystems) and a standard temperature protocol as previously described.24 GAPDH and NFKB2 p100 were used as control genes for normalization. Primer sequences are available on request.
DNase I footprinting and EMSA
DNase I footprinting assays were performed by incubating a 130-bp 32 P end-labeled RRM1 promoter DNA fragment (bp −190 to −61) with recombinant THAP domain of human THAP18 at 28°C for 20 minutes in 20 μL binding buffer (20 mM Tris, 100 mM NaCl, 0.1% NP40, 5% glycerol, 100 μg/mL BSA). After 1 minute of cleavage by DNase I (50 ng/mL) in the presence of CaCl2 (1 mM), the reaction was stopped with 5 μL of stop solution (1.5 M NaOAc, 250 mM EDTA), DNA was purified and analyzed on 10% Urea–polyacrylamide gel and autoradiographed. Sequence references were obtained by sequencing the identical PCR fragments with the Maxam and Gilbert sequencing method.45 Electrophoretic mobility shift assay (EMSA) experiments were performed as described previously,8 using the upstream THAP1-binding sequence (THABS, underlined) of the RRM1 promoter as probe (5′-CGAACTCGGCTTGCCCACACAAAACATGGTA-3′). A 50-fold molar excess of wild-type THABS or mutated THABS (mutTHABS) unlabeled oligonucleotides8 was used for competition experiments.
Chromatin immunoprecipitations (ChIPs)
ChIP assays were performed as previously described,25,26 using 5 μg control anti-Flag antibody (Sigma) or 5 μg of antibodies specific for NF-YB25 or THAP1. Semiquantitative PCRs were performed with the following primers: NF-YA 5′-GGAGGCCCGATTCCCCTTTG-3′, 5′-GGTCAGCGAGACCCGCCAAT-3′; RRM1 5′-CAGCGGGCTCTAGGTGCTAC-3′, 5′-CGGGGTAGGCTTCACAGACT-3′. For ChIP-qPCR assays, ChIP was performed as described27 and quantified in triplicate by qPCR using the percent of input method.28 In brief, the amount of genomic DNA coprecipitated with anti-THAP1 antibodies was calculated as a percent of total input the following way: ΔCT = CT (input) − CT (THAP1-IP); %input = 2ΔCT × 0.25 (0.25% of total input was used). The following primers were used for the ChIP-qPCR assays: qRRM1 5′-CCCATGGGAGAGGCGTAGT-3′, 5′-CAGACTGACAGGCGACGTGTA-3′; qNF-YA 5′-CCGGTACTGGAGCCAATCA-3′, 5′-GGATATTGGCTCCTCACACTCAC-3′.
RNA interference
Control GL2 luciferase and THAP1 SMARTpool and individual siRNA duplexes were purchased from Dharmacon (Lafayette, CO). Two successive transfections of HUVECs were performed at 24-hour intervals by incubating cells for 6 hours with siRNA duplexes at 20 nM final concentration in Oligofectamine and serum-free Opti-MEM-1 (Invitrogen). Samples for qPCR, Western blot, BrdU labeling, determination of cell numbers, FACS and TUNEL assays were taken at the indicated time points after the second siRNA transfection.
Results
Retroviral-mediated gene transfer of THAP1 in primary human ECs inhibits proliferation and G1/S cell-cycle progression
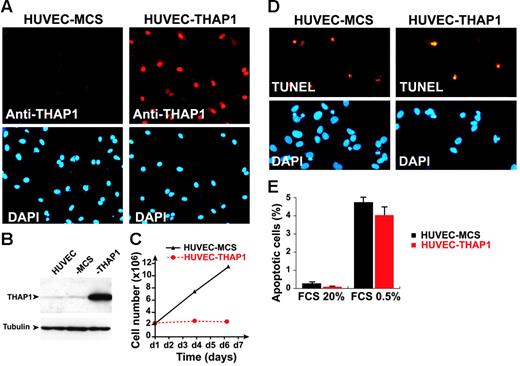
To better understand the function of THAP1 as a nuclear factor in the vasculature, we sought to ectopically express THAP1 in primary human vascular ECs using retroviral gene transfer. Primary HUVECs were transduced with pMLV-THAP1 or pMLV-MCS (as the negative infection control) retroviral expression vectors, and ectopic expression of THAP1 in transduced endothelial cells was verified by Western blotting and indirect immunofluorescence staining (Figure 1A-B). Overexpression of THAP1 was found to result in an inhibition of endothelial cell proliferation (Figure 1C). This effect was specific for the THAP1 gene since it was not observed after retroviral-mediated transfer of other genes, including the unrelated nuclear factor NF-HEV, the chemokines CCL21 and CXCL9 or the green fluorescent protein (data not shown). The loss of cell proliferation was not due to nonspecific toxic effects of THAP1 expression since the presence of floaters or non–trypan blue excluding cells in THAP1-expressing cultures was not observed and TUNEL assays failed to reveal enhanced apoptosis (Figure 1D). In addition, the levels of apoptosis were also similar when the cells were incubated in low serum media for 24 hours (Figure 1E). Thus, the antiproliferative effects of THAP1 on primary human endothelial cells were not dependent on induction of apoptosis.
Retroviral-mediated gene transfer of THAP1 inhibits EC proliferation. (A) Indirect immunofluorescence analysis of HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors, with anti-THAP1 polyclonal antibodies. Nuclei were counterstained with DAPI. (B) Western blot analysis of HUVEC-MCS and HUVEC-THAP1 with anti-THAP1 polyclonal antibodies or anti–αtubulin (loading control) mouse monoclonal antibody. (C) Analysis of cell proliferation in HUVEC-MCS and HUVEC-THAP1. Expression of THAP1 results in inhibition of EC proliferation. (D) TUNEL labeling of apoptotic nuclei in HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors, and incubated in low-serum media for 24 hours. Nuclei were counterstained with DAPI. (E) Apoptosis levels were quantified in HUVEC-MCS and HUVEC-THAP1 incubated in high-serum (20% FCS) or low-serum (0.5% FCS) media for 24 hours. TUNEL labeling of apoptotic nuclei was performed at day 4 after infection and at least 500 cells were counted for each condition. Results are mean and SD of 2 independent retroviral transduction experiments.
Retroviral-mediated gene transfer of THAP1 inhibits EC proliferation. (A) Indirect immunofluorescence analysis of HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors, with anti-THAP1 polyclonal antibodies. Nuclei were counterstained with DAPI. (B) Western blot analysis of HUVEC-MCS and HUVEC-THAP1 with anti-THAP1 polyclonal antibodies or anti–αtubulin (loading control) mouse monoclonal antibody. (C) Analysis of cell proliferation in HUVEC-MCS and HUVEC-THAP1. Expression of THAP1 results in inhibition of EC proliferation. (D) TUNEL labeling of apoptotic nuclei in HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors, and incubated in low-serum media for 24 hours. Nuclei were counterstained with DAPI. (E) Apoptosis levels were quantified in HUVEC-MCS and HUVEC-THAP1 incubated in high-serum (20% FCS) or low-serum (0.5% FCS) media for 24 hours. TUNEL labeling of apoptotic nuclei was performed at day 4 after infection and at least 500 cells were counted for each condition. Results are mean and SD of 2 independent retroviral transduction experiments.
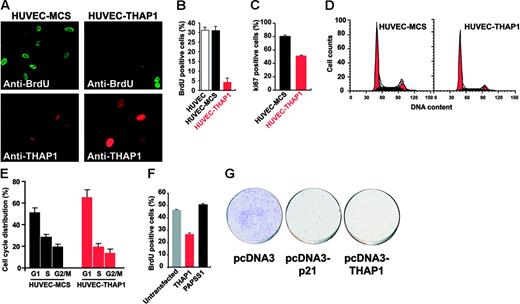
To further characterize the antiproliferative properties of THAP1, we analyzed cell-cycle progression in THAP1-expressing cells by staining cells for BrdU incorporation and nuclear proliferation marker Ki-67. We found that retroviral-mediated ectopic expression of THAP1 into HUVECs greatly inhibited DNA synthesis compared with uninfected cells or HUVECs transduced with pMLV-MCS control vector (Figure 2A). The percentage of cells in S-phase incorporating BrdU fell from approximately 30% in control HUVECs to 5% in cells expressing THAP1 (Figure 2B). Nuclear proliferation marker Ki-67, which stains cells in mid- to late G1 and S-G2/M cell-cycle phases, was also down-regulated (Figure 2C). Cell-cycle analysis of HUVEC-THAP1 cells by flow cytometry revealed a significant reduction in both the S- and G2/M-phase cell populations and a corresponding increase in the number of cells in G1 (Figure 2D-E), indicating that ectopic expression of THAP1 into primary HUVECs inhibits cell-cycle progression at the G1/S transition. Another difference between THAP1-expressing HUVECs and control HUVECs was seen in mitotic cells several days after the retroviral transduction; while the majority of mitotic figures in HUVECs transduced with pMLV-MCS control vector appeared morphologically normal, many of the mitotic figures in HUVECs transduced with THAP1 retroviral expression vector showed abnormalities (data not shown).
Ectopic expression of THAP1 into primary human ECs inhibits cell-cycle progression and blocks S-phase DNA synthesis. (A) Inhibition of DNA synthesis in ECs expressing THAP1. HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors were pulse-labeled with BrdU for 90 minutes. Cy2-conjugated anti-BrdU antibody was used to identify cells incorporating BrdU in the S phase of the cell cycle. Cells were visualized for BrdU incorporation (top panels) and THAP1 expression (bottom panels). (B) The graph shows the percentage of BrdU+ cells in HUVEC, HUVEC-MCS, and HUVEC-THAP1 cell populations. At least 500 cells were counted for each condition. Results are the mean of 2 independent retroviral transduction experiments. (C) Down-regulation of nuclear proliferation marker Ki-67 in ECs expressing THAP1. The graph shows the percentage of Ki-67+ cells in HUVEC-MCS and HUVEC-THAP1 cell populations. Ki-67 is a marker of cycling cells in late G1 and S-G2/M cell-cycle phases; therefore, reduction in the number of Ki-67+ cells in HUVEC-THAP1 indicates an inhibition of cell-cycle progression in ECs expressing THAP1. (D) Flow cytometry analysis of cell-cycle distribution in HUVEC-MCS and HUVEC-THAP1 cell populations. Black area represents cells in S phase, and red area represents cells in G1 and G2/M phases. (E) Ectopic expression of THAP1 into HUVECs inhibits cell-cycle progression at the G1/S transition. The graph shows the percentage of cells in G1, S, and G2/M phases of the cell cycle in HUVEC-MCS and HUVEC-THAP1 cell populations. Results are the mean of 2 independent retroviral transduction experiments. (F) Ectopic expression of THAP1 into human U2OS osteosarcoma cancer cells inhibits S-phase DNA synthesis. The graph shows the percentage of BrdU+ cells in control untransfected cells or in U2OS cell populations expressing THAP1 (% BrdU+ cells in the THAP1+ population) or the unrelated nuclear factor PAPSS1 (% BrdU+ cells in the PAPSS1+ population). Analysis was performed 48 hours after transfection. (G) Ectopic expression of THAP1 impairs growth of U2OS cells. Cells transfected with indicated pcDNA3 expression vectors were selected in neomycin for 14 days prior to crystal violet staining. Error bars in panels B, C, E, and F indicate SD.
Ectopic expression of THAP1 into primary human ECs inhibits cell-cycle progression and blocks S-phase DNA synthesis. (A) Inhibition of DNA synthesis in ECs expressing THAP1. HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors were pulse-labeled with BrdU for 90 minutes. Cy2-conjugated anti-BrdU antibody was used to identify cells incorporating BrdU in the S phase of the cell cycle. Cells were visualized for BrdU incorporation (top panels) and THAP1 expression (bottom panels). (B) The graph shows the percentage of BrdU+ cells in HUVEC, HUVEC-MCS, and HUVEC-THAP1 cell populations. At least 500 cells were counted for each condition. Results are the mean of 2 independent retroviral transduction experiments. (C) Down-regulation of nuclear proliferation marker Ki-67 in ECs expressing THAP1. The graph shows the percentage of Ki-67+ cells in HUVEC-MCS and HUVEC-THAP1 cell populations. Ki-67 is a marker of cycling cells in late G1 and S-G2/M cell-cycle phases; therefore, reduction in the number of Ki-67+ cells in HUVEC-THAP1 indicates an inhibition of cell-cycle progression in ECs expressing THAP1. (D) Flow cytometry analysis of cell-cycle distribution in HUVEC-MCS and HUVEC-THAP1 cell populations. Black area represents cells in S phase, and red area represents cells in G1 and G2/M phases. (E) Ectopic expression of THAP1 into HUVECs inhibits cell-cycle progression at the G1/S transition. The graph shows the percentage of cells in G1, S, and G2/M phases of the cell cycle in HUVEC-MCS and HUVEC-THAP1 cell populations. Results are the mean of 2 independent retroviral transduction experiments. (F) Ectopic expression of THAP1 into human U2OS osteosarcoma cancer cells inhibits S-phase DNA synthesis. The graph shows the percentage of BrdU+ cells in control untransfected cells or in U2OS cell populations expressing THAP1 (% BrdU+ cells in the THAP1+ population) or the unrelated nuclear factor PAPSS1 (% BrdU+ cells in the PAPSS1+ population). Analysis was performed 48 hours after transfection. (G) Ectopic expression of THAP1 impairs growth of U2OS cells. Cells transfected with indicated pcDNA3 expression vectors were selected in neomycin for 14 days prior to crystal violet staining. Error bars in panels B, C, E, and F indicate SD.
To determine whether the cell-cycle inhibitory properties of THAP1 are specific for primary ECs, we analyzed the effects of THAP1 ectopic expression in human epithelial cell lines, which express endogenous THAP1 at levels similar to those found in ECs (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). Transient transfection of the THAP1 plasmid into human U2OS osteosarcoma cancer cells (Figure 2F) or human 293T embryonic kidney cell line (data not shown) resulted in significant reduction of BrdU incorporation. In addition, ectopic expression of THAP1 significantly reduced the colony-forming ability of U2OS cells (Figure 2G), indicating that THAP1 overexpression inhibits proliferation of epithelial cancer cells.
Overexpression of THAP1 in primary human ECs down-regulates mRNA levels of critical cell-cycle regulators and pRB-E2F target genes
To gain further insight into the effects of THAP1 on EC growth regulatory pathways, DNA microarray experiments were performed after 1 (#1xTHAP1) or 2 (#2xTHAP1) consecutive transductions of HUVECs with pMLV-MCS or pMLV-THAP1 retroviral expression vectors. Although fold increase or decrease for each gene probe was significantly lower after the single retroviral transduction because of lower transduction efficiency (only 251 genes showing differential expression with P < .01 in the #1xTHAP1 experiment), this stringent analysis identified gene probes reproducibly affected by THAP1 expression in fully independent experiments (independent HUVEC primary cell cultures, independent transductions with independent retroviral supernatants, independent microarray analyses). Using a threshold of 1.5-fold increase or decrease, we identified 80 gene probes with decreased expression and 16 gene probes with increased expression in response to retroviral-mediated gene transfer of THAP1 into primary HUVECs (Table 1).
Genes regulated after ectopic expression of THAP1 in endothelial cells
| Unigene ID . | Gene symbol . | Gene name and description . | Fold change . | Previously identified as E2F target . | Previously identified as Rb target . |
|---|---|---|---|---|---|
| Genes down-regulated after ectopic expression of THAP1 in primary ECs | |||||
| Cell-cycle control | |||||
| Hs.374378 | CKS1B* | CDC28 protein kinase regulatory subunit 1B (CKS1) | −2.19 | Yes35 | Yes31 |
| Hs.520506 | FBXO5* | F-box protein 5 (APCcdh1 inhibitor Emil) | −1.83 | Yes32 | Yes32 |
| Hs.334562 | CDC2* | Cell division cycle 2, G1 to S and G2 to M | −1.81 | Yes33-36 | Yes29-31,36 |
| Hs.194698 | CCNB2* | Cyclin B2 | −1.66 | Yes34,36 | Yes29-31,36 |
| Hs.23960 | CCNBI* | Cyclin B1 | −1.61 | Yes34,36 | Yes29-31,36 |
| DNA replication/DNA repair | |||||
| Hs.234896 | GMNN* | Geminin, DNA replication inhibitor | −2.02 | No | Yes29 |
| Hs.198363 | MCM10 | MCM10 minichromosome maintenance-deficient 10 (S cerevisiae) | −2.01 | No | No |
| Hs.409065 | FEN1* | Flap structure-specific endonuclease 1 | −2 | Yes32-36 | Yes29,31,32,36 |
| Hs.202672 | DNMT1* | DNA (cytosine-5-)-methyltransferase 1 | −2 | Yes33 | Yes29,31 |
| Hs.5199 | HSPC150* | Similar to ubiquitin-conjugating enzyme Ubc13p (S cerevisiae) | −1.92 | Yes35,36 | Yes36 |
| Hs.383396 | RRM1* | Ribonucleotide reductase M1 polypeptide | −1.79 | Yes32,34-36 | Yes29-32,36 |
| Hs.438720 | MCM7* | MCM7 minichromosome maintenance deficient 7 (S cerevisiae) | −1.75 | Yes32-34 | Yes29-32 |
| Hs.489037 | ASK* | Activator of S phase kinase | −1.74 | Yes35 | Yes31 |
| Hs.433180 | PSF2 | DNA replication complex GINS protein PSF2 | −1.73 | No | No |
| Hs.226390 | RRM2* | Ribonucleotide reductase M2 polypeptide | −1.58 | Yes32,34 | Yes29-32 |
| Chromosome condensation and segregation | |||||
| Hs.99819 | USP16 | Ubiquitin-specific protease 16 (associated with mitotic chromosomes) | −3.37 | No | No |
| Hs.497741 | CENPF* | Centromere protein F, 350/400 kDa (mitosin) | −2.8 | Yes36 | Yes31,36 |
| Hs.75573 | CENPE* | Centromere protein E, 312 kDa | −2.15 | Yes35,36 | Yes31,36 |
| Hs.434953 | HMGB2* | High-mobility group box 2 (associated with mitotic chromosomes) | −2.09 | Yes32-34 | Yes29-32 |
| Hs.42650 | ZWINT* | Kinetochore protein ZW10 interactor | −1.78 | No | Yes31 |
| Hs.14559 | C10orf3 | Chromosome 10 open reading frame 3 (SMC-domain protein) | −1.72 | No | No |
| Hs.5719 | CNAP1 | Chromosome condensation-related SMC-associated protein 1 | −1.65 | No | No |
| Hs.350966 | PTTG1* | Securin (pituitary tumor-transforming 1) | −1.63 | Yes34-36 | Yes29,31,36 |
| Hs.521097 | PTTG3 | Pituitary tumor-transforming 3 | −1.62 | No | No |
| Hs.511755 | PTTG2 | Pituitary tumor-transforming 2 | −1.52 | No | No |
| Mitotic spindle checkpoint | |||||
| Hs.421956 | SPC25* | Kinetochore protein Spc25 homologue (S cerevisiae) | −2.04 | Yes36 | Yes29,31,36 |
| Hs.234545 | CDCA1 | Kinetochore protein Nuf2 (cell division cycle-associated 1) | −1.91 | No | No |
| Hs.533185 | MAD2L1* | Mitotic spindle assembly checkpoint protein MAD2 (MAD2-like 1) | −1.91 | Yes35,36 | Yes31,36 |
| Hs.36708 | BUB1B* | Mitotic checkpoint serine/threonine-protein kinase BUBR1 | −1.85 | Yes33,34,36 | Yes29,31,36 |
| Hs.414407 | KNTC2* | Kinetochore protein Hec1 (kinetochore-associated 2) | −1.79 | Yes35,36 | Yes31,36 |
| Hs.469649 | BUB1* | Mitotic checkpoint serine/threonine-protein kinase BUB1 | −1.76 | No | Yes29-31 |
| Hs.524947 | CDC20* | CDC20 cell division cycle 20 homologue (S cerevisiae) | −1.6 | Yes34 | Yes29-31 |
| Mitotic spindle assembly | |||||
| Hs.121028 | ASPM | Abnormal spindle (asp)-like, microcephaly associated (Drosophila) | −2.05 | No | No |
| Hs.514033 | SPAG5 | Mitotic spindle coiled-coil-related protein SPAG5 (MAP126) | −1.91 | No | No |
| Hs.77695 | DLG7* | HURP/KIAA0008, discs large homologue 7 (Drosophila) | −1.9 | No | Yes29,31 |
| Hs.104019 | TACC3 | Transforming, acidic coiled-coil-containing protein 3 | −1.8 | No | No |
| Hs.119324 | KIF22* | Kinesin family member 22 (KNSL4, kinesin-like 4) | −1.76 | Yes35,36 | Yes29,31,36 |
| Hs.90073 | CSEIL* | CSE1 chromosome segregation 1–like (yeast) | −1.63 | No | Yes31 |
| Hs.514527 | BIRC5* | Survivin (baculoviral IAP repeat-containing 5) | −1.62 | Yes36 | Yes31,36 |
| Hs.72550 | HMMR* | Hyaluronan-mediated motility receptor (RHAMM) | −1.6 | Yes35 | Yes29,31 |
| Hs.73625 | KIF20A* | Kinesin family member 20A (RAB6KIFL) | −1.59 | Yes36 | Yes29,30,36 |
| Hs.532793 | KPNB1* | Karyopherin (importin) beta 1 | −1.58 | Yes36 | Yes36 |
| Hs.69360 | KIF2C* | Kinesin family member 2C (KNSL6, kinesin-like 6) | −1.53 | Yes36 | Yes31,36 |
| Cytokinesis | |||||
| Hs.126774 | RAMP | RA-regulated nuclear matrix-associated protein | −1.69 | No | No |
| Hs.459362 | PRCI* | Protein regulator of cytokinesis 1 | −1.66 | Yes35,36 | Yes29,36 |
| Hs.62180 | ANLN* | Anillin, actin binding protein (scraps homologue, Drosophila) | −1.61 | Yes35,36 | Yes29,36 |
| Cell cycle/cell proliferation miscellaneous | |||||
| Hs.470654 | CDCA7 | Cell division cycle-associated 7 (c-myc target gene JPO1) | −2.55 | No | No |
| Hs.104741 | PBK | PDZ binding kinase (mitotic kinase TOPK) | −1.89 | No | No |
| Hs.434886 | CDCA5 | Cell division cycle-associated 5 | −1.88 | No | No |
| Hs.436187 | TRIP13* | Thyroid hormone receptor interactor 13 | −1.78 | No | Yes30,31 |
| Hs.239 | FOXM1* | Forkhead box M1 | −1.65 | No | Yes31 |
| Hs.89497 | LMNB1* | Lamin B1 | −1.65 | Yes33 | Yes29-31 |
| Hs.484813 | DEK* | DEK oncogene (DNA binding) | −1.61 | Yes32 | Yes32 |
| Hs.208912 | C22orf18 | Proliferation-associated nuclear element (PANE1) | −1.56 | No | Yes29,36 |
| Miscellaneous | |||||
| Hs.421907 | GLTSCR2 | Glioma tumor suppressor candidate region gene 2 | −4.41 | No | No |
| Hs.528306 | U1SNRNPBP | U11/U12 snRNP 35K | −3.77 | No | No |
| Hs.497636 | LAMB3 | Laminin, beta 3 | −2.91 | No | No |
| Hs.160411 | TSHR | Thyroid-stimulating hormone receptor | −2.55 | No | No |
| Hs.7432 | THAP1 | THAP domain-containing, apoptosis-associated protein 1 | −2.13 | No | No |
| Hs.248941 | TAF9 | TAF9 RNA polymerase II | −2.08 | No | No |
| Hs.472054 | C20orf42 | Chromosome 20 open reading frame 42 (kindlerin KIND1) | −1.86 | No | No |
| Hs.71827 | RRS1 | RRS1 ribosome biogenesis regulator homologue (S cerevisiae) | −1.84 | No | No |
| Hs.296310 | GJA4 | Gap junction protein, alpha 4, 37kDa (connexin 37) | −1.68 | No | No |
| Hs.472119 | MKKS | McKusick-Kaufman syndrome | −1.62 | No | No |
| Hs.166539 | ITGB3BP | Integrin beta 3-binding protein (beta3-endonexin, NRIF3) | −1.55 | No | No |
| Predicted proteins with unknown functions | |||||
| Hs.327252 | LZIC | Leucine zipper and CTNNB1P1 domain containing | −2.18 | No | No |
| Hs.388087 | ZCSL2 | Zinc finger, CSL domain-containing 2 | −2.07 | No | No |
| Hs.525764 | ARHGAP11A | Rho GTPase activating protein 11A | −2.03 | No | No |
| Hs.529778 | FKSG14* | Leucine zipper protein FKSG14 | −2.01 | Yes32 | Yes32 |
| Hs.508292 | FLJ10514 | Hypothetical protein FLJ10514 | −1.89 | No | No |
| Hs.404323 | FLJ10156 | Hypothetical protein FLJ10156 | −1.81 | No | No |
| Hs.121536 | FAM54A | Family with sequence similarity 54, member A | −1.79 | No | No |
| Hs.162717 | MGC15668 | Hypothetical protein MGC15668 | −1.7 | No | No |
| Hs.330663 | FLJ20641 | Hypothetical protein FLJ20641 | −1.66 | No | No |
| Hs.127767 | LOC130502 | Similar to CG14894-PA | −1.63 | No | No |
| Hs.426696 | FLJ20516 | Timeless-interacting protein | −1.62 | No | No |
| Hs.209715 | FLJ22624 | FLJ22624 protein | −1.6 | No | No |
| Hs.283532 | BM039 | Uncharacterized bone marrow protein BM039 | −1.58 | No | No |
| Hs.550564 | RPL7L1 | Ribosomal protein L7-like 1 | −1.53 | No | No |
| Hs.30696 | TCFL5 | Transcription factor-like 5 (basic helix-loop-helix) | −1.5 | No | No |
| Genes up-regulated after ectopic expression of THAP1 in primary EC | |||||
| Miscellaneous | |||||
| Hs.531081 | LGALS3 | Lectin, galactoside-binding, soluble, 3 (galectin 3) | 1.52 | No | No |
| Hs.198308 | WRB | Tryptophan-rich basic protein | 1.58 | No | No |
| Hs.387794 | CHGN | Chondroitin beta 1,4 N-acetylgalactosaminyltransferase | 1.6 | No | No |
| Hs.45140 | TMEM35 | Transmembrane protein 35 | 1.65 | No | No |
| Hs.505141 | FLJ14337 | Similar to peptidylprolyl isomerase A, isoform 1 | 1.67 | No | No |
| Hs.549129 | RP11-529110.4 | Deleted in a mouse model of primary ciliary dyskinesia | 1.69 | No | No |
| Hs.508835 | TEP1 | Telomerase-associated protein 1 | 1.7 | No | No |
| Hs.518731 | UCHL1 | Ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 1.71 | No | No |
| Hs.106511 | PCDH17 | Protocadherin 17 | 1.71 | No | No |
| Hs.492261 | TP53INP1 | Tumor protein p53 inducible nuclear protein 1 | 1.89 | No | No |
| Hs.126598 | TMEM45A | Transmembrane protein 45A | 1.96 | No | No |
| Hs.391561 | FABP4 | Fatty acid binding protein 4, adipocyte | 1.98 | No | No |
| Hs.386470 | NMB | Neuromedin B | 2.01 | No | No |
| Hs.517581 | HMOX1 | Heme oxygenase (decycling) 1 | 3.06 | No | No |
| Hs.289292 | FOXL2 | Forkhead box L2 | 4.21 | No | No |
| Hs.546296 | SECTM1 | Secreted and transmembrane 1 | 5.51 | No | No |
| Unigene ID . | Gene symbol . | Gene name and description . | Fold change . | Previously identified as E2F target . | Previously identified as Rb target . |
|---|---|---|---|---|---|
| Genes down-regulated after ectopic expression of THAP1 in primary ECs | |||||
| Cell-cycle control | |||||
| Hs.374378 | CKS1B* | CDC28 protein kinase regulatory subunit 1B (CKS1) | −2.19 | Yes35 | Yes31 |
| Hs.520506 | FBXO5* | F-box protein 5 (APCcdh1 inhibitor Emil) | −1.83 | Yes32 | Yes32 |
| Hs.334562 | CDC2* | Cell division cycle 2, G1 to S and G2 to M | −1.81 | Yes33-36 | Yes29-31,36 |
| Hs.194698 | CCNB2* | Cyclin B2 | −1.66 | Yes34,36 | Yes29-31,36 |
| Hs.23960 | CCNBI* | Cyclin B1 | −1.61 | Yes34,36 | Yes29-31,36 |
| DNA replication/DNA repair | |||||
| Hs.234896 | GMNN* | Geminin, DNA replication inhibitor | −2.02 | No | Yes29 |
| Hs.198363 | MCM10 | MCM10 minichromosome maintenance-deficient 10 (S cerevisiae) | −2.01 | No | No |
| Hs.409065 | FEN1* | Flap structure-specific endonuclease 1 | −2 | Yes32-36 | Yes29,31,32,36 |
| Hs.202672 | DNMT1* | DNA (cytosine-5-)-methyltransferase 1 | −2 | Yes33 | Yes29,31 |
| Hs.5199 | HSPC150* | Similar to ubiquitin-conjugating enzyme Ubc13p (S cerevisiae) | −1.92 | Yes35,36 | Yes36 |
| Hs.383396 | RRM1* | Ribonucleotide reductase M1 polypeptide | −1.79 | Yes32,34-36 | Yes29-32,36 |
| Hs.438720 | MCM7* | MCM7 minichromosome maintenance deficient 7 (S cerevisiae) | −1.75 | Yes32-34 | Yes29-32 |
| Hs.489037 | ASK* | Activator of S phase kinase | −1.74 | Yes35 | Yes31 |
| Hs.433180 | PSF2 | DNA replication complex GINS protein PSF2 | −1.73 | No | No |
| Hs.226390 | RRM2* | Ribonucleotide reductase M2 polypeptide | −1.58 | Yes32,34 | Yes29-32 |
| Chromosome condensation and segregation | |||||
| Hs.99819 | USP16 | Ubiquitin-specific protease 16 (associated with mitotic chromosomes) | −3.37 | No | No |
| Hs.497741 | CENPF* | Centromere protein F, 350/400 kDa (mitosin) | −2.8 | Yes36 | Yes31,36 |
| Hs.75573 | CENPE* | Centromere protein E, 312 kDa | −2.15 | Yes35,36 | Yes31,36 |
| Hs.434953 | HMGB2* | High-mobility group box 2 (associated with mitotic chromosomes) | −2.09 | Yes32-34 | Yes29-32 |
| Hs.42650 | ZWINT* | Kinetochore protein ZW10 interactor | −1.78 | No | Yes31 |
| Hs.14559 | C10orf3 | Chromosome 10 open reading frame 3 (SMC-domain protein) | −1.72 | No | No |
| Hs.5719 | CNAP1 | Chromosome condensation-related SMC-associated protein 1 | −1.65 | No | No |
| Hs.350966 | PTTG1* | Securin (pituitary tumor-transforming 1) | −1.63 | Yes34-36 | Yes29,31,36 |
| Hs.521097 | PTTG3 | Pituitary tumor-transforming 3 | −1.62 | No | No |
| Hs.511755 | PTTG2 | Pituitary tumor-transforming 2 | −1.52 | No | No |
| Mitotic spindle checkpoint | |||||
| Hs.421956 | SPC25* | Kinetochore protein Spc25 homologue (S cerevisiae) | −2.04 | Yes36 | Yes29,31,36 |
| Hs.234545 | CDCA1 | Kinetochore protein Nuf2 (cell division cycle-associated 1) | −1.91 | No | No |
| Hs.533185 | MAD2L1* | Mitotic spindle assembly checkpoint protein MAD2 (MAD2-like 1) | −1.91 | Yes35,36 | Yes31,36 |
| Hs.36708 | BUB1B* | Mitotic checkpoint serine/threonine-protein kinase BUBR1 | −1.85 | Yes33,34,36 | Yes29,31,36 |
| Hs.414407 | KNTC2* | Kinetochore protein Hec1 (kinetochore-associated 2) | −1.79 | Yes35,36 | Yes31,36 |
| Hs.469649 | BUB1* | Mitotic checkpoint serine/threonine-protein kinase BUB1 | −1.76 | No | Yes29-31 |
| Hs.524947 | CDC20* | CDC20 cell division cycle 20 homologue (S cerevisiae) | −1.6 | Yes34 | Yes29-31 |
| Mitotic spindle assembly | |||||
| Hs.121028 | ASPM | Abnormal spindle (asp)-like, microcephaly associated (Drosophila) | −2.05 | No | No |
| Hs.514033 | SPAG5 | Mitotic spindle coiled-coil-related protein SPAG5 (MAP126) | −1.91 | No | No |
| Hs.77695 | DLG7* | HURP/KIAA0008, discs large homologue 7 (Drosophila) | −1.9 | No | Yes29,31 |
| Hs.104019 | TACC3 | Transforming, acidic coiled-coil-containing protein 3 | −1.8 | No | No |
| Hs.119324 | KIF22* | Kinesin family member 22 (KNSL4, kinesin-like 4) | −1.76 | Yes35,36 | Yes29,31,36 |
| Hs.90073 | CSEIL* | CSE1 chromosome segregation 1–like (yeast) | −1.63 | No | Yes31 |
| Hs.514527 | BIRC5* | Survivin (baculoviral IAP repeat-containing 5) | −1.62 | Yes36 | Yes31,36 |
| Hs.72550 | HMMR* | Hyaluronan-mediated motility receptor (RHAMM) | −1.6 | Yes35 | Yes29,31 |
| Hs.73625 | KIF20A* | Kinesin family member 20A (RAB6KIFL) | −1.59 | Yes36 | Yes29,30,36 |
| Hs.532793 | KPNB1* | Karyopherin (importin) beta 1 | −1.58 | Yes36 | Yes36 |
| Hs.69360 | KIF2C* | Kinesin family member 2C (KNSL6, kinesin-like 6) | −1.53 | Yes36 | Yes31,36 |
| Cytokinesis | |||||
| Hs.126774 | RAMP | RA-regulated nuclear matrix-associated protein | −1.69 | No | No |
| Hs.459362 | PRCI* | Protein regulator of cytokinesis 1 | −1.66 | Yes35,36 | Yes29,36 |
| Hs.62180 | ANLN* | Anillin, actin binding protein (scraps homologue, Drosophila) | −1.61 | Yes35,36 | Yes29,36 |
| Cell cycle/cell proliferation miscellaneous | |||||
| Hs.470654 | CDCA7 | Cell division cycle-associated 7 (c-myc target gene JPO1) | −2.55 | No | No |
| Hs.104741 | PBK | PDZ binding kinase (mitotic kinase TOPK) | −1.89 | No | No |
| Hs.434886 | CDCA5 | Cell division cycle-associated 5 | −1.88 | No | No |
| Hs.436187 | TRIP13* | Thyroid hormone receptor interactor 13 | −1.78 | No | Yes30,31 |
| Hs.239 | FOXM1* | Forkhead box M1 | −1.65 | No | Yes31 |
| Hs.89497 | LMNB1* | Lamin B1 | −1.65 | Yes33 | Yes29-31 |
| Hs.484813 | DEK* | DEK oncogene (DNA binding) | −1.61 | Yes32 | Yes32 |
| Hs.208912 | C22orf18 | Proliferation-associated nuclear element (PANE1) | −1.56 | No | Yes29,36 |
| Miscellaneous | |||||
| Hs.421907 | GLTSCR2 | Glioma tumor suppressor candidate region gene 2 | −4.41 | No | No |
| Hs.528306 | U1SNRNPBP | U11/U12 snRNP 35K | −3.77 | No | No |
| Hs.497636 | LAMB3 | Laminin, beta 3 | −2.91 | No | No |
| Hs.160411 | TSHR | Thyroid-stimulating hormone receptor | −2.55 | No | No |
| Hs.7432 | THAP1 | THAP domain-containing, apoptosis-associated protein 1 | −2.13 | No | No |
| Hs.248941 | TAF9 | TAF9 RNA polymerase II | −2.08 | No | No |
| Hs.472054 | C20orf42 | Chromosome 20 open reading frame 42 (kindlerin KIND1) | −1.86 | No | No |
| Hs.71827 | RRS1 | RRS1 ribosome biogenesis regulator homologue (S cerevisiae) | −1.84 | No | No |
| Hs.296310 | GJA4 | Gap junction protein, alpha 4, 37kDa (connexin 37) | −1.68 | No | No |
| Hs.472119 | MKKS | McKusick-Kaufman syndrome | −1.62 | No | No |
| Hs.166539 | ITGB3BP | Integrin beta 3-binding protein (beta3-endonexin, NRIF3) | −1.55 | No | No |
| Predicted proteins with unknown functions | |||||
| Hs.327252 | LZIC | Leucine zipper and CTNNB1P1 domain containing | −2.18 | No | No |
| Hs.388087 | ZCSL2 | Zinc finger, CSL domain-containing 2 | −2.07 | No | No |
| Hs.525764 | ARHGAP11A | Rho GTPase activating protein 11A | −2.03 | No | No |
| Hs.529778 | FKSG14* | Leucine zipper protein FKSG14 | −2.01 | Yes32 | Yes32 |
| Hs.508292 | FLJ10514 | Hypothetical protein FLJ10514 | −1.89 | No | No |
| Hs.404323 | FLJ10156 | Hypothetical protein FLJ10156 | −1.81 | No | No |
| Hs.121536 | FAM54A | Family with sequence similarity 54, member A | −1.79 | No | No |
| Hs.162717 | MGC15668 | Hypothetical protein MGC15668 | −1.7 | No | No |
| Hs.330663 | FLJ20641 | Hypothetical protein FLJ20641 | −1.66 | No | No |
| Hs.127767 | LOC130502 | Similar to CG14894-PA | −1.63 | No | No |
| Hs.426696 | FLJ20516 | Timeless-interacting protein | −1.62 | No | No |
| Hs.209715 | FLJ22624 | FLJ22624 protein | −1.6 | No | No |
| Hs.283532 | BM039 | Uncharacterized bone marrow protein BM039 | −1.58 | No | No |
| Hs.550564 | RPL7L1 | Ribosomal protein L7-like 1 | −1.53 | No | No |
| Hs.30696 | TCFL5 | Transcription factor-like 5 (basic helix-loop-helix) | −1.5 | No | No |
| Genes up-regulated after ectopic expression of THAP1 in primary EC | |||||
| Miscellaneous | |||||
| Hs.531081 | LGALS3 | Lectin, galactoside-binding, soluble, 3 (galectin 3) | 1.52 | No | No |
| Hs.198308 | WRB | Tryptophan-rich basic protein | 1.58 | No | No |
| Hs.387794 | CHGN | Chondroitin beta 1,4 N-acetylgalactosaminyltransferase | 1.6 | No | No |
| Hs.45140 | TMEM35 | Transmembrane protein 35 | 1.65 | No | No |
| Hs.505141 | FLJ14337 | Similar to peptidylprolyl isomerase A, isoform 1 | 1.67 | No | No |
| Hs.549129 | RP11-529110.4 | Deleted in a mouse model of primary ciliary dyskinesia | 1.69 | No | No |
| Hs.508835 | TEP1 | Telomerase-associated protein 1 | 1.7 | No | No |
| Hs.518731 | UCHL1 | Ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 1.71 | No | No |
| Hs.106511 | PCDH17 | Protocadherin 17 | 1.71 | No | No |
| Hs.492261 | TP53INP1 | Tumor protein p53 inducible nuclear protein 1 | 1.89 | No | No |
| Hs.126598 | TMEM45A | Transmembrane protein 45A | 1.96 | No | No |
| Hs.391561 | FABP4 | Fatty acid binding protein 4, adipocyte | 1.98 | No | No |
| Hs.386470 | NMB | Neuromedin B | 2.01 | No | No |
| Hs.517581 | HMOX1 | Heme oxygenase (decycling) 1 | 3.06 | No | No |
| Hs.289292 | FOXL2 | Forkhead box L2 | 4.21 | No | No |
| Hs.546296 | SECTM1 | Secreted and transmembrane 1 | 5.51 | No | No |
The Unigene ID, gene symbol, gene name, and description are indicated for each gene as well as the fold changes obtained after 2 retroviral transductions of HUVECs with pMLV-MCS or pMLV-THAP1 expression vectors (#2xTHAPI experiment). The table includes only those genes reproducibly affected by THAP1 expression (P < .01) in the 2 fully independent microarray experiments (#lxTHAP1 and #2xTHAP1).
Genes previously identified as targets of the pRB/E2F pathway.
Strikingly, most of the genes encoding proteins with known functions that are down-regulated after THAP1 overexpression (54 distinct genes of 80) corresponded to genes encoding proteins associated with cell-cycle/cell proliferation (Table 1), indicating a highly selective nature of THAP1-mediated inhibition of gene expression. The list of down-regulated genes included genes with a well-established role in cell-cycle control, genes involved in DNA replication and DNA repair, and genes encoding proteins that function in central mitotic processes, such as chromosome condensation and segregation, mitotic spindle checkpoint, mitotic spindle assembly, and cytokinesis (Table 1). Interestingly, many of these genes have previously been found to be regulated by the pRB-E2F pathway (38 genes of 54; Table 1), exhibiting down-regulation upon expression of pRB and up-regulation upon overexpression of E2F.29-36
In addition to the cell-cycle–specific genes, a series of genes with diverse biologic roles were also identified as target genes down-regulated or up-regulated by THAP1, but these could not be grouped into any functional category (Table 1, Miscellaneous), and the effect of THAP1 on the regulation of these genes was not further studied.
To validate the microarray data, RNA samples from HUVECs transduced with pMLV-MCS or pMLV-THAP1 retroviral expression vectors were analyzed by qPCR, using specific primers for 15 distinct genes selected from Table 1. As shown in Figure 3, the qPCR data were in good agreement with the microarray results and showed that expression of all these genes was decreased more than 2-fold after retroviral-mediated gene transfer of THAP1 into primary HUVECs. Together, the microarray and qPCR data indicated that overexpression of THAP1 into primary HUVECs inhibits cell proliferation through coordinated repression of critical cell-cycle regulators and pRb/E2F target genes.
Quantitative real-time RT-PCR analysis of gene expression repressed in response to retroviral-mediated gene transfer of THAP1 into primary human ECs. RNA samples from HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors were analyzed by qPCR. The fold change in the mRNA levels for 15 selected genes (identified in the DNA microarray experiments; Table 1), between HUVEC-MCS and HUVEC-THAP1, was calculated. The mean and standard error for 2 independent data sets are shown (Q, ▪). For comparison, the fold changes obtained in the DNA microarray experiments (P < .01) are indicated (M, □). The levels of down-regulation observed in THAP1-expressing ECs were generally higher in the qPCR experiments than in the microarray experiments.
Quantitative real-time RT-PCR analysis of gene expression repressed in response to retroviral-mediated gene transfer of THAP1 into primary human ECs. RNA samples from HUVECs transduced with pMLV-MCS (HUVEC-MCS) or pMLV-THAP1 (HUVEC-THAP1) retroviral expression vectors were analyzed by qPCR. The fold change in the mRNA levels for 15 selected genes (identified in the DNA microarray experiments; Table 1), between HUVEC-MCS and HUVEC-THAP1, was calculated. The mean and standard error for 2 independent data sets are shown (Q, ▪). For comparison, the fold changes obtained in the DNA microarray experiments (P < .01) are indicated (M, □). The levels of down-regulation observed in THAP1-expressing ECs were generally higher in the qPCR experiments than in the microarray experiments.
Endogenous THAP1 is required for EC proliferation, S-phase DNA synthesis, and G1/S cell-cycle progression
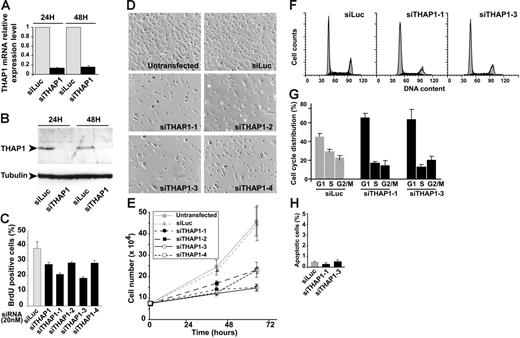
To determine whether endogenous THAP1 regulates EC proliferation and cell-cycle progression, we performed a knock-down of THAP1 in HUVECs using RNA interference with siRNAs. For an efficient depletion of THAP1, we used a THAP1 siRNA SMARTpool (siTHAP1) at a final concentration of 20 nM, which is known to avoid off-target effects of the siRNAs.37 Analysis of THAP1 expression by qPCR and Western blot at 24 hours and 48 hours after THAP1 siRNA transfection revealed up to 80% reduction of THAP1 mRNA and protein levels when compared with samples treated with a control luciferase siRNA (Figure 4A-B). We measured BrdU incorporation in cells treated with siTHAP1 or siLuc control siRNA and found that THAP1 knock-down resulted in inhibition of S-phase DNA synthesis (Figure 4C). Specificity of the effect was demonstrated by the use of 4 individual siRNAs targeted at different areas of the THAP1 coding region, which led to similar (siTHAP1-2, siTHAP1-4) or even higher (siTHAP1-1, siTHAP1-3) reductions of BrdU incorporation when compared with ECs treated with siLuc control siRNA (Figure 4C). Inspection of transfected cultures indicated that silencing of THAP1 resulted in less proliferation (Figure 4D). Quantification of the proliferation rate of ECs treated with the 4 individual THAP1 siRNAs revealed a significant inhibition of cell proliferation compared with untransfected ECs or ECs transfected with the siLuc control siRNA (Figure 4E). Cell-cycle analysis of ECs transfected with THAP1 siRNAs revealed a reduction in both the S- and G2/M-phase cell populations and a corresponding increase in the number of cells in G1 (Figure 4F-G), indicating that endogenous THAP1 is required for G1/S cell-cycle progression. TUNEL assays showed that knock-down of THAP1 did not enhance apoptosis levels (Figure 4H). Together, these data indicate that endogenous THAP1 is essential for EC proliferation and cell-cycle progression at the G1/S transition.
Silencing of endogenous THAP1 inhibits EC proliferation, S-phase DNA synthesis, and G1/S cell-cycle progression. (A) siRNA-mediated knock-down of THAP1 mRNA in primary human ECs. Levels of THAP1 mRNA were analyzed by qPCR at different time points after transfection of siTHAP1 or siLuc control siRNAs (20 nM final concentration). (B) siRNA-mediated silencing of THAP1 protein in primary human ECs. Levels of THAP1 and αtubulin proteins were analyzed by Western blot at different time points after transfection of siTHAP1 or siLuc control siRNAs (20 nM final concentration). (C) Knock-down of THAP1 in primary human ECs inhibits S-phase DNA synthesis. The graph shows the percentage of BrdU+ cells in HUVECs transfected with siLuc, siTHAP1 (pool), siTHAP1-1, siTHAP1-2, siTHAP1-3, or siTHAP1-4 siRNAs. At least 500 cells were counted for each condition. Results are the mean of 2 independent experiments performed 48 hours after siRNA transfection. (D,E) siRNA-mediated knock-down of THAP1 inhibits proliferation of primary human ECs. Representative photos (D) of HUVECs treated with siLuc or the 4 individual THAP1 siRNAs are shown. Cell count assay (E) showing inhibition of proliferation in ECs treated with the 4 individual THAP1 siRNAs compared with untreated cells or ECs treated with siLuc control siRNA. Results are the mean of 3 independent experiments. (F,G) Knock-down of THAP1 in primary human ECs inhibits G1/S cell-cycle progression. Flow cytometry analysis of cell-cycle distribution (F) in HUVECs transfected with siLuc, siTHAP1-1, or siTHAP1-3 siRNAs. Black area represents cells in S phase, and gray area represents cells in G1 and G2/M phases. The graph shows the percentage of cells in G1, S, and G2/M phases of the cell cycle (G) in HUVECs transfected with siLuc, siTHAP1-1, or siTHAP1-3 siRNAs. Results are the mean of 3 independent experiments performed 48 hours after siRNA transfection. (H) Apoptosis levels were quantified in HUVECs transfected with siLuc, siTHAP1-1, or siTHAP1-3 siRNAs. TUNEL labeling of apoptotic nuclei was performed 48 hours after siRNA transfection, and at least 500 cells were counted for each condition. Results are the mean of 3 independent experiments. Error bars in panels C, G, H indicate SD.
Silencing of endogenous THAP1 inhibits EC proliferation, S-phase DNA synthesis, and G1/S cell-cycle progression. (A) siRNA-mediated knock-down of THAP1 mRNA in primary human ECs. Levels of THAP1 mRNA were analyzed by qPCR at different time points after transfection of siTHAP1 or siLuc control siRNAs (20 nM final concentration). (B) siRNA-mediated silencing of THAP1 protein in primary human ECs. Levels of THAP1 and αtubulin proteins were analyzed by Western blot at different time points after transfection of siTHAP1 or siLuc control siRNAs (20 nM final concentration). (C) Knock-down of THAP1 in primary human ECs inhibits S-phase DNA synthesis. The graph shows the percentage of BrdU+ cells in HUVECs transfected with siLuc, siTHAP1 (pool), siTHAP1-1, siTHAP1-2, siTHAP1-3, or siTHAP1-4 siRNAs. At least 500 cells were counted for each condition. Results are the mean of 2 independent experiments performed 48 hours after siRNA transfection. (D,E) siRNA-mediated knock-down of THAP1 inhibits proliferation of primary human ECs. Representative photos (D) of HUVECs treated with siLuc or the 4 individual THAP1 siRNAs are shown. Cell count assay (E) showing inhibition of proliferation in ECs treated with the 4 individual THAP1 siRNAs compared with untreated cells or ECs treated with siLuc control siRNA. Results are the mean of 3 independent experiments. (F,G) Knock-down of THAP1 in primary human ECs inhibits G1/S cell-cycle progression. Flow cytometry analysis of cell-cycle distribution (F) in HUVECs transfected with siLuc, siTHAP1-1, or siTHAP1-3 siRNAs. Black area represents cells in S phase, and gray area represents cells in G1 and G2/M phases. The graph shows the percentage of cells in G1, S, and G2/M phases of the cell cycle (G) in HUVECs transfected with siLuc, siTHAP1-1, or siTHAP1-3 siRNAs. Results are the mean of 3 independent experiments performed 48 hours after siRNA transfection. (H) Apoptosis levels were quantified in HUVECs transfected with siLuc, siTHAP1-1, or siTHAP1-3 siRNAs. TUNEL labeling of apoptotic nuclei was performed 48 hours after siRNA transfection, and at least 500 cells were counted for each condition. Results are the mean of 3 independent experiments. Error bars in panels C, G, H indicate SD.
To determine whether THAP1 is expressed in proliferating ECs in vivo, we performed in situ hybridization and immunohistochemistry experiments, but we failed to detect expression, probably because endogenous THAP1 expression levels are below the detection limits of these techniques (data not shown). However, we succeeded in showing expression of endogenous THAP1 mRNA in ECs freshly purified from different human tissues (Figure S1), including rheumatoid arthritis synovium, a tissue associated with high levels of angiogenesis and EC proliferation. Therefore, THAP1 is likely to be expressed in proliferating ECs in vivo. Actually, the results obtained with proliferating and quiescent HUVECs indicated that THAP1 is expressed at slightly higher levels in proliferating ECs than in quiescent growth-arrested ECs (Figure S2).
Silencing of endogenous THAP1 inhibits expression of pRB/E2F cell-cycle target genes
To address the physiologic role of THAP1 in the regulation of pRB/E2F cell-cycle target genes, relative mRNA levels of these genes were quantified by qPCR in ECs treated with siTHAP1 or siLuc control siRNAs. Knock-down of THAP1 using either siTHAP1-1 or siTHAP1-3 siRNAs led to a significant decrease in the expression of the 8 tested pRB/E2F cell-cycle target genes (RRM1, Mad2, survivin, HMMR, RRM2, CDC2, cyclin B1, and DLG7) when compared with ECs treated with control siRNA (Figure 5A-B). There was no significant change in the expression level of actin between ECs treated with siTHAP1 and siLuc control siRNAs, indicating that THAP1 knock-down led to a specific reduction of mRNA levels of the pRB/E2F cell-cycle target genes RRM1, Mad2, survivin, HMMR, RRM2, CDC2, cyclin B1, and DLG7.
Knock-down of endogenous THAP1 in human primary ECs inhibits expression of pRB/E2F cell-cycle target genes RRM1, Mad2, survivin, HMMR, RRM2, CDC2, cyclin B1, and DLG7. (A,B) Expression level analysis by qPCR following siRNA-mediated knock-down of THAP1 in primary human ECs. RNA was isolated from ECs transfected with siTHAP1-1 (A), siTHAP1-3 (B), or siLuc siRNAs 48 hours after siRNA transfection, and used for qPCR analysis with the indicated human gene primers (control gene: actin; pRB/E2F cell-cycle target genes: RRM1, Mad2, survivin, HMMR, RRM2, CDC2, cyclin B1, and DLG7). NF-KB2 p100 was used as a control gene for normalization. The mean and standard error for 2 independent data sets are shown.
Knock-down of endogenous THAP1 in human primary ECs inhibits expression of pRB/E2F cell-cycle target genes RRM1, Mad2, survivin, HMMR, RRM2, CDC2, cyclin B1, and DLG7. (A,B) Expression level analysis by qPCR following siRNA-mediated knock-down of THAP1 in primary human ECs. RNA was isolated from ECs transfected with siTHAP1-1 (A), siTHAP1-3 (B), or siLuc siRNAs 48 hours after siRNA transfection, and used for qPCR analysis with the indicated human gene primers (control gene: actin; pRB/E2F cell-cycle target genes: RRM1, Mad2, survivin, HMMR, RRM2, CDC2, cyclin B1, and DLG7). NF-KB2 p100 was used as a control gene for normalization. The mean and standard error for 2 independent data sets are shown.
Endogenous THAP1 associates in vivo with the promoter of RRM1, a pRb/E2F cell-cycle target gene required for S-phase DNA synthesis
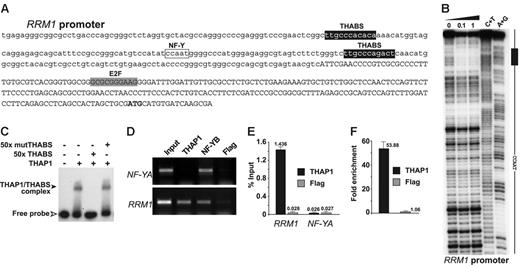
The presence of the sequence-specific, DNA-binding THAP domain in THAP18 suggested that THAP1 may bind directly to the promoters of some of the pRb-E2F cell-cycle–specific genes identified. In agreement with this possibility, we found candidate THAP1-binding sites (THABS)8 in the promoters of several genes: RRM1, RRM2, BIRC5/survivin, cyclin B1, and USP16. We selected the RRM1 gene for further in vitro and in vivo characterization of potential THAP1-promoter interactions. RRM1, a gene activated at the G1/S cell-cycle transition, encodes the ribonucleotide reductase M1 subunit, which is essential for S-phase DNA synthesis.38,39 The RRM1 promoter exhibited 2 consensus THABS approximately 100 nt upstream the 5′ end of the mRNA (Figure 6A). The upstream THABS sequence was found to be protected in DNase I footprinting assays (Figure 6B), suggesting direct binding of the THAP domain of THAP1 to these sites. In vitro EMSA protein/DNA-binding assays confirmed interaction of THAP1 with the RRM1 THABS motif and demonstrated the specificity of binding (Figure 6C). To examine a possible association of endogenous THAP1 with the RRM1 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) assays with cross-linked extracts from proliferating HUVECs, using anti-THAP1, anti-NF-YB, and anti-Flag antibodies (Figure 6D). As expected, the NF-YB transcription factor, used as a positive control,25 bound to the NF-YA and RRM1 promoters which contain CCAAT boxes (NF-Y binding sites). THAP1 bound to the endogenous RRM1 promoter, but not to the NF-YA promoter, which doesn't exhibit THABS motifs. Consistent with the specificity of our ChIP assay, the control anti-Flag antibody did not immunoprecipitate significant levels of the different promoters analyzed. ChIP-qPCR assays were performed to confirm and quantify the ChIP results. This revealed significant binding of endogenous THAP1 to the RRM1 promoter (1.4% of total input DNA precipitated) whereas binding of THAP1 to the NF-YA promoter was similar to background levels, obtained with anti-Flag antibodies on both the RRM1 and NF-YA promoters (Figure 6E). Quantification indicated an approximately 50-fold enrichment of THAP1 on the RRM1 promoter compared with the NF-YA promoter (Figure 6F). Taken together with the in vitro DNA-binding assays (Figure 6B-C), and the down-regulation of RRM1 mRNA observed after knock-down of endogenous THAP1 (Figure 5), these in vivo ChIP assays indicate that RRM1 is a direct transcriptional target of endogenous THAP1.
THAP1 binds to the RRM1 promoter in vitro and in vivo. (A) Sequence of the RRM1 promoter indicating the positions of THAP1 binding sites (THABS), E2F cell-cycle regulatory elements, and NF-Y/CCAAT box. The mRNA region is indicated in uppercase letters. The initiating methionine is shown in bold. (B) Dnase I footprinting analysis of RRM1 promoter region. Lane 0, partial DnaseI cleavage of DNA fragment without incubation with protein. Lanes 0.1 and 1, partial DnaseI cleavage after incubation with 0.1 μM or 1 μM THAP domain of human THAP1, respectively. Lanes C+T and A+G, Maxam-Gilbert chemical sequencing references (cleavage after pyrimidine C+T and purine A+G). (C) In vitro EMSA using the first THABS motif of RRM1 promoter and 0.1 μM THAP domain of human THAP1. A 50-fold molar excess of wild-type (THABS) and mutant (mutTHABS) cold competitor oligonucleotides were used to show specificity of binding. (D) Identification of endogenous THAP1 on the RRM1 promoter in vivo using ChIP assays. Cross-linked chromatin from proliferating HUVECs was subjected to immunoprecipitation with antibodies against THAP1, NF-YB (positive control), and Flag epitope (negative control). The NF-YA promoter was used as a positive control for the NF-YB transcription factor and a negative control promoter for THAP1. (E) ChIP-qPCR assays were used to quantify the amount of RRM1 or NF-YA promoter DNA precipitated by anti-THAP1 or anti-Flag antibodies. Immunoprecipitated DNA was quantified in triplicate by qPCR using the percent of input method (see “Materials and methods”). A representative experiment out of 3 is shown. (F) Fold enrichment of THAP1 on the RRM1 promoter was calculated by dividing the amount of RRM1 promoter DNA precipitated by anti-THAP1 antibodies to the amount of DNA precipitated from the NF-YA negative control promoter. No enrichment was observed with anti-Flag negative control antibodies. Error bars in panels E and F indicate SD.
THAP1 binds to the RRM1 promoter in vitro and in vivo. (A) Sequence of the RRM1 promoter indicating the positions of THAP1 binding sites (THABS), E2F cell-cycle regulatory elements, and NF-Y/CCAAT box. The mRNA region is indicated in uppercase letters. The initiating methionine is shown in bold. (B) Dnase I footprinting analysis of RRM1 promoter region. Lane 0, partial DnaseI cleavage of DNA fragment without incubation with protein. Lanes 0.1 and 1, partial DnaseI cleavage after incubation with 0.1 μM or 1 μM THAP domain of human THAP1, respectively. Lanes C+T and A+G, Maxam-Gilbert chemical sequencing references (cleavage after pyrimidine C+T and purine A+G). (C) In vitro EMSA using the first THABS motif of RRM1 promoter and 0.1 μM THAP domain of human THAP1. A 50-fold molar excess of wild-type (THABS) and mutant (mutTHABS) cold competitor oligonucleotides were used to show specificity of binding. (D) Identification of endogenous THAP1 on the RRM1 promoter in vivo using ChIP assays. Cross-linked chromatin from proliferating HUVECs was subjected to immunoprecipitation with antibodies against THAP1, NF-YB (positive control), and Flag epitope (negative control). The NF-YA promoter was used as a positive control for the NF-YB transcription factor and a negative control promoter for THAP1. (E) ChIP-qPCR assays were used to quantify the amount of RRM1 or NF-YA promoter DNA precipitated by anti-THAP1 or anti-Flag antibodies. Immunoprecipitated DNA was quantified in triplicate by qPCR using the percent of input method (see “Materials and methods”). A representative experiment out of 3 is shown. (F) Fold enrichment of THAP1 on the RRM1 promoter was calculated by dividing the amount of RRM1 promoter DNA precipitated by anti-THAP1 antibodies to the amount of DNA precipitated from the NF-YA negative control promoter. No enrichment was observed with anti-Flag negative control antibodies. Error bars in panels E and F indicate SD.
Discussion
In this manuscript, we demonstrate that the human THAP–zinc finger protein THAP1 is an endogenous physiologic regulator of EC proliferation. Silencing of THAP1 by RNA interference in human primary ECs resulted in inhibition of G1/S cell-cycle progression and down-modulation of several pRb/E2F cell-cycle target genes, including RRM1, a gene activated at the G1/S transition, essential for S-phase DNA synthesis. ChIP assays in proliferating ECs revealed that endogenous THAP1 associates in vivo with a consensus THAP1-binding site found in the RRM1 promoter, indicating that RRM1 is a direct target gene of THAP1, and providing important mechanistic insights about the role of endogenous THAP1 in the regulation of EC proliferation. Together, our results suggest that THAP1 may constitute a potential target for inhibition of EC proliferation in angiogenesis-dependent diseases, such as cancer and chronic inflammatory diseases.
THAP–zinc finger proteins represent a previously uncharacterized family of cellular factors with approximately 100 distinct members in model animal organisms,7,8 including zebrafish orthologue of cell-cycle regulator E2F6, and 5 C elegans proteins, LIN-36, LIN-15A, LIN-15B, HIM-17 and GON-14, interacting genetically with LIN-35/Rb, the C elegans orthologue of the tumor suppressor pRB.10-13,17 While several of these THAP proteins (LIN-36, LIN-15B) have been shown to function as inhibitors of G1/S cell-cycle transition and cell proliferation,12,14,16 our data indicate that human THAP1 may rather play a positive role in cell proliferation and G1/S cell-cycle progression. Interestingly, cell division defects were recently observed in the intestine, gonad, and vulva of gon14 null mutants, suggesting that C elegans THAP family member GON-14 may also function as a positive regulator of cell proliferation.17 Therefore, both in humans and model animal organisms, THAP–zinc finger proteins appear to be critical regulators of cell proliferation and cell-cycle progression.
The similar phenotypes observed after THAP1 overexpression through retroviral-mediated gene transfer or silencing through RNA interference suggest that THAP1 overexpression inhibits the function of endogenous THAP1 in cell proliferation and cell-cycle progression. Similar observations have been made in ECs for components of the Notch pathway; silencing or overexpression of the Notch ligand DLL4 in ECs led to a significant inhibition of proliferation,40,41 and overexpression or knock-out of Notch4 produced similar vascular phenotypes in mice.42 This indicated that there may be a window of appropriate Notch expression levels for EC proliferation. Our data suggest that an optimal range of THAP1 expression also appears to be critical for EC proliferation and cell-cycle progression. One could imagine that overexpression of THAP1 may inhibit the function of endogenous THAP1 protein complexes by disrupting the normal stoichiometry of these complexes.
Since large-scale gene expression profiling showed a selective modulation of pRB/E2F cell-cycle target genes in ECs overexpressing THAP1 (Table 1), we asked whether a similar modulation of pRb-E2F cell-cycle target genes could be observed in ECs transfected with THAP1 siRNAs. We found that silencing of endogenous THAP1 by RNA interference resulted in down-modulation of the 8 pRB/E2F cell-cycle target genes analyzed, RRM1, Mad2, survivin, HMMR, RRM2, CDC2, cyclin B1, and DLG7. Therefore, THAP1 may play a role in the activation or the maintenance of the activated state of cell-cycle–specific genes in proliferating cells. Interestingly, we identified potential THAP1-binding sites in the promoters of several genes (RRM1, RRM2, BIRC5/survivin, cyclin B1) down-modulated after THAP1 silencing. We selected RRM1, a critical pRb/E2F target gene activated at the G1/S transition,29-32,34-36 for further characterization, and we could demonstrate direct binding of the THAP domain of human THAP1 to the RRM1 promoter in vitro using EMSA and footprinting assays and association of endogenous THAP1 with the RRM1 promoter in vivo using quantitative ChIP assays in proliferating ECs. Together, these data indicate that RRM1 is a direct transcriptional target of THAP1, and suggest that THAP1 may associate with the promoters of other pRb/E2F cell-cycle target genes and modulate their activity. This model is supported by observations made in model animal organisms. In zebrafish and other fish species, the THAP-E2F6 fusion proteins8 are likely to associate with cell-cycle–specific promoters since this has clearly been shown for mouse and human E2F6 orthologues using ChIP assays.9 In C elegans, the THAP protein LIN-36 has been proposed to act in a transcriptional repressor complex with LIN-35/Rb and Efl-1/E2F4-5, to repress G1/S control genes.12,16
Genetic data obtained in C elegans suggest that THAP1 may act at the level of chromatin regulation. Indeed, several members of the C elegans THAP family (LIN-36, LIN-15B, LIN-15A, HIM-17, GON-14) have been found to interact genetically with known components of chromatin-modifying and/or chromatin-remodeling complexes, including members of the Rb and nucleosome remodeling deacetylase (NuRD) complexes and components of the Tip60/NuA4 histone acetyltransferase complex.10-13,17,43,44 Interestingly, the human THAP7 protein has also been found to interact with chromatin-modifying enzymes.19,20 Together, these observations suggest that THAP1 may function at the level of chromatin regulation, by recruiting chromatin-modifying and/or chromatin-remodeling complexes to specific DNA sites in target genes (ie, RRM1 promoter and potentially other pRb/E2F cell-cycle target genes), resulting in transcriptional activation of these target genes during cell proliferation.
In summary, the results presented in this manuscript are important because they provide the first link in humans among the THAP proteins, the cell cycle, and the pRB/E2F pathway. In addition, they suggest that THAP proteins may play important roles in the control of cell proliferation and cell-cycle progression in humans, similarly to what has been found in C elegans. Future studies should aim at determining whether other human THAP proteins play a role in cell proliferation, defining the composition of the endogenous THAP1 protein complexes and characterizing the potential links between THAP1 and E2F family members in the regulation of cell-cycle–specific promoters.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: the authors declare no competing financial interests.
Contribution: C.C. performed research and analyzed data (cell proliferation/cell-cycle analyses, apoptosis assays, antibody generation and validation, Western blot and immunofluorescence analyses, qPCR experiments, retroviral transduction, RNA interference, ChIP-qPCR assays); C.L. and M.C. performed research (ChIP assays); C.M. performed research (microarray data mining, bioinformatics); V.E. performed research (in vitro DNA-binding assays); E.L. performed research (qPCR experiments); V.L. performed research (microarray experiments at the IGR Microarray Platform); P.D. performed research (bioinformatics analyses at the IGR Microarray Platform); R.M. analyzed data (supervised ChIP assays); L.A. analyzed data; J.-P.G. performed research (Northern blot), designed research, analyzed data, and wrote the manuscript.
We are grateful to Dr R. A. Willemsen (Daniel den Hoed Cancer Center, Erasmus MC, Rotterdam, The Netherlands) for the gift of the pBullet retroviral expression vector and to the Laboratoire de Biothérapies (Institut National de la Santé et de la Recherche Médicale [INSERM] Institut Fédératif de Recherche (IFR)31–Ligue Régionale contre le Cancer, Toulouse, France), and Dr P. Bouille and R. Gayon (Endocube, Labège, France) for help with the retroviral transduction experiments. We thank F. Viala for iconography, and N. Ortega and S. Roga (Institut de Pharmacologie et de Biologie Structurale (IPBS)–Centre National de la Recherche Scientifique [CNRS], Toulouse, France) for help with HUVEC culture and qPCR experiments, respectively. Special thanks go to Prof F. Amalric (IPBS-CNRS, Toulouse, France) for stimulating discussions.
This work was supported by grants from Ligue Nationale contre le Cancer (Equipe labellisée), Agence Nationale de Recherche (ANR)–programme blanc “Cuboïdale” and Ministère de la Recherche Action Concerté Incitative-Cancéropôle Grand Sud-Ouest 2004 (Network ‘Angiogenesis and Invasion’).



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal