Abstract
Gene expression profiling of early eosinophil development shows increased transcript levels of proinflammatory cytokines, chemokines, transcription factors, and a novel gene, EGO (eosinophil granule ontogeny). EGO is nested within an intron of the inositol triphosphate receptor type 1 (ITPR1) gene and is conserved at the nucleotide level; however, the largest open reading frame (ORF) is 86 amino acids. Sucrose density gradients show that EGO is not associated with ribosomes and therefore is a noncoding RNA (ncRNA). EGO transcript levels rapidly increase following interleukin-5 (IL-5) stimulation of CD34+ hematopoietic progenitors. EGO RNA also is highly expressed in human bone marrow and in mature eosinophils. RNA silencing of EGO results in decreased major basic protein (MBP) and eosinophil derived neurotoxin (EDN) mRNA expression in developing CD34+ hematopoietic progenitors in vitro and in a CD34+ cell line model. Therefore, EGO is a novel ncRNA gene expressed during eosinophil development and is necessary for normal MBP and EDN transcript expression.
Introduction
Eosinophils are tissue-dwelling hematopoietic cells that likely play a role in parasitic immunity and allergic disease, such as asthma.1 Activated eosinophils secrete toxic basic proteins such as major basic protein (MBP) and are postulated to cause bronchial hyperreactivity, damage of the bronchial mucosa, and remodeling of the airways.1 Mice lacking eosinophils fail to show hallmarks of asthma such as airway hyperresponsiveness, tissue remodeling, and mucous metaplasia.2,3 A more complete understanding of eosinophil development could lead to drugs targeting eosinophil progenitors prior to migration to the asthmatic lung.
Eosinophils develop in the bone marrow from hematopoietic stem cells and migrate mainly to the gut or to sites of inflammation. Eosinophils, neutrophils, and monocytes have a common progenitor in the myeloid pathway of development. The combinatorial interactions of several transcription factors, including GATA-1, PU.1, and the CCAAT enhancer binding proteins, c/EBPα and ϵ, are important to eosinophil development.4–7 High levels of PU.1 specify myeloid differentiation by antagonizing GATA-1 in the earliest stages of stem cell commitment.8 In particular, a high-affinity GATA-1 binding site within the GATA-1 promoter appears to be critical for eosinophil development; deletion of this binding site in mice specifically abolishes the entire eosinophil lineage.9 During later stages of eosinophil development, an intermediate level of GATA-1 in synergy with PU.1 directs the formation of the eosinophil lineage by activating dual binding sites in the MBP promoter.10–12 GATA-1 also activates the eotaxin receptor CC chemokine receptor-3 (CCR3) promoter and the interleukin-5 (IL-5) receptor α (IL-5Rα) gene.13 The CCAAT enhancer binding protein, c/EBPα, is important in early myeloid development, whereas c/EBPϵ plays a later role.14 Mouse knockouts of c/EBPϵ affect both neutrophil and eosinophil development at the myelocyte to metamyelocyte stage.14 Other genes involved in eosinophil development include the helix-loop-helix transcription factors, Id 1 and 2, and FOG (friend of GATA). FOG inhibits eosinophil development by interaction with GATA-1.15 Id 1 inhibits eosinophil development, whereas Id 2 enhances both neutrophil and eosinophil development.16 All of these transcription factors are used in general myeloid development; therefore, eosinophil development is regulated by fine tuning of combinatorial expression levels of transcription factors.
This complex interplay of transcription factors is influenced by the cytokines IL-3, granulocyte-macrophage colony-stimulating factor (GM-CSF), and particularly the Th2 cytokine, IL-5. CD34+ hematopoietic cells cultured in IL-5 are exclusively eosinophils after several weeks of culture.17 Furthermore, transgenic mice overexpressing IL-5 have massive eosinophilia.18–20 However, mouse IL-5 knockouts still have basal levels of eosinophils but do not develop eosinophilia when infected by helminths or challenged with aeroallergen.21,22 Inhalation of IL-5 in human asthmatics causes increased eosinophil numbers and airway hyperreactivity.23,24 Furthermore, a subset of mouse bone marrow cells expressing IL-5Rα are eosinophil progenitors.25 Therefore, IL-5 is the most important cytokine in eosinophil development, but alternative developmental pathways also exist.
In this study, we show that EGO is a novel, nested, noncoding RNA (ncRNA), gene expressed during eosinophil development and in mature eosinophils. ncRNA accounts for at least half of transcribed genes in mammals and has been increasingly implicated in playing a functional role in biology26–29 ncRNAs are often found nested in the introns or in 3′ untranslated regions of coding genes. We show that EGO is an ncRNA involved in regulating MBP and eosinophil derived neurotoxin (EDN) transcript expression.
Materials and methods
Cell isolation and culture
Bone marrow was obtained by informed consent in accordance with the Declaration of Helsinki from healthy volunteers after permission from the University of Utah International Review Board. CD34+ cells were enriched to 60%-80% purity from mononuclear cord blood or bone marrow mononuclear cells using the Midi Macs System (Miltenyi Biotech, Auburn, CA) and the CD34+ Human Selection kit (StemSep, Stem Cell Technologies Inc., Vancouver, BC). Eosinophils were isolated from peripheral blood granulocytes using negative selection with CD16 (Miltenyi). Cells were cultured in RPMI with penicillin/streptomycin (P/S), 10% fetal calf serum (FCS) (Hyclone, Logan, UT), and 5 ng/mL IL-5 (R&D Systems, Minneapolis, MN) or frozen in 10% dimethyl sulfoxide (DMSO) for later use. Cells were cultured 0.5 × 106 per well in a 48-well flat-bottom plate. CD34+ cells were stimulated with the following cytokines: 2 U/mL epoietin-α, 50 ng/mL stem cell factor (SCF), 20 ng/mL GM-CSF, 50 ng/mL M-CSF, 20 ng/mL GM-CSF, 50 ng/mL G-CSF, or 20 ng/mL IL-3 (R&D Systems). TF-1 cells were grown in RPMI, 10% FCS, P/S, and 5 ng/mL IL-5 or 2 U/mL epoietin-α (Amgen, Thousand Oaks, CA).
Microarray
CD34+ cells were isolated from the umbilical cord blood (UCB) of 4 placentas (nos. 17, 18, 4, and 23). Sample nos. 17 and 18 were split in half and cultured in IL-5 (5 ng/mL) or epoietin-α (2 U/mL). Sample no. 4 was cultured in IL-5, and sample no. 23 was cultured in epoietin-α. Samples were not pooled. Following 24 hours of culture, total RNA was isolated and amplified using the Riboamp RNA amplification kit (Arcturus, Sunnyvale, CA). Labeling and hybridization of amplified antisense RNA to Affymetrix (Santa Clara, CA) HG-U133A DNA chips was done in triplicate according to the manufacturer's instructions. Data were analyzed using GC-robust multiarray analysis (RMA) and ranked with nonparametric statistics.30
Quantitative RT-PCR
Total RNA was isolated using the Qiagen Rneasy kits. First strand cDNA was synthesized using the Superscript III Kit (Invitrogen, Carlsbad, CA). Polymerase chain reaction (PCR) conditions were 2 mM dNTPs, 0.5 μM primers, 1/150 000 dilution of Sybr Green (Invitrogen) I, 0.2 UAmplitaq (Applied Biosystems, Foster City, CA), 0.1 μg Taqstart antibody (Clontech, Mountain View, CA), 4 mM PCR buffer (Idaho Technology, Salt Lake City, UT), and 4 μL 10× diluted cDNA in a 20-μL reaction. PCR was done in triplicate on a Roche LightCycler, and averages and standard errors were plotted. Second derivative maxima of triplicates are within a half cycle of each other or were repeated. Parameters were 94°C for 0 seconds, 60°C for 20 seconds, 40 cycles. All reactions were normalized to an α-tubulin control31 with the exception of mature eosinophil cDNA, which was normalized to a GAPDH control. Efficiency of the α-tubulin primers is 2.0, and efficiency of all other primers is assumed to be 2.0. Second derivative maxima were used from LightCycler Data Analysis v3.5 to quantitate RNA levels. Primers for EGO-A are BCOF1 (cttctcctccaggccatacc) and 222314F(ccattgtgtagccccg). Primers for EGO-B are EGOsplR2 (ccatcgtgcctgatagaa) and BCOF1. Primers for granule proteins are EDNF2 (caccatggttccaaaactgttca), EDNR2(gtttttccatcgccgtt), MBPF(gggattgcggtacttataca), MBPR (atgggctcagctagtt), GAPDHF(tctctgctcctcctgtt), and GAPDHR(caagcttcccgttctca). Primers to measure silencing of EGO are 222314F8(aggaattatgattgtggggt) and BCO39547R3 (ggtatggcctggaggagaag). Other primers include GATA-1ex3F (ggactctcctccccag) and GATA-1ex3R(ctgaattgagggggct) and GAPDHF (tctctgctcctcctgtt) and GAPDHR (caagcttcccgttctca). All amplicons were a single band on agarose gels verified by DNA sequencing.
Northern blot
Poly A+ RNA (0.5-1 μg) (Qiagen, Oligotex kit, Valencia, CA) was electrophoresed on a 1% agarose MOPS (3-(N-morpholino)propanesulfonic acid) formaldehyde gel alongside Millennium Markers (Applied Biosystems). Markers were stained in 1× Tris Borate EDTA (TBE) and Sybr Gold (Invitrogen). Gels were transferred to nitrocellulose membranes in 6× SSC (or to GeneScreen for EGO-B) and hybridized overnight at 42° in 50% formamide to radiolabeled random primed EGO-A or B PCR products washed 3× in 48° 2XSSC and 2× in 55°C 0.2XSSC, then exposed to film.
Sucrose gradients
Approximately 7 × 106 CD34+ cells were cultured for 24 hours in 5 ng/mL IL-5. Cells were disrupted by Dounce homogenization and layered on a 10%-50% sucrose gradient. Gradients were centrifuged for 4 hours at 182 348 g (38 000 rpm) in a swinging bucket SW40Ti rotor. 0.5 mL fractions were collected from the bottom, and total RNA was isolated by phenol/chloroform purification and ethanol precipitation. OD260 readings were taken on each fraction to determine the location of the ribosomal peaks. RNA derived from fractions was reverse transcribed to cDNA and real-time PCR was performed as described. Normalization to tubulin was not done due to the differential quantities of tubulin in fractions containing ribosomes.
Multiple tissue expression
Multiple tissue human cDNA panels I and II were purchased from Clontech. Bone marrow cDNA was isolated from human bone marrow obtained by informed consent from healthy volunteers. Bone marrow and CD34+ cell cDNA were normalized to average α-tubulin. Clontech panel values and RNA levels were calculated as described in Figure 6.
Plasmid construction
pSilencer 2.0-U6 (Applied Bioscience) was digested with HindIII and blunt end cloned to Mlu I/AseI digested pEGFP-C1 (Clontech). The HindIII site was not lost. The multiple cloning sites in the enhanced green fluorescent protein (EGFP) gene were removed by a BglII/Bam HI digest and religation. This plasmid is the pSil Neg. Subsequently, the sequence 5′ gatcccaatagaaccgcaagaaaacaactcgagttgttttcttgcggttctatttttttggaaa3′ was cloned into the BamHI/HindIII sites to create the shRNA, plasmid pSil 20-2, targeting EGO-A and B.
siRNA: CD34+ cells
Frozen cells were reconstituted dropwise with media and allowed to recover for one hour at 37°C, 5% CO2 in 10 parts media prior to centrifugation. Endotoxin-free short hairpin plasmid DNA (1.0 μg) (Qiagen Endofree maxi kit) was electroporated into 1-2 × 106 CD34+ cells using the CD34+ Nucleofector kit and a Nucleofector device (Amaxa Biosystems, Gaithersburg, MD) using program U8. Cells were allowed to recover for 15 minutes in serum and antibiotic-free media at 37°C, 5% CO2. Cells were put in a 48-well flat-bottom plate and RPMI, 10% FCS, and P/S were immediately added. After 18 hours 5 ng/mL IL-5 was added (time = 0′), and EGFP+ cells were purified on a FACS Vantage cell sorter to at least 90% purity. Approximately 100 000 cells were cultured in a single well of a 96-well round-bottom plate. At the 24-hour time point (to measure EGO silencing) or the 5-day time point (to measure MBP silencing) total RNA was isolated.
TF-1 cells
Cells were grown in RPMI 1640, 10% FCS, and IL-5 (5 ng/mL). Approximately 5 × 106 freshly cultured cells were electroporated on program T3 using the V Nucleofector kit with 1-5 μg shDNA (Amaxa Biosystems). Cells were sorted for more than 90% pure GFP positive within 24 hours as described, and RNA was isolated.
Accession numbers
Results
Gene profiling of early eosinophil development
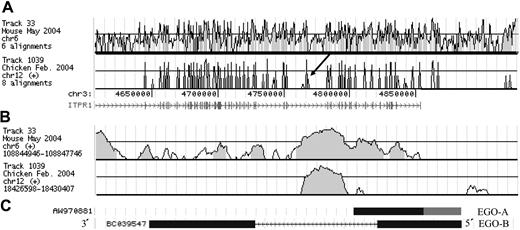
Gene-expression profiling identified transcripts expressed during early eosinophil development. Affymetrix HG-U133 A human microarray chips were hybridized with labeled antisense RNA amplified from CD34+ human hematopoietic stem cells derived from unpooled donors and cultured for 24 hours in IL-5, an eosinophil specific cytokine, or epoietin-α as a control. Arrays were normalized by GC-RMA (www.bioconductor.org), and increased transcript levels were analyzed using rank based statistics.30 The top 38 ranked genes have transcript levels at least 4-fold higher in IL-5–treated cells compared to control (Table 1). Several interleukins, interleukin receptors, and chemokine receptor transcripts are increased in response to IL-5: CXCL13, IL6, IL1 family members, as well as IL12B and IL21R have increased mRNA levels. PTGS2 (COX-2), a gene involved in inflammatory prostaglandin synthesis, also is differentially up-regulated. In addition, 2 transcription factors, WT1 and HEY1, increase in transcriptional expression following IL-5 treatment. One novel transcript, (Affymetrix 222314_x_at) is nested within an intron on the opposite strand of the inositol triphosphate receptor type1 (ITPR1) gene (Figure 1A). This transcript is of particular interest because of its inducible expression to high transcript levels, conservation, and lack of a large open reading frame (ORF). This gene was named EGO.
Microarray data of top 38 IL-5–induced transcripts
| Probe set* . | Gene . | Description . | Fold change† . |
|---|---|---|---|
| 205242_at | CCL13 | chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) | 85 |
| 205207_at | IL6 | interleukin 6 (interferon, beta 2) | 25 |
| 220322_at | IL1F9 | interleukin 1 family, member 9 | 17 |
| 201860_s_at | PLAT | plasminogen activator, tissue | 13 |
| 210072_at | CCL19 | chemokine (C-C motif) ligand 19 | 9 |
| 206336_at | CXCL6 | chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | 10 |
| 207533_at | CCL1 | chemokine (C-C motif) ligand 1 | 8 |
| 204470_at | CXCL1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating) | 9 |
| 207901_at | IL12B | interleukin 12B (natural killer cell stimulatory factor 2) | 9 |
| 204748_at | PTGS2 | prostaglandin-endoperoxide synthase 2 (COX-2) | 9 |
| 209278_s_at | TFPI2 | tissue factor pathway inhibitor 2 | 9 |
| 221658_s_at | IL21R | interleukin 21 receptor | 7 |
| 219424_at | EBI3 | Epstein-Barr virus-induced gene 3 (IL12R family) | 8 |
| 209774_x_at | CXCL2 | chemokine (C-X-C motif) ligand 2 (IL8RB) | 6 |
| 206432_at | HAS2 | hyaluronan synthase 2 | 8 |
| 44783_s_at | HEY1 | hairy/enhancer-of-split related with YRPW motif 1 (transcription factor) | 6 |
| 211538_s_at | HSPA2 | heat shock 70-kDa protein 2 | 5 |
| 210511_s_at | INHBA | inhibin, beta A (activin A, activin AB alpha polypeptide) | 9 |
| 210001_s_at | SOCS1 | suppressor of cytokine signaling 1 | 5 |
| 207608_x_at | CYP1A2 | cytochrome P450, family 1, subfamily A, polypeptide 2 | 5 |
| 210029_at | INDO | indoleamine-pyrrole 2,3 dioxygenase | 6 |
| 219255_x_at | IL17RB | interleukin 17 receptor B | 6 |
| 204014_at | DUSP4 | dual specificity phosphatase 4 | 6 |
| 219258_at | FLJ20516 | timeless-interacting protein | 5 |
| 222314_x_at | ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 (intron of ITPR1, EGO) | 4 |
| 208581_x_at | MT1X | metallothionein 1X | 4 |
| 207526_s_at | IL1RL1 | interleukin 1 receptor-like 1 | 6 |
| 205599_at | TRAF1 | TNF receptor-associated factor 1 | 5 |
| 204580_at | MMP12 | matrix metallopeptidase 12 (macrophage elastase) | 6 |
| 203562_at | FEZ1 | fasciculation and elongation protein zeta 1 (zygin I) | 6 |
| 215223_s_at | SOD2 | superoxide dismutase 2, mitochondrial | 4 |
| 205114_s_at | CCL3 | chemokine (C-C motif) ligand 3 | 5 |
| 209369_at | ANXA3 | annexin A3 | 4 |
| 203881_s_at | DMD | dystrophin (muscular dystrophy, Duchenne and Becker types) | 5 |
| 206067_s_at | WT1 | Wilms tumor 1 (zinc finger, transcription factor) | 4 |
| 209277_at | TFPI2 | Tissue factor pathway inhibitor 2 | 5 |
| 206881_s_at | LILRA3 | leukocyte immunoglobulin-like receptor, subfamily A, member 3 | 4 |
| 202071_at | SDC4 | syndecan 4 (amphiglycan, ryudocan) | 4 |
| Probe set* . | Gene . | Description . | Fold change† . |
|---|---|---|---|
| 205242_at | CCL13 | chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) | 85 |
| 205207_at | IL6 | interleukin 6 (interferon, beta 2) | 25 |
| 220322_at | IL1F9 | interleukin 1 family, member 9 | 17 |
| 201860_s_at | PLAT | plasminogen activator, tissue | 13 |
| 210072_at | CCL19 | chemokine (C-C motif) ligand 19 | 9 |
| 206336_at | CXCL6 | chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | 10 |
| 207533_at | CCL1 | chemokine (C-C motif) ligand 1 | 8 |
| 204470_at | CXCL1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating) | 9 |
| 207901_at | IL12B | interleukin 12B (natural killer cell stimulatory factor 2) | 9 |
| 204748_at | PTGS2 | prostaglandin-endoperoxide synthase 2 (COX-2) | 9 |
| 209278_s_at | TFPI2 | tissue factor pathway inhibitor 2 | 9 |
| 221658_s_at | IL21R | interleukin 21 receptor | 7 |
| 219424_at | EBI3 | Epstein-Barr virus-induced gene 3 (IL12R family) | 8 |
| 209774_x_at | CXCL2 | chemokine (C-X-C motif) ligand 2 (IL8RB) | 6 |
| 206432_at | HAS2 | hyaluronan synthase 2 | 8 |
| 44783_s_at | HEY1 | hairy/enhancer-of-split related with YRPW motif 1 (transcription factor) | 6 |
| 211538_s_at | HSPA2 | heat shock 70-kDa protein 2 | 5 |
| 210511_s_at | INHBA | inhibin, beta A (activin A, activin AB alpha polypeptide) | 9 |
| 210001_s_at | SOCS1 | suppressor of cytokine signaling 1 | 5 |
| 207608_x_at | CYP1A2 | cytochrome P450, family 1, subfamily A, polypeptide 2 | 5 |
| 210029_at | INDO | indoleamine-pyrrole 2,3 dioxygenase | 6 |
| 219255_x_at | IL17RB | interleukin 17 receptor B | 6 |
| 204014_at | DUSP4 | dual specificity phosphatase 4 | 6 |
| 219258_at | FLJ20516 | timeless-interacting protein | 5 |
| 222314_x_at | ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 (intron of ITPR1, EGO) | 4 |
| 208581_x_at | MT1X | metallothionein 1X | 4 |
| 207526_s_at | IL1RL1 | interleukin 1 receptor-like 1 | 6 |
| 205599_at | TRAF1 | TNF receptor-associated factor 1 | 5 |
| 204580_at | MMP12 | matrix metallopeptidase 12 (macrophage elastase) | 6 |
| 203562_at | FEZ1 | fasciculation and elongation protein zeta 1 (zygin I) | 6 |
| 215223_s_at | SOD2 | superoxide dismutase 2, mitochondrial | 4 |
| 205114_s_at | CCL3 | chemokine (C-C motif) ligand 3 | 5 |
| 209369_at | ANXA3 | annexin A3 | 4 |
| 203881_s_at | DMD | dystrophin (muscular dystrophy, Duchenne and Becker types) | 5 |
| 206067_s_at | WT1 | Wilms tumor 1 (zinc finger, transcription factor) | 4 |
| 209277_at | TFPI2 | Tissue factor pathway inhibitor 2 | 5 |
| 206881_s_at | LILRA3 | leukocyte immunoglobulin-like receptor, subfamily A, member 3 | 4 |
| 202071_at | SDC4 | syndecan 4 (amphiglycan, ryudocan) | 4 |
Affymetrix probe set IDs.
Fold change relative to epoietin-induced transcripts.
Conservation and gene structure of EGO. (A) Vista tracks of mouse and chicken on the UCSC browser showing conservation of the ITPR-1 region. The arrow indicates the EGO region. ITPR-1 exons are shown as vertical lines. The y axis ranges from 50%-100% percent identity. (B) Vista tracks of mouse and chicken on the UCSC browser at the same magnification as panel C. (C) The structure of EGO transcripts is shown. The representative cDNA clones (BCO35947 and AW970881) from BLAT (http://genome.ucsc.edu) are shown in black. The putative 5′ end of EGO-A is shown in gray. Note that EGO transcripts are shown 3′ to 5′ because they are on the minus strand of genomic DNA.
Conservation and gene structure of EGO. (A) Vista tracks of mouse and chicken on the UCSC browser showing conservation of the ITPR-1 region. The arrow indicates the EGO region. ITPR-1 exons are shown as vertical lines. The y axis ranges from 50%-100% percent identity. (B) Vista tracks of mouse and chicken on the UCSC browser at the same magnification as panel C. (C) The structure of EGO transcripts is shown. The representative cDNA clones (BCO35947 and AW970881) from BLAT (http://genome.ucsc.edu) are shown in black. The putative 5′ end of EGO-A is shown in gray. Note that EGO transcripts are shown 3′ to 5′ because they are on the minus strand of genomic DNA.
Conservation of EGO
Vista tracks on the UCSC (University of Santa Cruz) browser (http://genome.lbl.gov/vista/index.shtml) show that conservation of the region within the large intron of ITPR1, which includes the EGO gene, is high among human, mouse, and chicken (Figure 1A,B). However, the highest region of homology, close to 90% identity, is in the intron of EGO-B, suggesting an additional undiscovered gene or a regulatory region (Figures S2 and S3, available on the Blood website; see the Supplemental Figures link at the top of the online article). The 3′ portion of EGO-A has approximately 75% identity between mouse and human (Figure 1B). However, no ORFs over 86 amino acids are present in EGO-A or B, and amino acids are not conserved between mouse and human. Furthermore, ncRNA genes are often found within introns and untranslated regions of coding genes.27,28 ncRNA genes are often inferred by the lack of large ORFs or of conserved amino acid sequence. Therefore, the hypothesis that EGO is an ncRNA gene was investigated further.
EGO gene structure
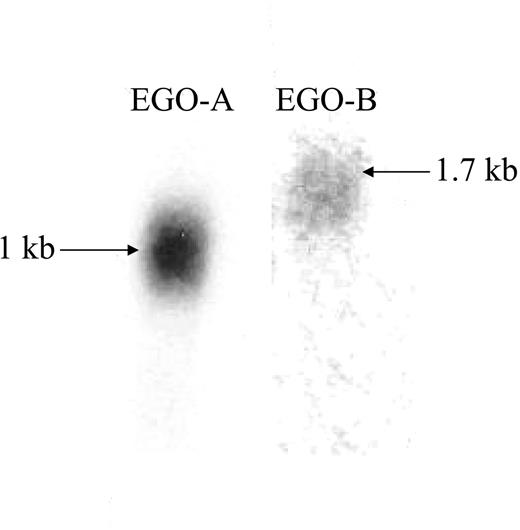
Two splice variants of the EGO transcript were found using a BLAT Search (May 2004 Assembly http://genome.ucsc.edu) (Figure 1C). The splice variant found by array analysis is a 535-base pair cDNA (GenBank accession no. AW970881), which was named EGO-A (Figure 1C). In addition, a 2 exon splice variant, EGO-B (GenBank accession no. BC039547) is 1460 bases in length (Figure 1C). However, Northern blot analysis using RNA isolated from TF-1 cells, a CD34+ erythroleukemic cell line, and a probe to a unique region of EGO-A revealed a 1-kb polyadenylated transcript, suggesting the reported cDNA is truncated (Figure 2 and Figure S1). RT-PCR data are consistent with EGO-A extending over 300 bases toward the 5′ end of the transcript (additional bases in the poly A tail may add 100-200 bases) (Figure S2). Northern blot using a probe complementary to exon 2 of EGO-B (Figure 2 and Figure S1) shows an approximately 1700-base polyadenylated transcript, which is consistent with GenBank data for BC039547 cDNA. Sequence data show that the 5′ splice junction for EGO-B is 46 bases upstream from the reported BC039547 cDNA splice site (4767665-476655 TTCTATCAG …. GCACGATGGT). Based on the single-stranded probes in the Affymetrix arrays, both EGO-A and B are sense on the- strand of genomic DNA, the opposite strand from ITPR1 transcription. The multiple overlapping transcripts derived from both strands in this region are characteristic of an RNA forest.27 Therefore, at least 2 EGO splice variants are transcribed within an intron on the opposite strand from ITPR1.
Northern blots of EGO transcripts. Northern blots of TF-1 cell poly A+ RNA hybridized with radiolabeled probe specific for panel A, EGO-A (1 kb), and panel B. EGO-B (1.7 kb). Sequenced splice junction of EGO-B is 4767665 TTCTATCAG…. GCACGATGGT 4766655 and 5′ end of EGO as determined by PCR is 4768306 TTCAAACAG (Figure S2).
Northern blots of EGO transcripts. Northern blots of TF-1 cell poly A+ RNA hybridized with radiolabeled probe specific for panel A, EGO-A (1 kb), and panel B. EGO-B (1.7 kb). Sequenced splice junction of EGO-B is 4767665 TTCTATCAG…. GCACGATGGT 4766655 and 5′ end of EGO as determined by PCR is 4768306 TTCAAACAG (Figure S2).
Sucrose density gradient analysis of ncRNA
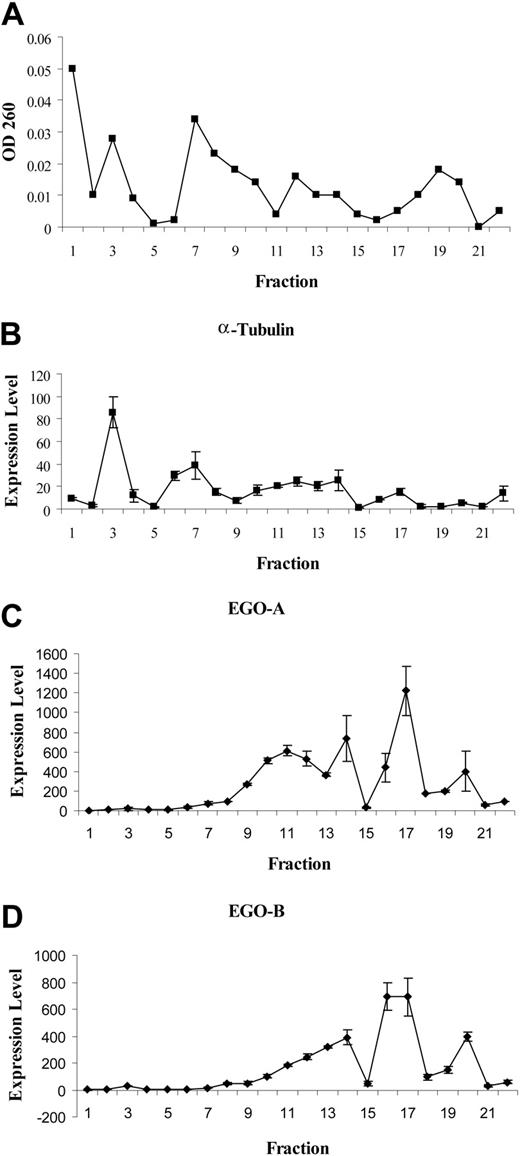
The hypothesis that EGO is an ncRNA was tested by measuring its association with ribosomes by sucrose density gradient sedimentation. Sucrose density gradient sedimentation was performed on lysates from CD34+ UCB cells stimulated for 24 hours with IL-5 to determine if EGO RNA is in the ribosomal fraction. Absorbance at 260 nm per fraction gives a characteristic plot in which ribosomes are concentrated at the bottom of the sucrose gradient. In this gradient, UV absorbance in fractions 1 and 3 are indicative of polysomes, and fraction 7 corresponds to single ribosomes (Figure 3A). Real-time RT-PCR was performed on each fraction for α-tubulin mRNA and for EGO-A and B transcript expression. As expected, α-tubulin transcripts are mainly associated with polysomes in fraction 3 (Figure 3B). In contrast, both EGO-A and B transcripts levels are more abundant in the lighter fractions of the gradient, especially in fraction 17 (Figure 3C,D). These data show that EGO transcripts are not associated with ribosomes in UCB CD34+ cells in vitro after stimulation for 24 hours with IL-5.
Real-time Q-RT-PCR of CD34+ RNA from sucrose density gradient fractions. (A) OD260. (B) α-tubulin. (C) EGO-A. (D) EGO-B. Expression level as fold change is shown on the y axis. Standard errors of PCR triplicates are shown.
Real-time Q-RT-PCR of CD34+ RNA from sucrose density gradient fractions. (A) OD260. (B) α-tubulin. (C) EGO-A. (D) EGO-B. Expression level as fold change is shown on the y axis. Standard errors of PCR triplicates are shown.
EGO gene expression
The inducible expression of EGO-A and B RNA was investigated by real-time Q-RT-PCR (quantitative reverse transcriptase polymerase chain reaction) of CD34+ cells derived from UCB or bone marrow and stimulated with cytokines over a time course. EGO-A and B transcripts derived from CD34+ UCB cells are highly expressed, more than 20-fold above the initial time point, at 6 hours after IL-5 addition, and expression is reduced to baseline levels by 72 hours after stimulation (Figure 4A,B). The specificity of IL-5–induced EGO RNA expression was investigated by real-time Q-RT-PCR of UCB CD34+ cells grown on the hematopoietic cytokines: epoietin-α, SCF, M-CSF/GM-CSF, IL-3, or GM-CSF/G-CSF. All cytokines except SCF cause a slight increase in EGO-A and B expression, suggesting that EGO transcription also may be involved in other lineages (Figure 4A,B). A positive control, MBP mRNA, is increasingly expressed from 72 hours to 2 weeks of IL-5 stimulation of CD34+ UCB cells but is not expressed in epoietin-α-stimulated cells (Figure 4C). Expression of EGO-A and B in IL-5–stimulated CD34+ bone marrow cells follows a temporal pattern similar to UCB CD34+ cells (Figure 4D). These results show that EGO-A and B RNA are expressed coordinately very early and briefly in eosinophil development in vitro in both UCB and bone marrow–derived CD34+ cells stimulated with IL-5.
Real-time Q-RT-PCR of transcript expression following cytokine stimulation of CD34+ cells. (A) EGO-A, UCB CD34+ cells. (B) EGO-B, UCB CD34+ cells. (C) MBP transcripts following IL-5 or epoietin-α stimulation of UCB CD34+ cells. (D) EGO-A and B transcripts following IL-5 stimulation of bone marrow CD34+ cells. EGO-A, solid line. EGO-B, dashed line. Standard errors of PCR triplicates are shown.
Real-time Q-RT-PCR of transcript expression following cytokine stimulation of CD34+ cells. (A) EGO-A, UCB CD34+ cells. (B) EGO-B, UCB CD34+ cells. (C) MBP transcripts following IL-5 or epoietin-α stimulation of UCB CD34+ cells. (D) EGO-A and B transcripts following IL-5 stimulation of bone marrow CD34+ cells. EGO-A, solid line. EGO-B, dashed line. Standard errors of PCR triplicates are shown.
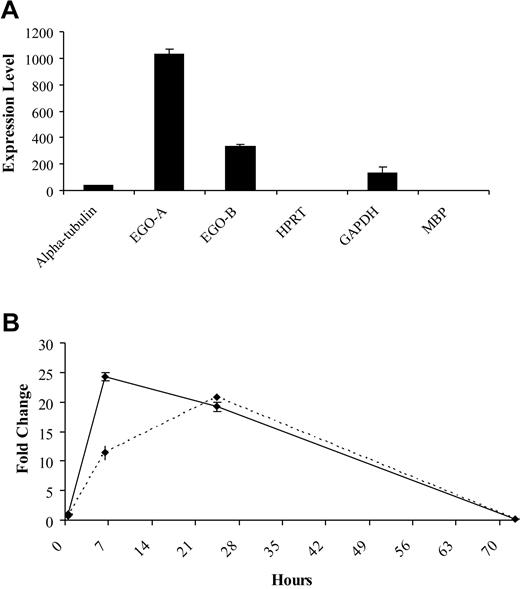
Expression levels of EGO in mature peripheral blood eosinophils were investigated. Although overall levels of RNA in mature eosinophils are low, EGO-A and B are expressed approximately 1000-fold and 300-fold higher, respectively, than the transcripts for the housekeeping genes, HPRT (hypoxanthine guanine phosphoribosyl transferase), α-tubulin, and the granule protein, MBP. GAPDH (glycerol aldehyde phosphate dehydrogenase) mRNA is expressed at slightly higher levels, about 100-fold higher than HPRT (Figure 5A). Eosinophils also were stimulated with IL-5 and cultured for various time points. EGO-A and B RNA are increased nearly 25-fold relative to GAPDH between 6 and 24 hours of IL-5 stimulation (Figure 5B). Therefore, EGO is present and inducible in the mature eosinophil.
Expression of EGO in peripheral blood eosinophils. (A) Real-time Q-RT-PCR of unstimulated peripheral blood eosinophils. (B). Real-time Q-RT-PCR of IL-5–stimulated peripheral blood eosinophils for EGO-A, solid line and EGO-B, dashed line. Standard errors of PCR triplicates are shown.
Expression of EGO in peripheral blood eosinophils. (A) Real-time Q-RT-PCR of unstimulated peripheral blood eosinophils. (B). Real-time Q-RT-PCR of IL-5–stimulated peripheral blood eosinophils for EGO-A, solid line and EGO-B, dashed line. Standard errors of PCR triplicates are shown.
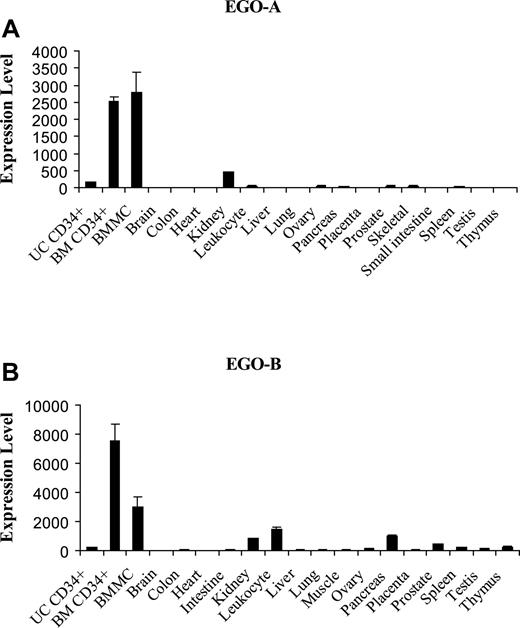
The relative expression of EGO in a panel of human tissue types was investigated by real-time Q-RT-PCR. Tissue expression of both EGO-A and B RNA is 2000-7000 higher in bone marrow mononuclear cells and bone marrow CD34+ cells than in brain, the tissue with the lowest expression (Figure 6A,B). Expression of both EGO transcripts also is high in kidney. EGO-B RNA is highly expressed in leukocytes and pancreas relative to EGO-A. The relatively high expression of EGO in bone marrow is consistent with a role for EGO in developing hematopoietic stem cells in vivo.
Real-time Q-RT-PCR of EGO transcripts derived from human tissues. (A) Fold expression levels of EGO-A in various tissues relative to brain. (B) Fold expression levels of EGO-B relative to brain. CD34+ cells and bone marrow mononuclear cell RNA was normalized to the average level of α-tubulin in the tissue panel. UC CD34+, umbilical cord CD34+; BM CD34+, bone marrow CD34+; BMMC, bone marrow mononuclear cells. Standard errors of PCR triplicates are shown.
Real-time Q-RT-PCR of EGO transcripts derived from human tissues. (A) Fold expression levels of EGO-A in various tissues relative to brain. (B) Fold expression levels of EGO-B relative to brain. CD34+ cells and bone marrow mononuclear cell RNA was normalized to the average level of α-tubulin in the tissue panel. UC CD34+, umbilical cord CD34+; BM CD34+, bone marrow CD34+; BMMC, bone marrow mononuclear cells. Standard errors of PCR triplicates are shown.
The role of EGO in eosinophil granule protein mRNA expression
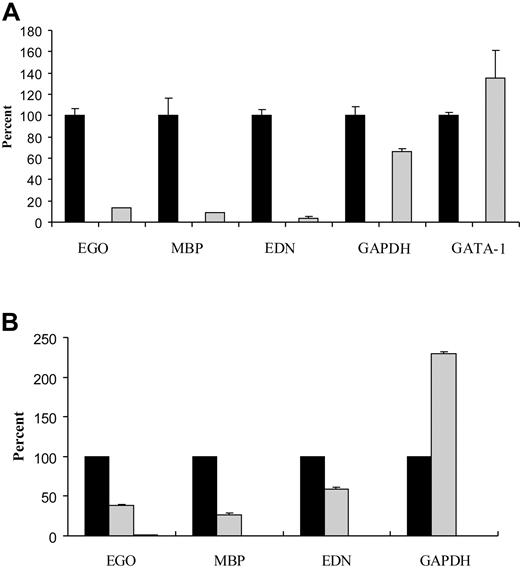
RNA silencing experiments were initiated in order to test the hypothesis that EGO expression influences eosinophil differentiation. TF-1 is a CD34+ erythroleukemic cell line that expresses eosinophil granule protein mRNA and can be growth stimulated with various cytokines, including IL-5. A short hairpin RNA (shRNA) expression plasmid containing either scrambled control sequences (pSil Neg) or sequences simultaneously targeting both variants of EGO (pSil 20-2) for silencing were transfected into TF-1 cells. Plasmids also contained an EGFP reporter gene. TF-1 cells were sorted to at least 90% purity for EGFP+ cells 24 hours after transfection. Real-time Q-RT-PCR of RNA isolated from EGFP+ cells was used to quantitate silenced levels of EGO RNA using primers spanning the putative RNA cut site and-tubulin primers as a control. Levels of EGO are diminished to 13% (87% knockdown) in the cells transfected with the shRNA plasmid directed at EGO transcripts compared to the negative control (Figure 7A). Levels of MBP and EDN mRNA, which are constitutively expressed in TF-1 cells, also were measured by Q-RT-PCR (eosinophil peroxidase mRNA is not expressed). In cells in which EGO transcripts have been silenced, levels of MBP mRNA are 9% of control, and EDN mRNA levels are 4% of control levels; however, GAPDH mRNA levels and the GATA-1 transcription factor mRNA levels remain relatively unchanged (Figure 7A). Repeated experiments in TF-1 cells show similar results with variations in transcript levels (Table 2). This suggests that diminished EGO RNA levels decrease transcript levels of the eosinophil granule proteins MBP and EDN.
EGO RNA silencing decreases EDN and MBP transcript levels. (A) TF-1 cells transfected with negative control plasmid, pSil Neg (black), or shRNA targeting both EGO transcripts, pSil 20-2 (gray). Results shown are from experiment 5 in Table 2. (B) UCB CD34+ cells transfected with negative control plasmid, pSil Neg (black), or shRNA targeting both EGO transcripts, pSil 20-2 (gray). Standard errors of PCR triplicates are shown.
EGO RNA silencing decreases EDN and MBP transcript levels. (A) TF-1 cells transfected with negative control plasmid, pSil Neg (black), or shRNA targeting both EGO transcripts, pSil 20-2 (gray). Results shown are from experiment 5 in Table 2. (B) UCB CD34+ cells transfected with negative control plasmid, pSil Neg (black), or shRNA targeting both EGO transcripts, pSil 20-2 (gray). Standard errors of PCR triplicates are shown.
EGO silencing in TF-1 cells
| Plasmid . | EGO . | MBP . | EDN . | GAPDH . | GATA-1 . |
|---|---|---|---|---|---|
| Experiment 1 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | nd |
| pSil 20-2 | 13 | 30 | 7 | 113 | nd |
| Experiment 2 | |||||
| pSil Neg | 100 | 100 | nd | 100 | nd |
| pSil 20-2 | 40 | 64 | nd | 73 | nd |
| Experiment 3 | |||||
| pSil Neg | 100 | 100 | 100 | nd | nd |
| pSil 20-2 | 35 | 21 | 61 | nd | nd |
| Experiment 4 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | 100 |
| pSil 20-2 | 32 | 85 | 75 | 76 | 97 |
| Experiment 5 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | 100 |
| pSil 20-2 | 13 | 9 | 4 | 66 | 135 |
| Experiment 6 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | 100 |
| pSil 20-2 | 46 | 74 | 16 | 85 | 99 |
| Plasmid . | EGO . | MBP . | EDN . | GAPDH . | GATA-1 . |
|---|---|---|---|---|---|
| Experiment 1 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | nd |
| pSil 20-2 | 13 | 30 | 7 | 113 | nd |
| Experiment 2 | |||||
| pSil Neg | 100 | 100 | nd | 100 | nd |
| pSil 20-2 | 40 | 64 | nd | 73 | nd |
| Experiment 3 | |||||
| pSil Neg | 100 | 100 | 100 | nd | nd |
| pSil 20-2 | 35 | 21 | 61 | nd | nd |
| Experiment 4 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | 100 |
| pSil 20-2 | 32 | 85 | 75 | 76 | 97 |
| Experiment 5 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | 100 |
| pSil 20-2 | 13 | 9 | 4 | 66 | 135 |
| Experiment 6 | |||||
| pSil Neg | 100 | 100 | 100 | 100 | 100 |
| pSil 20-2 | 46 | 74 | 16 | 85 | 99 |
Percentage of transcripts in silenced cells compared to controls.
nd indicates not determined.
CD34+ cells derived from UCB are a better model of eosinophil differentiation than TF-1 cells, because IL-5–stimulated CD34+ UCB cells develop into eosinophils, whereas TF-1 cells simply proliferate in response to a variety of cytokines. CD34+ UCB cells express MBP mRNA at 72 hours after IL-5 stimulation and have immature granules at 1 week; however, electroporated cells are fragile and have incomplete development, dying after 5 days. CD34+ cells were transfected with shRNA plasmids, sorted for more than 90% pure EGFP+ cells less than 24 hours after transfection, and grown for 5 days on IL-5. Silencing reduced EGO transcripts to 38% of control (62% knockdown) at 24 hours after IL-5 stimulation (Figure 6B). RNA was isolated and Q-RT-PCR performed to quantitate granule protein mRNA levels. MBP mRNA levels are reduced to 27% in EGO-silenced cells as compared to controls, and levels of EDN mRNA are 59% of control levels (Figure 6B). In contrast, GAPDH levels are increased in response to EGO silencing (Figure 6B). GATA-1 is expressed at very low levels in CD34+ cells and cannot be reliably measured. These data suggest that EGO RNA is necessary for normal MBP and EDN transcript levels during eosinophilopoiesis.
Discussion
Genes expressed in early eosinophil development were investigated by transcriptional profiling of CD34+ cells stimulated with IL-5. Increases in inflammatory cytokines, cytokine receptors, and chemokine ligand transcript expression were shown. Included in the differentially expressed proinflammatory molecules are: IL6, IL1F9, CCL19, CCL1, CCL6, PTGS2 (COX-2), IL12B, IL21R, EBI3, IL1RL1, and IL17RB. IL12B promoter polymorphism has been linked with asthma severity and atopic dermatitis,32,33 and IL21R is associated with rheumatoid arthritis.34 Of particular interest is the increased expression of the HEY1 (hairy enhancer of split) transcription factor. HEY1 is reported to physically interact with GATA-1 to decrease its activity.35 An intermediate level of GATA-1 is instrumental in eosinophil development; therefore, HEY1 may be the effector molecule that maintains these critical levels. Furthermore, our results show that EGO, a transcript nested within an intron on the opposite strand of ITPR1, also increases in response to IL-5. However, ITPR1 mRNA levels do not increase following IL-5 stimulation (Table 1). RNA silencing, sucrose density gradient, and gene expression experiments show that EGO is an ncRNA necessary for normal eosinophil granule protein transcript levels of MBP and EDN.
EGO is nested within a conserved intron of the ITPR1 gene. At least half of transcripts in mammals are noncoding, and many ncRNAs are found in introns or in the 3′ untranslated regions of coding genes.27,28 Approximately 158 coding nested genes have been identified in the human genome.36 Conservation of the ITPR1 intron containing EGO is higher than most ITPR1 exons, with up to 90% identity among mouse, human, and chicken (http://genome.lbl.gov/vista). EGO-A and the 5′ exon of EGO-B have up to 75% identity between human and mouse, reflecting evolutionary pressure and a possible functional role for these transcripts. However, the highest conservation is within the intron of EGO-B, suggesting an additional transcript or a regulatory region (Figure 1). Q-RT-PCR shows that the intron of EGO-B is highly expressed, but not inducible with IL-5 (Figure S2), further suggesting an undiscovered transcript in this region. Also, in support of an additional transcript, Northern blot data shows a 2-kb transcript when probed with the intron of EGO-B (Figure S3). These data show that EGO resides in a highly transcribed region of the genome and has likely been retained by natural selection for a functional role.
The lack of a large ORF and poor amino acid conservation of small open reading frames of fewer than 100 amino acids suggest that EGO is an ncRNA. Furthermore, EGO transcripts are not associated with ribosomes in vitro, although translated mRNA, such as α-tubulin, is present in the polyribosome fraction of the sucrose gradient, as expected. EGO-A and B are present in less dense, nonribosomal fractions of the sucrose gradient. As most, if not all, RNAs are associated with proteins, it is likely that EGO-A and B are bound to proteins. Although the translationally controlled expression of a small protein at discrete intervals in hematopoietic development is possible, the simplest explanation for the data is that EGO does not code for a protein.
Tissue-specific expression and inducible expression of ncRNA suggests functionality. Expression of EGO RNA is rapidly and transiently increased following IL-5 addition to CD34+ hematopoietic cell culture derived from UCB, bone marrow, or mature eosinophils. EGO RNA is very highly expressed in bone marrow; however, EGO transcript levels were not high in thymus cDNA, suggesting that EGO does not have a role in lymphoid development. The high tissue-specific expression of EGO in bone marrow suggests a role in development from bone marrow hematopoietic stem cells.
The functional role of EGO was investigated by RNA silencing. TF-1, a CD34+ cell line that is cytokine growth dependent, and CD34+ UCB cells were used to evaluate the effect of EGO RNA silencing on eosinophil development. TF-1 cells express EGO and granule protein mRNA constitutively during growth on a variety of cytokines. Silencing of EGO transcripts in TF-1 and CD34+ cells decreases the level of MBP and EDN mRNA; however, GATA-1 levels were not affected in TF-1 cells, suggesting a GATA-1–independent mechanism. The mechanism of EGO's action is not understood; however, EGO may be acting as a siRNA. Alternatively, EGO could influence transcription or protect mRNA from degradation via an RNA-protein complex. Therefore, EGO is an ncRNA expressed during IL-5 stimulation of CD34+ UCB cells, present in bone marrow mononuclear cells, and regulates granule protein MBP and EDN mRNA levels.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Special thanks to Tenley Schofield for help with the manuscript and Bob Palais for help with mathematics, and to the following for help with cord blood collection: Jessica Dustin, Drew Durtschi, Stephen Gleich, Leigh Cornu, Jeff Norris, and Lyo Ohnuki. Thanks to the University of Utah core flow cytometry facility, especially Hawk Namsen.
Financial support was from National Institutes of Health (NIH) grant RO1AI9728.
National Institutes of Health
Authorship
Contribution: L.W. was responsible for experimental design, performed experiments, and wrote the manuscript; C.C. performed experiments; A.G. supplied technical help with flow cytometry and isolated eosinophils; M.S.E. was responsible for cord blood tissue procurement; L.K. was responsible for bone marrow tissue procurement; B.W. provided microarray analysis and extensive intellectual contribution; J.S. provided intellectual contribution; D.D. performed the microarray experiment; and G.G. provided extensive intellectual contribution and financial support.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lori A. Wagner, 30 North 1900 East School of Medicine, Department of Dermatology, University of Utah, Salt Lake City, UT, 84132; e-mail: lori.wagner@hsc.utah.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal