Abstract
Mantle-cell lymphoma (MCL) is genetically characterized by the translocation t(11;14)(q13;q32) and a high number of secondary chromosomal abnormalities. To identify genes inactivated in this lymphoma, we examined 5 MCL cell lines following a strategy previously described in tumors with microsatellite instability that is based on the combined inhibition of the nonsense-mediated mRNA decay pathway and gene-expression profiling. This approach, together with the design of a conservative algorithm for analysis of the results, allowed the identification of 3 genes carrying premature stop codons. These genes were p53 with a mutation previously described in JEKO-1, the leukocyte-derived arginine aminopeptidase (LRAP) gene in REC-1 that showed a new splicing isoform generating a premature stop codon, and RB1 in UPN-1 that contained an intragenic homozygous deletion resulting in a truncated transcript and total loss of protein expression. The new LRAP isoform was detected also in 2 primary MCLs, whereas inactivating intragenic deletions of RB1 were found in the primary tumor from which UPN-1 was derived and 1 additional blastoid MCL. These tumors carried a concomitant inactivation of p53, whereas p16INK4a was wild type. These results indicate for the first time that RB1 may be inactivated in aggressive MCL by intragenic deletions.
Introduction
Mantle-cell lymphoma (MCL) is as a distinct type of B-cell non-Hodgkin lymphoma genetically characterized by the t(11;14)(q13;q32) translocation that leads to the dysregulation of cyclin D1.1 In addition to the constitutive overexpression of cyclin D1, MCL shows frequent alterations of genes involved in the DNA damage response pathway and in the G1/S-phase checkpoint.2 The dysregulation of the DNA damage response pathway may facilitate the chromosomal instability observed in these tumors. In that sense, inactivation of the tumor-suppressor genes (TSGs) p53 or ATM has been frequently observed in MCL with high number of chromosome abnormalities.3–5 Oncogenic alterations in the cell cycle machinery include the inactivation of INK4a/ARF and the amplification of potential oncogenes such as BMI-1 or CDK4.6–8 All these alterations play an important pathogenetic role in MCL, probably deregulating the cell cycle control by overcoming the suppressor effect that retinoblastoma 1 (RB1) performs in the G1/S-phase transition. In addition to these molecular oncogenic events, conventional cytogenetic and comparative genomic hybridization (CGH) studies have revealed a high number of secondary chromosomal abnormalities in MCL. However, only a few target genes of these genetic alterations have been identified.9,10
The nonsense-mediated mRNA decay (NMD) pathway is an eukaryotic cellular mRNA surveillance mechanism that rapidly degrades mRNA transcripts containing premature termination codons (PTCs).11,12 This mechanism of control prevents the accumulation of truncated proteins generated by translation of mRNAs that carry PTCs due to nonsense mutations, splicing errors, or inaccuracies during the transcription process. A physiological posttranscriptional regulatory role of this pathway has also been suggested.12 The molecular mechanisms involved in NMD are not fully understood. The exon-exon junctions of processed mRNAs are labeled by protein complexes that will be subsequently removed by the ribosomal machinery during the first cycle of translation. When a pre-mRNA that contains a PTC is processed, translation stops at this codon and all the protein complexes bound to exon-exon junctions distal to this point remain on the transcript. The presence of these complexes seems to provide a signal for the recruitment of new protein complexes that will result in the degradation of the abnormal mRNA. Recently, a new experimental strategy named gene identification by NMD inhibition (GINI) has been proposed for the identification of genes carrying mutations that would cause premature protein termination and destabilization of the mutant transcript by NMD. This method is based on the inhibition of the NMD pathway, which selectively would stabilize mutant transcripts degraded by this pathway, and the subsequent detection of the stabilized transcripts by gene expression profiling using microarrays.13 This approach has been used successfully to identify tumor-suppressor genes in tumor cell lines with microsatellite instability,14,15 but it has not been applied to study other types of tumor cell lines.
In this study, we have investigated whether the GINI strategy may help to identify potential TSGs inactivated in MCL cell lines. This approach has led us to discover that inactivation of RB1 occurs in a subset of aggressive mantle-cell lymphomas.
Materials and methods
The present study was approved by the Hospital Clinic of Barcelona institutional review board. Informed consent was provided in accordance with the Declaration of Helsinki.
Nonsense-mediated mRNA decay pathway inhibition, real-time polymerase chain reaction, and microarray hybridization
Five MCL cell lines (UPN-1, JEKO-1, REC-1, HBL-2, and GRANTA-519) previously characterized genetically were used in the study.16,17 The inhibition of the NMD pathway was performed according to a modified protocol previously described.13,14 The cells were treated with emetine (E; Sigma, Steinheim, Germany) (5 μM for 10 hours) or with emetine followed by incubation with actinomycin D (A; Sigma, Steinheim, Germany) (10 μM for 1, 3, and 5 hours; E + A). Mock-treated cells were used as reference. Total RNA was extracted from each cell line and experimental condition. The RNA from the mock-treated cells, emetine-treated cells, and a pool of emetine and actinomycin D–treated cells was hybridized onto HG-U133 Plus 2.0 oligonucleotide microarrays (Affymetrix, Santa Clara, CA). The sample preparation and processing were performed following the protocols described in the Affymetrix GeneChip Expression Analysis Manual (Affymetrix). The time course and drug treatment conditions of these experiments were established after a preliminary study using JEKO-1 cell line that harbors a known nonsense mutation of p53 gene and REC-1 that has a wild-type p53.16 These cell lines were treated at different time points with emetine or emetine followed by actinomycin D. Total RNA extracted from the cells during this time course was subjected to a real-time polymerase chain reaction (PCR) analysis of p53 mRNA. The primers and probe used to amplify and quantify the p53 cDNA were provided by Assay-on-Demand Gene Expression Products (Hs 00153349_m1; Applied Biosystems, Foster City, CA). The β-glucuronidase gene (GUSB; Applied Biosystems) was used as endogenous control as recommended by the manufacturer.
Gene-mutation analysis
Total RNA was isolated from the 5 MCL cells lines and 32 primary tumors by TRIZOL reagent (Invitrogen, Carlsbad, CA) following the manufacturer's protocols. The patients had given their informed consent and the study was approved by the hospital review board. The quality of all RNA samples was verified using the Bioanalyzer 2100 (Agilent, Palo Alto, CA). cDNA was generated from 1 μg total RNA using random hexamers from SuperScript Synthesis System for RT-PCR First-Strand following the manufacturer's specifications (Invitrogen).
Overlapping PCR reactions were designed to amplify the full-length cDNA of RB1 and leucocyte-derived arginine aminopeptidase (LRAP) following a step-down PCR strategy. Conditions for all fragments were one step at 94°C for 5 minutes; 2 cycles at 94°C for 30 seconds, at 62°C for 30 seconds, and at 72°C for 1 minute; 2 cycles with annealing at 60°C; 2 cycles with annealing at 58°C; and 30 cycles with annealing at 56°C. The first fragment of RB1 cDNA was amplified in the presence of 5% dimethyl sulfoxide.
The gene mutation screening in primary tumors was performed using denaturing high-pressure liquid chromatography (DHPLC, WAVE System; Transgenomic, Omaha, NE) as described previously.18 PCR fragments showing unique abnormal chromatogram profiles by the WAVE analysis were purified using a GenElute PCR Clean-Up kit (Sigma, St Louis, MO) and sequenced using cycle sequencing with BigDye Terminator Chemistry (Applied Biosystems). Sequencing reactions were run on an ABI-3100 automated sequencer (Applied Biosystems).
Genomic DNA was extracted from primary tumor tissues and cell lines using proteinase K/RNAse treatment and phenol-chloroform extraction. Multiplex PCR reactions were designed to amplify exons 1, 2, 3, 4, 5, 17, or 18 from RB1 together with β-actin, used as the internal control of the PCR reaction. Genomic DNA extracted from formalin-fixed and paraffin-embedded tissues was amplified in a multiplex PCR reaction with primers designed to amplify smaller fragments from RB1 exon 1 or 18 together with β-actin. PCR conditions were the same as described in the second paragraph of the present section. P53 mutations were analyzed following previously described protocols.19 Primer sequences are available on request.
Single nucleotide polymorphism microarray mapping assay
For the SNP mapping array analysis, we followed the Affymetrix GeneChip Mapping 100 k Assay Manual (Affymetrix). Briefly, 250 ng genomic DNA was digested with XbaI or HindIII, followed by adapter ligation, PCR amplification, PCR product fragmentation, end biotin labeling, and hybridization to the GeneChip Human Mapping 100K Set. Data analysis was performed using the GCOS, GTYPE, and the CNAT applications recommended by Affymetrix.
Western blot and immunohistochemistry analysis
Protein expression was examined by Western blot. Total protein was extracted from 107 cells following previously described protocols.20 Forty μg total cellular protein was run per lane on a 6% sodium dodecyl sulfate (SDS)–polyacrylamide gel and electroblotted to a nitrocellulose membrane (Amersham, Freiburg, Germany). After blocking, the membrane was incubated with the anti-RB1 monoclonal antibody (clone G3-245; BD Biosciences Pharmingen, Erembodegem, Belgium) that recognizes an epitope between amino acids 332 to 344 of the human retinoblastoma protein. The antibody was used at a final dilution of 1:100 following previously described protocols.20
Retinoblastoma protein expression was examined by immunohistochemistry on formalin-fixed paraffin-embedded sections using a monoclonal antibody against pRB1 (clone G3-245; BD Biosciences Pharmingen). Anti-RB1 was used at a 1:50 dilution and the immunoreaction was developed with the EnVision+System Peroxidase (DAB) method (DAKO, Carpinteria, CA). The antigen retrieval was done by 10 mmol citrate buffer in a pressure cooker. The slides were counterstained with hematoxylin. Immunohistochemical images were acquired using an Olympus BX51 clinical microscope and DP70 digital camera and software with objective lens UPlanFl 40×/0.75 (Olympus, Tokyo, Japan).
Microarray analysis and algorithm designed to identify potential TSGs
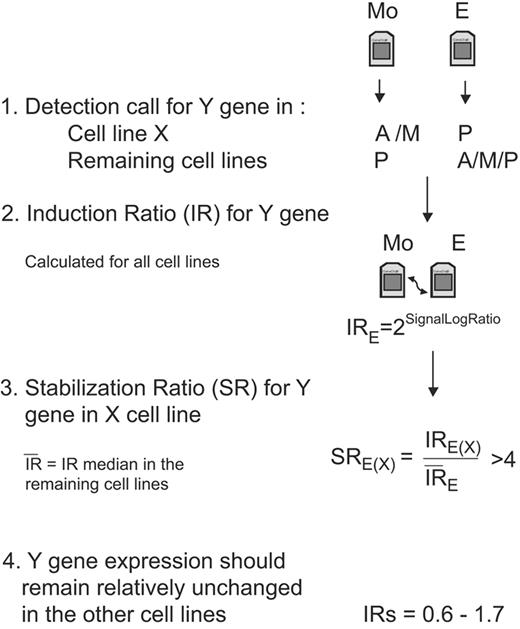
The GINI approach has successfully been tested in cancer cell lines with microsatellite instability that carry a high number of mutations.14,15 Cells with a functional DNA repair machinery should accumulate fewer nonsense mutations than cells with disabled DNA repair function. We therefore analyzed multiple cell lines and designed a very conservative algorithm to narrow the search of potentially inactivated genes and to avoid false-positive candidates (Figure 1). Our algorithm was based on the hypothesis that a true candidate gene in a specific cell line should show very low levels of mRNA in the presence of an active NMD pathway. Moreover, we considered an improbable event that a potential TSG would carry a PTC in more than one cell line.
Diagram of the algorithm steps to select candidate genes in a specific cell line. For simplification, we represent the emetine treatment and the analysis of only one gene in the flowchart, but the same filters and analysis should be applied for the actinomycin D treatment and for the rest of the genes. The initial step to select a candidate gene (Y gene) in a specific cell line (X cell line) is to obtain the detection called present “P,” absent “A,” or marginal “M” calculated by the GCOS software. This first filter allows selection of genes that are specifically down-regulated in one cell line in the presence of a functional NMD pathway (before drug treatment) compared to the other mock-treated cell lines. Afterward, the level of induction for each gene is quantified in each cell line, and these IRs are used to calculate the SR. This SR should allow differentiating between an induction due to stabilization of a transcript carrying a PTC in a specific cell line from a general transcriptional induction due to the drug treatment or because of the accumulation of common alternative splicings that could occur in several cell lines. We establish the arbitrary cutoff of SR more than 4. The final filter will select only genes that in addition to having a high SR in a specific cell line have an mRNA level that remains mainly unmodified after drug treatment in the other cell lines (IR= 06-1.7).
Diagram of the algorithm steps to select candidate genes in a specific cell line. For simplification, we represent the emetine treatment and the analysis of only one gene in the flowchart, but the same filters and analysis should be applied for the actinomycin D treatment and for the rest of the genes. The initial step to select a candidate gene (Y gene) in a specific cell line (X cell line) is to obtain the detection called present “P,” absent “A,” or marginal “M” calculated by the GCOS software. This first filter allows selection of genes that are specifically down-regulated in one cell line in the presence of a functional NMD pathway (before drug treatment) compared to the other mock-treated cell lines. Afterward, the level of induction for each gene is quantified in each cell line, and these IRs are used to calculate the SR. This SR should allow differentiating between an induction due to stabilization of a transcript carrying a PTC in a specific cell line from a general transcriptional induction due to the drug treatment or because of the accumulation of common alternative splicings that could occur in several cell lines. We establish the arbitrary cutoff of SR more than 4. The final filter will select only genes that in addition to having a high SR in a specific cell line have an mRNA level that remains mainly unmodified after drug treatment in the other cell lines (IR= 06-1.7).
The microarray data were normalized using a global scaling strategy and a target intensity of 150 was applied. We used the detection call and the signal value generated by the GCOS version 1.4 software as qualitative and quantitative measurements of the transcript levels detected by each probe set. The primary Cel files have been deposited in the public repository Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) with the series record (GSE6728). Our algorithm to select the best candidate genes includes stringent filters based on the comparison of the results in the 5 MCL cells lines. These comparisons should distinguish between a general gene expression induction by the drug treatment and specific increase of mRNA levels due to stabilization of mutated transcripts in the presence of an inactivated NMD pathway. In addition, the comparison between cell lines should also exclude frequent splicing variants that generate PTC.
The gene selection strategy required first that for a specific cell line the genes were considered “nonpresent” (the detection call could be “absent” or “marginal”) in mock-treated cells but called “present” after drug treatment. Moreover, the genes should be called “present” in the rest of mock-treated cells. For the genes that passed these qualitative criteria, we established an additional quantitative filter for all cell lines and drug treatments. This filter, named mRNA induction ratio (IR), compared the gene expression levels of a specific treated cell line versus the levels of the respective mock-treated cells. This IR was defined as 2SignalLogRatio using the signal log ratio calculated by GCOS for each drug condition (E and E + A) and considering the mock-treated cells as baseline. Then, the IRs of all genes and drug conditions in each cell line were compared with the respective IRs in the other 4 cell lines. This comparison was named stabilization ratio (SR) and was calculated as the ratio of the IR in a specific cell line versus the IR average of the other 4 cell lines. A good candidate gene in a specific cell line was considered when the SR for both drug treatments was higher than 4 and, in addition, the gene expression was relatively unchanged by the treatment in the other 4 cell lines (IR: 0.6 and 1.7, corresponding to a variation of < 75%).
Results
NMD inactivation stabilizes p53 mRNA in JEKO-1
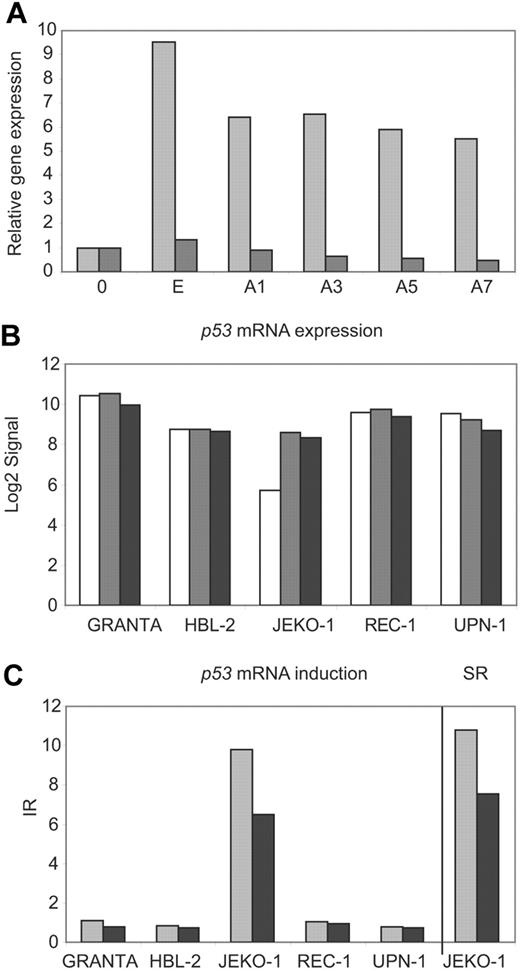
The GINI strategy has been useful to identify transcripts with PTCs in tumor cell lines carrying microsatellite instability but has not been applied in other types of neoplasms. To determine whether inhibition of the NMD pathway could be a valid strategy for identifying inactivated genes in MCL cell lines, we first investigated the potential stabilization of the p53 mRNA transcript by inhibition of the NMD in the JEKO-1 cell line that carries an inactivation of p53 by a gross deletion of one allele and the presence of a premature stop codon in exon 4 in the other allele.16 The REC-1 cell line carrying a wild-type p53 was used as a negative control. p53 mRNA levels were quantified in both cell lines by real-time PCR during a time course after emetine and emetine plus actinomycin D treatment (Figure 2A). The p53 transcripts were more than 5 fold higher in JEKO-1 after the emetine treatment compared to mock-treated cells. This p53 mRNA increase was also detected after transcription inhibition by actinomycin D treatment. On the contrary, p53 transcripts did not show any increase in REC-1 following drug treatments (Figure 2A). These results were used to design the experimental conditions for the GINI strategy and the whole genome microarray analysis in the 5 MCL cell lines.
p53 mRNA is stabilized in JEKO-1 after NMD inhibition. (A) p53 mRNA levels were analyzed by real-time PCR in nontreated cells (0) and after emetine (E) or emetine plus actinomycin D treatment during a time course (A1, A3, A5 and A7) in JEKO-1 (▩) and REC-1 (■). The expression levels were related to mock-treated cells (0). (B) After microarray normalization, the signals obtained with the GCOS software for p53 were log2 transformed and plotted for mock-treated (□), emetine-treated (▩), and emetine plus actinomycin D–treated (■) cells. (C) The p53 IRs for each cell line were plotted together with the p53 SR in JEKO-1 for both treatments, emetine (▩) and emetine plus actinomycin D (■).
p53 mRNA is stabilized in JEKO-1 after NMD inhibition. (A) p53 mRNA levels were analyzed by real-time PCR in nontreated cells (0) and after emetine (E) or emetine plus actinomycin D treatment during a time course (A1, A3, A5 and A7) in JEKO-1 (▩) and REC-1 (■). The expression levels were related to mock-treated cells (0). (B) After microarray normalization, the signals obtained with the GCOS software for p53 were log2 transformed and plotted for mock-treated (□), emetine-treated (▩), and emetine plus actinomycin D–treated (■) cells. (C) The p53 IRs for each cell line were plotted together with the p53 SR in JEKO-1 for both treatments, emetine (▩) and emetine plus actinomycin D (■).
To search for potential transcripts stabilized by the pharmacological inhibition of the NMD in the MCL cell lines, we initially applied a previously described algorithm that compared the expression profile of mock- and drug-treated cells.21 More than 300 probe sets passed the required cutoffs defined to filter candidate genes. However, in JEKO-1 the algorithm identified 32 probe sets better positioned than the positive control p53. On the other hand, sequencing analysis of selected genes in this list showed that all of them corresponded to false-positive genes with a wild-type sequence or different gene isoforms due to nonproductive splicing variants. Therefore, to minimize the number of false-positive genes in the analysis, we designed the more stringent algorithm described in “Materials and methods” (Figure 1). Using this method, only 6 probe sets were selected (Table S1). Four of them represented annotated genes and 2 were not annotated. Of note, 2 of the probe sets recognized the positive control p53 in JEKO-1, confirming the value of the algorithm. p53 mRNA levels were very low and the transcript was called absent in mock-treated JEKO-1 cells, whereas it was called present and showed higher levels in the remaining mock-treated cell lines (Figure 2B). After NMD inhibition, there was a marked increase in p53 mRNA levels in JEKO-1 cells and the transcript was called present, whereas in the other 4 cell lines the p53 transcript levels remained mainly unchanged after drug treatment (Figure 2B). These results suggest that the high levels of p53 mRNA in JEKO-1–treated cells were not due to an induction by the drug treatment, since this effect did not occur in any of the other cell lines. Consistent with that, the stabilization ratio for p53 transcript generated by our algorithm was very high in JEKO-1 (SRE= 11.2 and SRE + A= 7.5) (Figure 2C).
RB1 inactivation in MCL cell lines
The tumor-suppressor gene retinoblastoma 1 (RB1) was the second gene that passed our stringent filters. RB1 mRNA was called absent in UPN-1 mock-treated cells, whereas it showed high levels of expression in the other 4 mock-treated cell lines (Figure 3A). However, after the inhibition of the NMD by drug treatment, the RB1 mRNA transcript showed a marked stabilization in UPN-1 (SRE= 17.7 and SRE + A= 17.8). In the other 4 cell lines, the RB1 mRNA levels remained essentially constant after drug treatment (Figure 3B).
RB1 mRNA is stabilized in UPN-1 after NMD inhibition. (A) Normalized log2 transformed signals generated by GCOS for RB1 were plotted for all cell lines and treatments (mock, □; emetine, ▩; emetine plus actinomycin D, ■). (B) The RB1 induction ratios (IRs) for each cell line were plotted together with the RB1 stabilization ratio (SR) in UPN-1 (E, ▩; E + A, ■).
RB1 mRNA is stabilized in UPN-1 after NMD inhibition. (A) Normalized log2 transformed signals generated by GCOS for RB1 were plotted for all cell lines and treatments (mock, □; emetine, ▩; emetine plus actinomycin D, ■). (B) The RB1 induction ratios (IRs) for each cell line were plotted together with the RB1 stabilization ratio (SR) in UPN-1 (E, ▩; E + A, ■).
To confirm the potential RB1 gene alteration, we amplified the whole RB1 cDNA in 8 overlapping fragments as described in “Materials and methods.” Only 2 PCR fragments covering exons 20 through 27 could be amplified using mRNA isolated from emetine UPN-1–treated cells, but all 8 fragments were amplified in the control sample (data not shown). To identify the exons involved in this apparent deletion, we designed exon-specific PCRs to amplify from genomic DNA the exons present in the first fragment (exons 1, 2, and partially exon 3) and in the sixth fragment (containing exons 17, 18, and 19). Only exons 18 and 19 were amplified, suggesting that the deletion encompasses at least exons 1 through 17, both included (Figure 4A). Supporting these results, the hybridization of genomic DNA from UPN-1 onto the Human Mapping 100K set SNP microarray (Affymetrix) showed a homozygous deletion of all 6 SNPs present in the array that were located between exons 2 and 5 of the RB1 locus. However, the SNP rs198563 located close to exon 18 showed a signal equivalent to 2 allelic copies, indicating that this gene region was not deleted (Figure 4B; Table S2). The SNP array analysis revealed that the chromosomal region 13q14-qter in UPN-1 was gained and corresponded to a uniparental trisomy. This 13q14-qter gain in UPN-1 was confirmed by conventional CGH.
RB1 is inactivated in UPN-1 and in the original primary MCL. (A) Multiplex PCR amplifications of RB1 exons 1, 17, and 18 performed with genomic DNA from UPN-1 cells are shown. Although β-actin (β-act) was amplified in both UPN-1 and in the control (C+), only exon 18 of RB1 was amplified in UPN-1. (B) A section of the Log2Ratio graph corresponding to the 13q14 region generated by the CNAT application is showed. All SNPs are displayed in blue color indicating that the log2 ratio values were higher than 0. A zoom view of a small region containing negative log2 ratio values show that it correspond to the RB1 locus. Two maps are shown, one represents all RB1 exons by gray vertical lines, and the second represents the SNPs located in the region. In red are indicated the SNPs that are deleted with signal log2 ratio value lower than −1, and conserved SNPs are shown in black. (C) pRB1 expression was analyzed by Western blot in 5 cell lines. Equal amount of total protein was loaded in each lane. A section of the Coomassie staining gel is showed at the bottom. (D) Immunohistochemical staining of pRB1 in the primary tumor from which UPN-1 was established. Tumor cells are negative, whereas endothelial cells, considered as internal positive controls, showed positivity (inset). (E) A multiplex PCR reaction was designed to amplify exon 1 and 18 of RB1 together with β-actin. Both RB1 exons were amplified when we used a DNA control (C+), but when we used DNA extracted from the primary tumor from which the UPN-1 cell line was established (UPN-1 case) only the RB1 exon 18 was amplified.
RB1 is inactivated in UPN-1 and in the original primary MCL. (A) Multiplex PCR amplifications of RB1 exons 1, 17, and 18 performed with genomic DNA from UPN-1 cells are shown. Although β-actin (β-act) was amplified in both UPN-1 and in the control (C+), only exon 18 of RB1 was amplified in UPN-1. (B) A section of the Log2Ratio graph corresponding to the 13q14 region generated by the CNAT application is showed. All SNPs are displayed in blue color indicating that the log2 ratio values were higher than 0. A zoom view of a small region containing negative log2 ratio values show that it correspond to the RB1 locus. Two maps are shown, one represents all RB1 exons by gray vertical lines, and the second represents the SNPs located in the region. In red are indicated the SNPs that are deleted with signal log2 ratio value lower than −1, and conserved SNPs are shown in black. (C) pRB1 expression was analyzed by Western blot in 5 cell lines. Equal amount of total protein was loaded in each lane. A section of the Coomassie staining gel is showed at the bottom. (D) Immunohistochemical staining of pRB1 in the primary tumor from which UPN-1 was established. Tumor cells are negative, whereas endothelial cells, considered as internal positive controls, showed positivity (inset). (E) A multiplex PCR reaction was designed to amplify exon 1 and 18 of RB1 together with β-actin. Both RB1 exons were amplified when we used a DNA control (C+), but when we used DNA extracted from the primary tumor from which the UPN-1 cell line was established (UPN-1 case) only the RB1 exon 18 was amplified.
The protein expression analysis of UPN-1 by Western blot showed a complete absence of pRB1 in this cell line consistent with the homozygous deletion of the gene (Figure 4C). The REC-1 cell line that has a hemizygous deletion of 13q14 showed reduced levels of pRB1 compared to the other cell lines with no alteration in this chromosome region (Figure 4C).
To determine whether the RB1 inactivation observed in UPN-1 was acquired during the cell line establishment, we studied the original mantle-cell lymphoma biopsy of the patient from whom the cell line was derived. The primary tumor of this patient had been diagnosed as a blastoid MCL. The pRB1 expression was examined by immunohistochemistry in tissue sections from this biopsy and, as shown in Figure 4D, tumor cells were completely negative for pRB1, whereas normal endothelial and reactive cells showed strong nuclear staining. Furthermore, we extracted genomic DNA from this biopsy and were able to amplify RB1 exon 18 but not RB1 exon 1 in a multiplex PCR reaction (Figure 4E). These results indicate that the RB1 inactivation by an intragenic homozygous deletion already occurred in the primary blastoid MCL.
The initial characterization of the UPN-1 cell line detected the 856G→A (Glu286Lys) mutation in p53.22 We analyzed if the inactivation of p53 also occurred in the primary MCL or whether it took place during the cell line establishment. The amplification and sequencing of DNA extracted from paraffin-embedded tissue of the initial biopsy confirmed that p53 inactivation also occurred in the primary MCL (data not shown). Although UPN-1 has an allelic loss of 9p13, we detected relatively high levels of p16INK4a mRNA in the expression array analysis suggesting that the gene is not deleted in this MCL cell line.17
RB1 inactivation in primary MCL
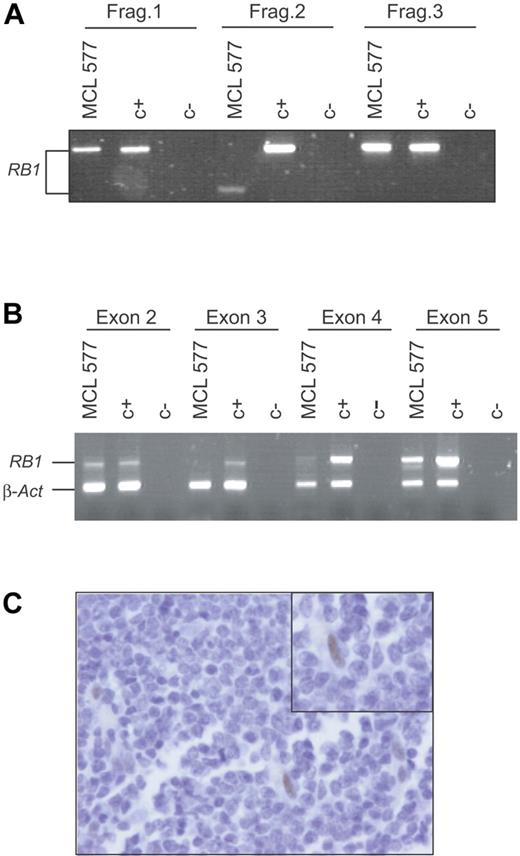
To determine whether other primary MCLs could have RB1 inactivations, we analyzed a series of 32 primary MCLs, including 22 typical and 10 blastoid variants. The full-length RB1 coding cDNA was amplified and a mutation screening was performed using a DHPLC system as described in “Materials and methods.” Twelve of 32 cases analyzed showed deletion of 13q14 by CGH.9 One patient showed an abnormally shorter RB1 cDNA-amplified fragment (Figure 5A). The sequence of this fragment revealed that exons 3 and 4 were missing and the new cDNA would codify for a smaller truncated protein. An amplification of the genomic DNA of this tumor using a multiplex PCR showed a normal signal of exons 2 and 5, but the amplification signal of exons 3 and 4 was virtually absent suggesting the presence of a homozygous deletion of these 2 exons (Figure 5B). To confirm this RB1 inactivation, we examined the pRB1 expression by immunohistochemistry in tumor tissue sections. Tumor cells did not show pRB1 expression, whereas occasional endothelial and reactive cells, considered as internal controls, were strongly positive (Figure 5C). Altogether, these results indicate that RB1 was also inactivated in this primary MCL tumor by an intragenic homozygous deletion resulting in an anomalous transcript and lack of protein expression. Of interest, this MCL was diagnosed as a blastoid variant that carried a homozygous deletion of p53.9 The INK4a locus of this tumor was examined previously and shown to be in germ line.7 To summarize, 2 of 11 aggressive blastoid MCLs in our series, including the primary tumor from which UPN-1 was established, showed concomitant inactivation of RB1 and p53 with a maintained INK4a/ARF locus.
RB1 is inactivated in primary MCL. (A) Three of 8 PCR fragments designed to amplify the full-length RB1 cDNA are shown. Fragment 2, involving exon 2, 3, 4, and 5, amplified from a primary MCL (MCL 577), was shorter than the one amplified from the control (C+). (B) Genomic DNA from this primary MCL (MCL 577) and normal tissue (C+) were used in a multiplex PCR reaction in order to amplify exon 2, 3, 4, and 5 of RB1 together with β-actin. Only exons 2 and 5 were amplified in the primary MCL (MCL 577). (C) Immunohistochemical analysis of pRB1 in the primary tumor (MCL 577). A number of normal cells were positively stained (inset), but the tumor cells were negative.
RB1 is inactivated in primary MCL. (A) Three of 8 PCR fragments designed to amplify the full-length RB1 cDNA are shown. Fragment 2, involving exon 2, 3, 4, and 5, amplified from a primary MCL (MCL 577), was shorter than the one amplified from the control (C+). (B) Genomic DNA from this primary MCL (MCL 577) and normal tissue (C+) were used in a multiplex PCR reaction in order to amplify exon 2, 3, 4, and 5 of RB1 together with β-actin. Only exons 2 and 5 were amplified in the primary MCL (MCL 577). (C) Immunohistochemical analysis of pRB1 in the primary tumor (MCL 577). A number of normal cells were positively stained (inset), but the tumor cells were negative.
LRAP isoform not previously described is expressed in MCL
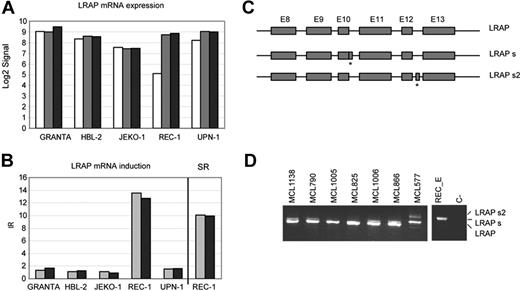
The third gene identified as a potential candidate carrying a premature stop codon was leukocyte-derived arginine aminopeptidase (LRAP). In REC-1 the LRAP transcript showed a marked stabilization (SRE= 10.1 and SRE + A= 9.94) after NMD inhibition (Figure 6A-B). In order to identify the cause of this stabilization, we amplified and sequenced the full cDNA derived from emetine-treated REC-1 cells. The study of the cDNA identified a new exon, not previously described, between exons 12 and 13 (LRAPS2, Figure 6C). The mRNA generated as a consequence of this alternative splicing contains a premature stop codon. After drug treatment, REC-1 showed expression of this new variant only.
LRAP mRNA is stabilized in REC-1 after NMD inhibition. (A) Normalized signals generated by GCOS for LRAP were log2 transformed and plotted for all cell lines and treatments (mock, □; E, ▩; E + A, ■). (B) The LRAP induction ratios (IRs) for each cell line were plotted together with the LRAP stabilization ratio (SR) in REC-1 (E, ▩; E + A, ■). (C) Schematic view of the LRAP splicing identified in REC-1 (LRAPS2) together with the normal variant (LRAP) and with a previously reported splicing (LRAPS). PTCs are labeled with asterisks. (D) Identification in primary MCLs of the LRAP splicing described in REC-1 emetine-treated cells.
LRAP mRNA is stabilized in REC-1 after NMD inhibition. (A) Normalized signals generated by GCOS for LRAP were log2 transformed and plotted for all cell lines and treatments (mock, □; E, ▩; E + A, ■). (B) The LRAP induction ratios (IRs) for each cell line were plotted together with the LRAP stabilization ratio (SR) in REC-1 (E, ▩; E + A, ■). (C) Schematic view of the LRAP splicing identified in REC-1 (LRAPS2) together with the normal variant (LRAP) and with a previously reported splicing (LRAPS). PTCs are labeled with asterisks. (D) Identification in primary MCLs of the LRAP splicing described in REC-1 emetine-treated cells.
To determine whether this new splicing isoform was also present in primary tumors, we analyzed the cDNA of 14 MCL (Figure 6D). Twelve patients (86%) showed expression of the normal variant. In 5 of the cases this was the only isoform detected, but in 7 patients low levels of an additional shorter splicing variant previously described were also observed.23 The 2 cases without expression of the normal variant showed expression of the new isoform recognized in UPN-1 together with the previously described short variant (Figure 6D).
Discussion
Inhibition of the nonsense-mediated mRNA decay pathway followed by microarray analysis has been used successfully in cancer cell lines with microsatellite instability, including colon and prostate cell lines.14,15 However, this approach has never been applied to other types of tumor cell lines. The accumulation of nonsense mutations that could lead to premature termination codons would be expected to occur at lower rate in cells with functional DNA repair machinery. For that reason, we designed a very conservative algorithm and analyzed multiple cell lines to minimize the number of false-positive genes. This algorithm was based on the hypothesis that a potential tumor-suppressor gene inactivated in a specific cell line by premature stop codons should show very low levels of mRNA in the presence of an active NMD pathway (mock-treated cells), whereas it would be expressed in other cells with a wild-type gene configuration. We also considered that it would be very unlikely that a tumor-suppressor gene is inactivated in more than one cell line by premature stop codons. In addition, the use of multiple cell lines allowed us to filter out nonproductive splicings present in several cell lines. The application of this method led to the selection of only 6 probe sets, 4 of them representing 3 annotated genes. Of note, 2 of these probe sets correspond to the positive control p53 supporting the value of the method. The 2 other genes that passed the criteria to select candidate genes were RB1 and LRAP in UPN-1 and REC-1 cell lines, respectively. The first was inactivated by a partial intragenic homozygous deletion, and the second showed an uncommon alternative splicing variant not previously described. The 2 probe sets that did not correspond to annotated genes interrogate the expression of potential cDNAs that code for hypothetical proteins that do not show any particular functional motif. Since a potential candidate gene that could be inactivated in a specific cell line by a premature stop codon might also be inactivated by homozygous deletion or hypermethylation in other cell lines, we relaxed our first filter to include this scenario. Thus, we considered that a potential inactivated gene in a specific cell line should be considered “present” in at least 3 of 4 remaining cell lines. When we applied this new filter, 2 new probe sets were selected. Only one of them corresponded to an annotated gene (CDC42EP3) (data not shown). This gene contains an upstream open reading frame (uORF), one of the putative features that might predispose a gene as a potential target of the NMD pathway12 and could explain its stabilization after emetine treatment. However, we sequenced the whole coding region of the gene and no mutations were identified.
Mantle-cell lymphoma is characterized by genetic alterations in cyclin D1 and other cell cycle regulatory genes involved in G1/S-phase transition, senescence, apoptosis, and DNA damage response. Among the genes that play a role in the G1/S-phase transition, genes involved in the p16INK4a/CDK4/pRB pathway stand out.2 The tumor-suppressor gene p16INK4a is inactivated mainly by homozygous deletion, particularly in highly proliferative and aggressive MCL variants. Moreover, amplification and overexpression of CDK4 and BMI-1 occur in a reduced fraction of MCL samples without p16INK4a inactivation.6,7 Curiously, although RB1 is a key element of this cell cycle regulatory pathway, its inactivation has been described only by Southern blot analysis in very few hematologic neoplasms.24–27 In a previous Southern blot study using a DNA probe located at 3′ end of the cDNA, we did not identify RB1 gross deletions in MCL.20 Other authors have reported monoallelic deletions of 13q14 including in some cases the RB1 locus.28–30 Moreover, protein expression analysis and functional experiments on the binding properties of pRB had suggested that RB1 was not inactivated in MCL.31 These studies included a relatively low number of cases, however, and were not designed to identify subtle changes such as point mutations in the whole RB1 sequence or small deletions that could not be detected by Southern blot. In the present study, we have identified for the first time that RB1 is inactivated in an MCL cell line (UPN-1). Although this cell line has been analyzed by conventional CGH and bacterial artificial chromosome (BAC) arrays, the fact that RB1 is inactivated by a small intragenic homozygous deletion explains why this abnormality had remained unreported. In fact, the analysis of UPN-1 by hybridization of the 100K set SNPs arrays has allowed us to identify that the 13q14-qter shows an uniparental trisomy, a phenomenon recently described in other chromosomal regions in several MCL cell lines.32,33
In this study, we were able to investigate the primary diagnostic biopsy of the MCL from which the UPN-1 cell line was derived. Thus, we have confirmed that RB1 inactivation occurred already in the primary MCL and it was not related to the establishment of the cell line, suggesting that this inactivation played an important role in the pathogenesis of the tumor. In addition, the search of RB1 mutation in a series of primary MCLs led us to the identification of an additional blastoid MCL with a similar intragenic homozygous deletion of the RB1 gene that was also associated with a total lack of protein expression. These results suggest that RB1, like other elements of the p16INK4a/CDK4/pRB pathway, is also inactivated in a small subset of aggressive MCL. This alteration occurs in cases with wild-type p16INK4a, in agreement with the idea that oncogenic alteration of more than one member of this pathway does not provide a biologic advantage to the tumor cells. Furthermore, the 2 primary MCLs with RB1 inactivation are blastoid variants and both cases harbored a concomitant inactivation of p53, supporting the hypothesis that there is pressure to inactivate the 2 main tumor-suppressor pathways represented by p16INK4a/CDK4/RB1 and p14ARF/MDM2/p53.7 Of interest, an intragenic deletion of RB1 was described in a blast crisis of a so called “intermediately differentiated lymphoma,” now considered to be the same entity as MCL.24 This previous finding would reinforce the idea that inactivation of RB1 could play a role in the progression of a subset of blastoid MCL.
LRAP is a newly identified aminopeptidase that is localized in the endoplasmatic reticulum (ER).34,35 This enzyme could play an important role in the regulation of several biologic processes including antigen processing by trimming peptides presented to MHC class I molecules in the ER. Moreover, it has been postulated that low or deregulated expression of aminopeptidases, including LRAP, could lead to an impaired antigen processing favoring the escape of tumors cells to the immune surveillance.36 The full-length cDNA contains 19 exons and codifies for a protein with aminopeptidase activity, but 2 truncated proteins produced by alternative splicing lack enzymatic activity.23,34 In the present work, we have identified LRAP as the third potential candidate to carry a premature stop codon in REC-1. The sequencing of the cDNA has led us to identify a new exon not described previously. The mRNA generated as a consequence of this alternative splicing codifies for a truncated protein that lacks the last 350 amino acids. We identified this alternative splicing isoform also in 2 primary MCLs that did not show expression of the normal variant. Further investigations are required to determine if this truncated form lacks enzymatic activity as is the case in the other shorter variants previously described.
In conclusion, our study demonstrates that the GINI strategy can identify genes harboring premature termination codons in tumor cell lines without microsatellite instability. Multiple cell lines and stringent filters, however, should be used in the analysis to precisely select the candidate genes. Using this approach, we have identified for the first time that RB1 is inactivated by intragenic homozygous deletions in a subset of blastoid MCL with concomitant p53 alterations but wild-type p16INK4a.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC ection 1734.
Acknowledgments
This work was supported by Comisión Interministerial de Ciencia y Tecnología (CICYT) SAF 05-5855, Lymphoma Research Foundation MCL Research Grantee, the European Union Contract LSHC-CT 2004-503351, Redes Temáticas de Investigación Cooperativa de Cáncer from the Instituto de Salud Carlos III, and Generalitat de Catalunya (2005SGRO870). A.R. is supported by the Interdisciplinary Center for Clinical Research (IZKF), University of Wuerzburg, Germany.
We thank Iracema Nayach and Montse Sánchez for their excellent technical assistance. Sequencing analysis was performed using the serveis Cientifico-Técnics of the University of Barcelona.
Authorship
Contribution: M.P. and L.P. performed the cell line experiments and primary MCL tissue analysis; P.J. performed the microarray analysis; S.B. analyzed the chromosomal alterations of the tumors by SNP arrays; V.R. and J.B. provided cell lines and tissue samples; E.C. and A.R. selected and reviewed the MCL samples; M.P., P.J., and E.C. wrote the paper; P.J., M.P., and E.C. designed the whole study. All authors reviewed critically the final version of the paper. E.C. and P.J. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pedro Jares, Genomics Unit, IDIBAPS, c/Villarroel 170, 08036 Barcelona, Spain; e-mail: pjares@clinic.ub.es.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal