Abstract
New Zealand black (NZB) mice with autoimmune and B lymphoproliferative disease (B-LPD) are a model for human chronic lymphocytic leukemia (CLL). A genomewide linkage scan of the NZB loci associated with lymphoma was conducted in F1 backcrosses of NZB and a control strain, DBA/2. Of 202 mice phenotyped for the presence or absence of LPD, surface maker expression, DNA content, and microsatellite polymorphisms, 74 had disease. The CD5+, IgM+, B220dim, hyperdiploid LPD was linked to 3 loci on chromosomes 14, 18, and 19 that are distinct from previously identified autoimmunity-associated loci. The region of synteny with mouse D14mit160 is the human 13q14 region, associated with human CLL, containing microRNAs mir-15a16-1. DNA sequencing of multiple NZB tissues identified a point mutation in the 3′ flanking sequence of the identical microRNA, mir-16-1, and this mutation was not present in other strains, including the nearest neighbor, NZW. Levels of miR-16 were decreased in NZB lymphoid tissue. Exogenous miR-16 delivered to an NZB malignant B-1 cell line resulted in cell-cycle alterations and increased apoptosis. Linkage of the mir-15a/16-1 complex and the development of B-LPD in this spontaneous mouse model suggest that the altered expression of the mir-15a/16-1 is the molecular lesion in CLL.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most common hematologic malignancy in the Western world;1 the molecular etiology and pathogenesis of CLL remains unknown, but antigen exposure may play a role.2 Normal B-cell development and differentiation as well as CLL leukemogenesis are thought to be a multistep process. Thus, CLL may represent an accumulation of several genetic alterations. CLL shows the highest incidence of familial leukemia and is believed to be polygenetic.3 Although the cell of origin remains unknown, it is thought by some to arise in a CD5+ B-cell splenic marginal zone subpopulation in which the immunoglobulin genes encode for polyreactive IgM autoantibodies.4,5 Molecular cytogenetic abnormalities reported associated with CLL include 13q14 deletions.6,7 A minimally deleted region (MDR) has been described, and several genes in this region have been sequenced, but no mutations in protein-coding regions have been identified.8 However, mutations in mir-16 have been described,9 as well as a reduction in the levels of expression in some patients with CLL.10 The New Zealand black (NZB) mouse has been studied extensively as a model to investigate autoimmune diseases such as systemic lupus erythematosus (SLE)11,12 as well as a model for the B-cell lymphoproliferative disorder CLL.13 In both the NZB and human CLL, the disease is late appearing and, similar to a subset of patients with CLL, NZB mice develop autoimmune hemolytic anemia (AIHA). As NZB mice age, they develop a monoclonal lymphoproliferative expansion characterized by increased numbers of CD5+ B220dull B cells that are hyperdiploid.14 In addition, the NZB malignant CD5+ B clones frequently have increased IL-10 production and development of CLL-like clones is associated with elevated IL-10 in crosses of NZB with DBA/2.15 In contrast, the DBA/2 strain has very few CD5+ B cells and does not demonstrate an age-related expansion of these cells. In the present report, we used crosses of these 2 strains to determine the genetic loci linked to the development of lymphoproliferative disorder (LPD).
Microsatellite or simple sequence-length polymorphisms (SSLPs) are widely used in mouse genetics because they are numerous, highly polymorphic, and widely dispersed throughout the genome;16 these SSLPs have been used to create dense linkage maps for at least 12 inbred strains of mice. The commercial availability of primers and the polymerase chain reaction (PCR)–based identification of SSLP chromosome markers make it feasible to systematically search the entire mouse genome for linkage. In the present study, we performed a genomewide scan to identify loci for murine LPD, and additional direct DNA sequencing analysis was conducted on a locus on mouse chromosome 14 with synteny to human 13q14.
Materials and methods
Mouse colony
A mouse-breeding colony was established within the FDA facility at the NIH campus, and all protocols were approved by the Animal Use Committee. NZB and DBA/2 (DBA) male and female mice were purchased from the Jackson Laboratories (Bar Harbor, ME). These mice and their progeny were housed under uniform conditions in the disease-free animal rooms at the FDA (Bethesda, MD). Male and female NZB and DBA/2 mice were mated to produce a group of F1 mice (NZB × DBA/2). F1 mice were then backcrossed to NZB mice to produce the population of [(NZB × DBA/2) F1 × NZB]. Animals were aged in single-sex groups of 10. A total of 202 F1 backcross (F1BC) animals were phenotyped, of which 74 were positive microscopically for the presence of B-cell LPD within the spleen. Additional subsequent crosses between NZB and DBA/2 mice were performed involving 230 additional study animals.
Autopsy
At the time of these studies, the mice were between 21 and 27 months of age. Animals were bled under light anesthesia (methoxyflurane [Metofane]; Pitman-Moore, Mundelein, IL) followed by cervical dislocation, peritoneal lavage, and autopsy. The animals′ total weight, sex, state of health, and spleen weight were noted. Sections were obtained from the sternum, heart, lungs, salivary glands, liver, spleen, both kidneys, lymph nodes, and intestines for histologic evaluation.
Histopathology
Tissue samples for microscopic examination were formalin fixed, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E; EPL, Vienna, VA). Spleens were systematically reviewed for overall size, pattern of the periarteriolar lymphoid sheath (PALS), number and size of germinal centers (GCs), degree of extramedullary hematopoiesis (EMH), and whether GCs were inactive or reactive. LPD was classified as previously reported.17–19 Histology was performed using a Zeiss Axiophot microscope (Carl Zeiss Inc, Thornwood, NY) using a 5 × objective with a 0.16 numerical aperture. The appearance of the bridging of 2 GCs by marginal zone cells was defined as minimal LPD or early spleen marginal zone lymphoma. Moderate involvement was infiltration of the red pulp by these marginal zone cells. Marked involvement was complete replacement of normal splenic architecture. A 40 × objective (0.5-1.0 numerical aperture) was used for nuclear morphology analysis. Photomicrographs were produced with a Cool Snap model HQ camera (Princeton Instruments, Trenton, NJ) and the software used for acquisition was IPLab (BD Biosciences, Rockville, MD).
Blood
Blood was obtained from mice anesthetized with methoxyflurane via brachial artery exsanguination or cardiac puncture. This blood was used for a complete blood count (CBC), reticulocyte count, blood film, and serum immunoglobulin levels (Analytics, Gaithersburg, MD). These results were compared with normal mouse values, and cut-off values were determined (Table 1).
Association of NZB phenotype and LPD
| Phenotype . | N . | Non-LPD (mean ± SD) . | N . | LPD (mean ± SD) . |
|---|---|---|---|---|
| Spleen | ||||
| Total | 128 | 391 ± 500 | 74 | 932 ± 937 |
| More than 500 (mg) | 31 | 1016 ± 699 | 41 | 1459 ± 975* |
| WBC | ||||
| Total | 106 | 6.7 ± 5.0 | 64 | 17.1 ± 34.0 |
| More than 20 (×106 cells/L) | 3 | 29.0 ± 5.2 | 11 | 65.3 ± 34.0* |
| RBC | ||||
| Total | 110 | 6.5 ± 2.0 | 64 | 5.8 ± 1.8 |
| Less than 4 (×109 cells/L) | 11 | 2.5 ± 0.5 | 7 | 2.3 ± 0.8 |
| MVC | ||||
| Total | 110 | 51.4 ± 12.0 | 64 | 53.5 ± 12.2 |
| More than 60 (fL) | 17 | 75.0 ± 11.6 | 11 | 76.0 ± 12.3 |
| IgM | ||||
| Total | 127 | 586 ± 206 | 71 | 577 ± 233 |
| More than 700 (mg/L) | 50 | 784.0 ± 43.5 | 29 | 814.4 ± 31.1 |
| RBC reticulocytes | ||||
| Total | 116 | 17.1 ± 29.1 | 70 | 17.2 ± 24.5 |
| More than 20* (%) | 24 | 68.1 ± 27.4 | 19 | 50.1 ± 26.3 |
| Phenotype . | N . | Non-LPD (mean ± SD) . | N . | LPD (mean ± SD) . |
|---|---|---|---|---|
| Spleen | ||||
| Total | 128 | 391 ± 500 | 74 | 932 ± 937 |
| More than 500 (mg) | 31 | 1016 ± 699 | 41 | 1459 ± 975* |
| WBC | ||||
| Total | 106 | 6.7 ± 5.0 | 64 | 17.1 ± 34.0 |
| More than 20 (×106 cells/L) | 3 | 29.0 ± 5.2 | 11 | 65.3 ± 34.0* |
| RBC | ||||
| Total | 110 | 6.5 ± 2.0 | 64 | 5.8 ± 1.8 |
| Less than 4 (×109 cells/L) | 11 | 2.5 ± 0.5 | 7 | 2.3 ± 0.8 |
| MVC | ||||
| Total | 110 | 51.4 ± 12.0 | 64 | 53.5 ± 12.2 |
| More than 60 (fL) | 17 | 75.0 ± 11.6 | 11 | 76.0 ± 12.3 |
| IgM | ||||
| Total | 127 | 586 ± 206 | 71 | 577 ± 233 |
| More than 700 (mg/L) | 50 | 784.0 ± 43.5 | 29 | 814.4 ± 31.1 |
| RBC reticulocytes | ||||
| Total | 116 | 17.1 ± 29.1 | 70 | 17.2 ± 24.5 |
| More than 20* (%) | 24 | 68.1 ± 27.4 | 19 | 50.1 ± 26.3 |
Significant difference between non-LPD and LPD, χ2 analysis P ≤ .001 (BD Biosciences, San Jose, CA).
N indicates number of mice.
Flow cytometric analysis
Single-cell spleen and peritoneal suspensions were obtained. Spleen suspensions were further treated with ammonium chloride lysis. The cells were analyzed for both surface markers and DNA content (cell cycle). For surface markers, cells (1 × 106) were stained with FITC-conjugated goat anti-IgM, anti-CD5 conjugated with phycoerythrin (PE), anti-B220 conjugated with PE-Cy5 (TriChrome), or isotype controls (Caltag, Burlingame, CA). Unstained cells, isotype controls, and singly and doubly stained cells in conjunction with AutoComp software were used to set compensation. A FACScan flow cytometer (BD Biosciences, San Jose, CA) was used to collect 2 × 104 events. Lysis II and CellQuest software (BD Biosciences) and Mod Fit (Verity Software House, Topsham, ME) were used for data analysis and display. For cell-cycle analysis, cell suspensions (1 × 106 cells) were stained with the FITC anti-IgM reagent, fixed in cold 70% ETOH, and stored overnight at 4°C. At the time of analysis, the ethanol-fixed cells were incubated with propidium iodide (PI; 50 μg/mL) and ribonuclease (1 mg/mL) for 30 minutes at 37°C and analyzed immediately on a FACScan flow cytometer using CELLFIT software (BD Biosciences) for both the acquisition and analysis of 100 000 events. Chicken erythrocyte and calf thymus nuclei were used as linearity controls. A DNA index was derived by the CELLFIT software.
DNA extraction for genotyping
DNA was extracted from nonpathologic tissues, usually from the liver. DNA was purified by the standard phenol/chloroform method followed by the QIAamp tissue kit (Qiagen Inc, Chatsworth, CA). The samples were resuspended in TE (10 mM Tris-Cl [pH 7.6] and 1 mM EDTA) at a concentration close to 1 μg/μL.
Primer pair selection
Microsatellite oligonucleotide primer pairs were purchased from Research Genetics (Huntsville, AL). SSLPs were typed by determining the size of the PCR products obtained with individual primer sets. The identification of a polymorphic locus in the 2 inbred strains (NZB and DBA/2) was first determined. Of 450 primers analyzed, 75 were deemed informative with detectable size difference in PCR product size between the 2 parental strains. Representative informative primers included D1Mit68, D1Mit303, D1Mit49, D1Mit33, D2Mit15, D2Mit21, D3Mit60, D4Mit95, D4Mit259, D4 Mit 9, D4Mit37, D4Miit232, D5Mit294, D5Mit81, D5Mit8, D5Mit101, D6MIT84, D6Mit123, D6Mit188, D6Mit10, D6Mit25, D6Mit15, D7nds5, D7Mit39, D8Mit4, D8Mit6, D8Mit137, D9Mit205, D9Mit45, D9Mit182, D9Mit52, D11Mit229, D11Mit29, D11Mit99, D11Mit50, D12Mit56, D12Mit35, D12Mit51, D13Mit221, D13Mit202, D13Mit110, D13Mit130, D13Mit78, D14Mit98, D14Mit129, D14Mit5, D14Mit160, D15Mit85, D16Mit131, D16Mit118, D17Mit176, D17Mit93, D18Mit61, D18Mit62, D18Mit4, D19Mit35, and D19Mit6.
PCR conditions
PCR reactions were performed using reagents and methods from Invitrogen (Carlsbad, CA) with Taq enzyme (Roche Diagnostics, Indianapolis, IN). A total of 35 cycles were used of 2 minutes at 94°C, 2 minutes at 56 to 62°C, and 30 seconds at 72°C. Reactions were 20 μL vol covered with oil in a microtiter plate. A hot start of 2 minutes at 98°C was followed by the addition of enzyme at 85°C. The PCR products were resolved on a 3% Metaphor (Cambrex Corp, Rockland, ME) agarose gel stained with ethidium bromide. For the 75 loci chosen for this study, the longest distance between a marker and an autosomal locus was 15.8 cM. The X chromosome was not evaluated.
Linkage analysis
For all linkage analysis studies using the informative primer pairs, 3 DNA controls were used: NZB parental, DBA/2 parental, and F1 (NZB × DBA/2) DNA. The F1 would show the presence of both parental bands, whereas the F1BC could show either both bands or only the NZB band, and the F2 could be homozygous for either parent or heterozygous. The samples were scored for heterozygosity versus homozygosity, and the results were analyzed for expected distribution using chi-square analysis. In additional studies, only 3 loci were re-examined: D14Mit160, D18Mit4, and D19Mit6.
Sequencing and designing primers for mirv15a/16-1
Using the miRBase Sequence Database20 and the nomenclature established,21,22 the stem-loop and mature sequences as well as the base coordinates were obtained for each of the following: hsa-mir-16-1 and hsa-mir-15a (both human); and mmu-mir-16-1 and mmu-mir-15a (both mouse). Using the National Center for Biotechnology Information (NCBI) BLAST 2 Sequences (http://www.ncbi.nlm.nih.gov), regions of homology were found among the mir-15a/16-1 regions between Homo sapiens chromosome 13 and Mus musculus chromosome 14. For amplification of mouse chromosome 14 region containing mmu-mir-15a and mmu-mir-16-1 (determined using the Ensemble Genome Browser; http://www.ensembl.org), the following primers were produced by the NJMS Molecular Resource Facility to create a 502–base pair amplicon: 5′cctggtatgcagtggtaaggc 3′(forward primer), 5′ctattgaggtgctaggag 3′(reverse primer).
DNA extraction, PCR, and DNA sequencing
Sources of DNA were obtained from the spleen, liver, kidney, bone marrow, or lymph node from NZB/NJ, NZW, DBA/2J, C57Bl/6J, NOD/SCID/DR1, and SJL strains obtained from The Jackson Laboratories. DNA was extracted using the Qiagen DNeasy Tissue Kit, and amplified via PCR on the GeneAmp PCR system 9600 (Applied Biosystems, Foster City, CA, USA) at an annealing temperature of 55°C for 35 cycles. The PCR products were purified using the Qiagen QIAquick PCR Purification Kit and sequenced using 4-color fluorescent sequencing reactions on the Applied Biosystems model 3130xl sequencer. Using NCBI's BLAST 2, the DNA sequences obtained were compared to reference sequences. The sequence obtained in NZB mice has been reported to GenBank as accession no. EF042973.
Functional analysis of miR-16 RNA
Tissue and cells were obtained and placed in TRizol (Invitrogen) and the RNA was extracted. Real-time PCR was used to quantitate miR-16 in tissues23 using the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems) and the reverse transcriptase (RT) 391miR-16 5 × RT primer. The RT reaction was run on the GeneAmp PCR System 9600 for 1 cycle at 16°C for 30 minutes, 42°C for 30 minutes, and 85°C for 5 minutes. Following the stem-loop RT reaction, reverse transcription PCR was performed using the TaqMan 2 × Universal PCR Master Mix (Applied Biosystems) and the TaqMan MicroRNA Assay TM 391 miR-16 20 × Real Time primers (Applied Biosystems), and run on the Applied Biosystems 7500 Real Time PCR System at 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds, and 60°C for 60 seconds. Change in gene expression was reported as change in the ΔCT compared with the glyceraldehyde-3-phosphare dehydrogenase (GAPDH) control (TaqMan Rodent GAPDH Control; Applied Biosystems) and a relative quantification study was performed on the RT products according to the manufacturer's protocol. Northern analysis was also performed, and the level of expression reported as normalized femtomoles (normalized to the median of tRNA signals).
To study the effects of exogenous miR-16, a total of 1 to 4 million cells from an NZB malignant cell line, were transfected with a microRNA mimic using the using the Amaxa Nucleofector II (Amata Inc, Gaithersburg, MD). Experimental groups included untreated, electroporated, and transfected (miR-16 or control mimic) cells. Cells were transfected with 3 μg of miRIDIAN Mimic mmu-miR-16 (Dharmacon-C-310112-02) or 3 μg of miRIDIAN Mimic Negative Control no. 1(Dharmacon-CN-001000-01; Dharmacon, Lafayette, CO) according to the manufacturer's protocol using program G-16, and solution T. Cells were plated at 0.5 × 106/mL and incubated at 37°C for 24 hours. Following incubation, flow cytometry was used to detect cell-cycle changes and apoptosis by staining the DNA with PI (Calbiochem, La Jolla, CA). For PI staining, 1 × 106 cells were stained with hypotonic PI (0.05 mg/mL PI and 0.1% Triton X-100). Fluorescence-activated cell sorter (FACS) data were acquired on Becton Dickinson FACS Calibur. Acquisition was done using CELLQUEST software (Becton Dickinson), and analysis was performed using ModFit LT software.
Results
Phenotype of F1 backcrosses
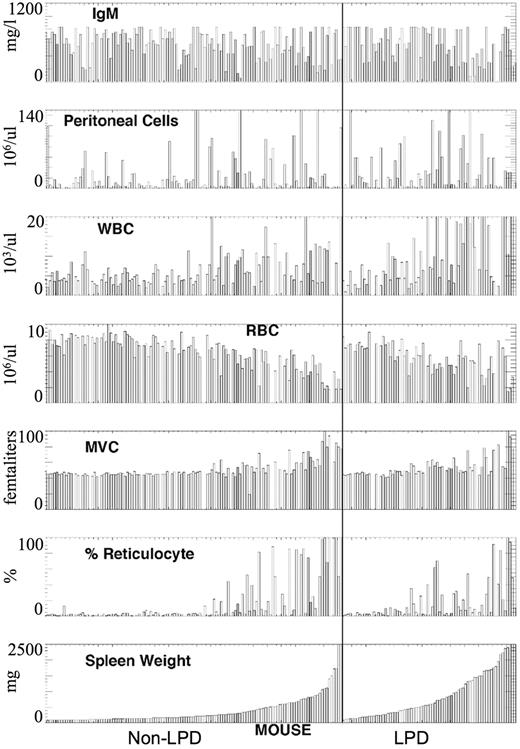
Based on the results of the phenotypic analysis of 202 mice examined, the backcross mice were divided on the basis of the presence or absence of LPD (non-LPD and LPD; Figure 1; Table 1). Enlarged spleens were present in both LPD and non-LPD mice; however, greatly enlarged spleens were more frequently seen in LPD mice (55% vs 24%; Table 1). Based on red blood cell (RBC) count, mean corpuscular volume (MCV), and percentage of reticulocytes, there was no significant difference in the presence of autoimmune hemolytic anemia in LPD versus non-LPD mice. However, leukocytosis was more frequent in the LPD mice (17% vs 3%; Table 1), and lymphocytosis was confirmed by review of the blood film. Peritoneal cell count and serum IgM mean levels were nearly identical and do not suggest a disease related difference. Table 2 suggests that there is an increase in aneuploidy in the LPD F1BC animals.
Phenotypic characterization of F1 backcross of NZB × DBA/2. The mice were divided into LPD (right panel) and non-LPD (left panel) based on splenic histopathology. These values were ranked ordered in each group based on splenic weight. Data are presented for serum IgM level (IgM), peritoneal cell count, white blood count (WBC), RBC, MCV, and percentage of reticulocytes.
Phenotypic characterization of F1 backcross of NZB × DBA/2. The mice were divided into LPD (right panel) and non-LPD (left panel) based on splenic histopathology. These values were ranked ordered in each group based on splenic weight. Data are presented for serum IgM level (IgM), peritoneal cell count, white blood count (WBC), RBC, MCV, and percentage of reticulocytes.
Association of LPD and aneuploidy
| Cell type . | Total mice, no. . | Non-LPD mice, no. . | LPD mice, no. . |
|---|---|---|---|
| Spleen | |||
| Diploid | 135 | 93 | 42 |
| Aneuploid | 67 | 35 | 32* |
| Peritoneum | |||
| Diploid | 85 | 50 | 35 |
| Aneuploid | 90 | 59 | 31 |
| Cell type . | Total mice, no. . | Non-LPD mice, no. . | LPD mice, no. . |
|---|---|---|---|
| Spleen | |||
| Diploid | 135 | 93 | 42 |
| Aneuploid | 67 | 35 | 32* |
| Peritoneum | |||
| Diploid | 85 | 50 | 35 |
| Aneuploid | 90 | 59 | 31 |
Significant difference between non-LPD and LPD, χ2 analysis P ≤ .02.
Histopathology
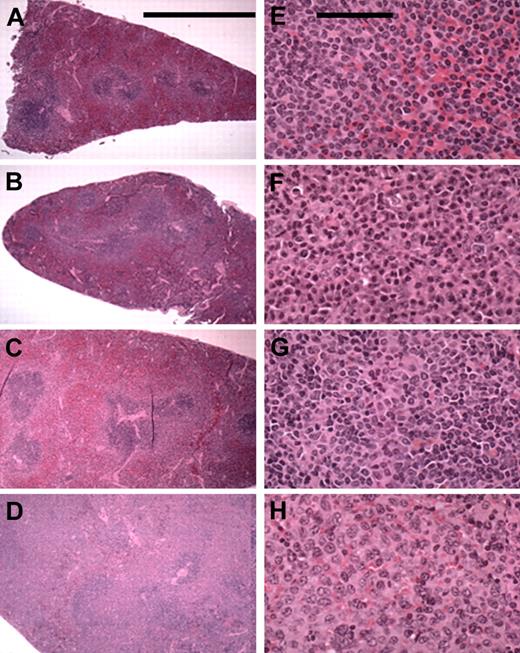
In all, 37% (74 of 202) of the mice demonstrated LPD. A total of 94% (67 of 71) of the splenic murine lymphomas were classified as marginal zone lymphomas (MZLs). Various stages of marginal zone involvement could be seen (Figure 2): early marginal zone hyperplasia (Figure 2A-B); the confluence of 2 or more GCs; massive infiltration of the red pulp (Figure 2B-C); and the entire replacement of the spleen (Figure 2D). Higher-power views of tumor-involved areas showed cellular details (Figure 2E-H). The marginal zone cells were characterized as monocytoid lymphocytes with clumped chromatin and a slight nuclear indentation (Figure 2E-F). The nuclear pattern became more vesicular in some mice (Figure 2G-H). This cellular spectrum suggests heterogeneity and marginal zone transformation. Other lymphocytic neoplasms (< 5%) identified in mice with LPD included were lymphoplasmocytic lymphoma (2 mice) centroblastic lymphoma (1 mouse) and small lymphocytic lymphoma (1 mouse).
Splenic MZL in F1BC NZB mice. The left column (A-D) contains low-power (5.5 × objective; bar is 5 cm) photomicrographs of selected mice spleens showing the various stages of MZL seen: early marginal zone hyperplasia and the confluence of 2 or more GCs (A-B); moderate and massive infiltration of the red pulp (C-D). The right column (E-H) shows selected areas of involvement at a higher power (43 × objective; bar is 100 μm) for cellular detail.
Splenic MZL in F1BC NZB mice. The left column (A-D) contains low-power (5.5 × objective; bar is 5 cm) photomicrographs of selected mice spleens showing the various stages of MZL seen: early marginal zone hyperplasia and the confluence of 2 or more GCs (A-B); moderate and massive infiltration of the red pulp (C-D). The right column (E-H) shows selected areas of involvement at a higher power (43 × objective; bar is 100 μm) for cellular detail.
Flow cytometric analysis
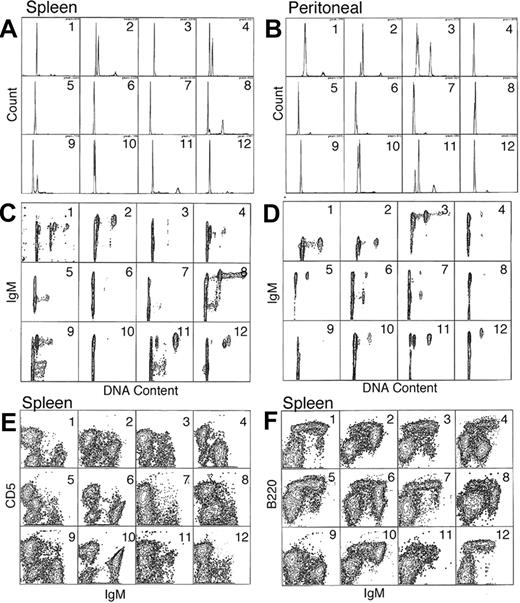
Flow cytometric data from 12 representative F1BC mice with LPD as defined by histopathology are presented in Figure 3. Hyperdiploidy was detected in both spleen and peritoneal cavity cells (Figure 3A-B), but not all mice with LPD had detectable aneuploidy. Further, the hyperdiploid cells were IgM+ B cells (Figure 3C-D), confirming that the LPD was due to a B-cell expansion. In the spleens shown in this figure, 75% (9 of 12) of the mice had hyperdiploid B-cell clones, whereas in the peritoneal cells, 50% (6 of 12) of the mice had no detectable hyperdiploid cells. Thus hyperdiploid cells may be present in the spleen yet undetectable in the matched peritoneum (mice nos. 4 and 12, Figure 3A and B).
Flow cytometric surface and cell-cycle analysis. A total of 12 representative mice with LPD were examined and their analysis presented in order in panels A-F. The mice are numbered from 1 to 12, and data from the same animal are shown for both spleen and peritoneal lavage cells (A-D). (A-B) Single-parameter DNA content histograms for spleen and peritoneal cells. (C-D) Two-parameter IgM versus DNA content (FL2-A vs FL1-H) for spleen and peritoneal cells (E-F) Expression of IgM versus CD5 (FL1-H vs FL2-H) and IgM versus B220 (FL1 vs FL3-H), respectively, for spleen cells. Examples of splenic hyperdiploidy can be seen in panel A, mice nos. 2, 4, 8, and 9. Further resolution of these aberrant subpopulations can be better appreciated in the corresponding panel C. The smaller aneuploid populations seen in mice nos. 11 and 12 can be better appreciated in the 2-color analysis shown in panel C.
Flow cytometric surface and cell-cycle analysis. A total of 12 representative mice with LPD were examined and their analysis presented in order in panels A-F. The mice are numbered from 1 to 12, and data from the same animal are shown for both spleen and peritoneal lavage cells (A-D). (A-B) Single-parameter DNA content histograms for spleen and peritoneal cells. (C-D) Two-parameter IgM versus DNA content (FL2-A vs FL1-H) for spleen and peritoneal cells (E-F) Expression of IgM versus CD5 (FL1-H vs FL2-H) and IgM versus B220 (FL1 vs FL3-H), respectively, for spleen cells. Examples of splenic hyperdiploidy can be seen in panel A, mice nos. 2, 4, 8, and 9. Further resolution of these aberrant subpopulations can be better appreciated in the corresponding panel C. The smaller aneuploid populations seen in mice nos. 11 and 12 can be better appreciated in the 2-color analysis shown in panel C.
The amount of hyperdiploid B-cell clones varied among the individual mice. For instance, in mouse no. 12 there is evidence of a small splenic hyperdiploid clone in the G2M peak. In contrast, mouse no. 4 clearly demonstrates a dominant hyperdiploid B clone in the spleen that is lacking in the peritoneal cells (Figure 3B,D). A 3-color analysis of surface-marker (Figure 3E-F) expression indicates that the aneuploid population is within the IgM+, CD5dull+, B220dim+ population.
Genomic screening
In order to establish linkage between disease phenotype and genetic loci, informative polymorphic microsatellite regions were identified. To establish a linkage map, 450 SSLPs were screened. Only 75 primers were informative with a minimum coverage of 15.8 cM. A total of 202 F1BC animals were phenotyped. Of these, 37% (74 of 202) were found to have LPD upon histopathologic evaluation of their spleens. Of these 74 mice, 67 were subjected to a genomewide linkage analysis. Liver DNA from LPD-positive animals was evaluated for these 75 informative SSLP loci. Of the 75 informative loci evaluated in diseased F1BC mice, deviation from expected frequencies were observed at 3 distinct loci. The P values as determined by chi-square analysis were all 0.02 or less. These loci are located on mouse chromosomes 14, 18, and 19 (D14Mit160, D18Mit4, and D19Mit6), with deviations toward homozygosity for NZB.
Candidate gene linkage
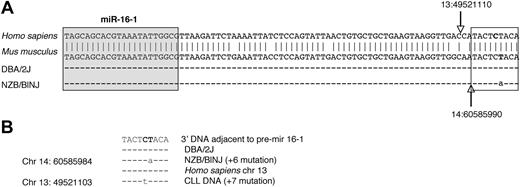
To further establish linkage to candidate loci, a region of mouse chromosome 14, approximately 11 Mb centromeric to the D14Mit160 location, was sequenced in both the NZB and the DBA/2 strain (Figure 4). There was a point mutation in the NZB in the flanking region 3′ to the stem loop structure of the pre–mir-16-1. This mutation was not found in the DBA2 or other mouse strains (tissues/cell lines from C57Bl/6, SJLJ, NZW, Balb/c, and NOD/SCID; data not shown). This mutation was observed in multiple tissue sources of DNA derived from the NZB as well as DNA from 2 in vitro cell lines established from NZB, the LNC (an NZB-derived malignant B-cell line)24 and 3C2 (an NZB-derived CD8+ CTL line;25 Figure 4; data not shown). In both the NZB sequence and that reported in a human CLL,9 the mutation is in a nearly identical location in the 3′ flanking region of mir-16-1 (Figure 4C).
Point mutation in 3′ DNA adjacent to pre–mir-16-1 region in NZB. (A) Nucleotide sequence comparison of the region of mouse chromosome (chr) 14 and human chr 13 on which miRNA mir-16-1 is located. The top sequence is the database reference sequence20 (antisense strand) in Homo sapiens for hsa-mir-16-1 with sense strand base coordinates (NCBI36) 13:49 521 099 to 13:49 521 187 The second sequence is the antisense of the database reference sequence in Mus musculus for mmu-mir-16-1 with the sense strand base coordinates (NCBIM36) 14:60 585 981 to 60 586 067. The sequence homology between the mir-16-1 region (including the 3′ flanking region) of Mus musculus (chr 14) and of Homo sapiens (chr 13) is shown (vertical arrows indicate the end of the pre–mir-16-1 in humans vs mouse). The third and fourth rows are sequence comparisons of splenic DNA from DBA/2J and NZB/BlNJ mice. The sequences are identical to the reference sequence, except for a T → A point mutation (on the antisense strand; A → T point mutation at base 60 585 990 on the sense strand of chr 14) in the NZB/BlNJ. In addition, DNA extracted from DBA/2J (5 weeks old) liver, NZW (5 weeks old) spleen, Balb/C B-cell lymphoma cell line (CH27), C57BL/6 (4 months old) kidney, SJL/J B cell lymphoma line (NJ117), and NOD/SCID (7 months old) liver showed no point mutations, whereas DNA from NZB/BlNJ spleen, liver (14 months old), kidney (15 months old), T-cell lymphoma line (3C2), and malignant B1 cell line (LNC) all had the same point mutation (data not shown). The precursor stem-loop structure sequences (pre-miRNA) are based on established nomenclature.20 In the mouse samples (pre–mmu-mir) for both pre–mir-15a (accession no. MI0000564) and pre–mir-16-1(MI0000565) and mature sequences for both miR-15a (MIMAT0000526) and 16-1 (MIMAT0000527) were also compared, and no other mutation was found in these regions (data not shown). The shaded boxed region is the mature miR-16, and the unshaded boxed region is the 3′ flanking region, which is further compared in panel B. (B) Sequence comparison between human and mouse 3′ adjacent to pre–mir-16-1. The point mutations in NZB/BINJ splenic DNA and in the reported DNA of patients with CLL9 are indicated. The NZB/BINJ splenic DNA shows an A → T mutation at the 60 585 984 base on chromosome 14, which is 6 bases from the end of the mmu-mir-16-1 sequence. CLL DNA has a reported G → A mutation at the 49 521 103 base on chromosome 13, 7 bases from the hsa-mir-16-1 sequence.9
Point mutation in 3′ DNA adjacent to pre–mir-16-1 region in NZB. (A) Nucleotide sequence comparison of the region of mouse chromosome (chr) 14 and human chr 13 on which miRNA mir-16-1 is located. The top sequence is the database reference sequence20 (antisense strand) in Homo sapiens for hsa-mir-16-1 with sense strand base coordinates (NCBI36) 13:49 521 099 to 13:49 521 187 The second sequence is the antisense of the database reference sequence in Mus musculus for mmu-mir-16-1 with the sense strand base coordinates (NCBIM36) 14:60 585 981 to 60 586 067. The sequence homology between the mir-16-1 region (including the 3′ flanking region) of Mus musculus (chr 14) and of Homo sapiens (chr 13) is shown (vertical arrows indicate the end of the pre–mir-16-1 in humans vs mouse). The third and fourth rows are sequence comparisons of splenic DNA from DBA/2J and NZB/BlNJ mice. The sequences are identical to the reference sequence, except for a T → A point mutation (on the antisense strand; A → T point mutation at base 60 585 990 on the sense strand of chr 14) in the NZB/BlNJ. In addition, DNA extracted from DBA/2J (5 weeks old) liver, NZW (5 weeks old) spleen, Balb/C B-cell lymphoma cell line (CH27), C57BL/6 (4 months old) kidney, SJL/J B cell lymphoma line (NJ117), and NOD/SCID (7 months old) liver showed no point mutations, whereas DNA from NZB/BlNJ spleen, liver (14 months old), kidney (15 months old), T-cell lymphoma line (3C2), and malignant B1 cell line (LNC) all had the same point mutation (data not shown). The precursor stem-loop structure sequences (pre-miRNA) are based on established nomenclature.20 In the mouse samples (pre–mmu-mir) for both pre–mir-15a (accession no. MI0000564) and pre–mir-16-1(MI0000565) and mature sequences for both miR-15a (MIMAT0000526) and 16-1 (MIMAT0000527) were also compared, and no other mutation was found in these regions (data not shown). The shaded boxed region is the mature miR-16, and the unshaded boxed region is the 3′ flanking region, which is further compared in panel B. (B) Sequence comparison between human and mouse 3′ adjacent to pre–mir-16-1. The point mutations in NZB/BINJ splenic DNA and in the reported DNA of patients with CLL9 are indicated. The NZB/BINJ splenic DNA shows an A → T mutation at the 60 585 984 base on chromosome 14, which is 6 bases from the end of the mmu-mir-16-1 sequence. CLL DNA has a reported G → A mutation at the 49 521 103 base on chromosome 13, 7 bases from the hsa-mir-16-1 sequence.9
miR-16 function analysis in NZB
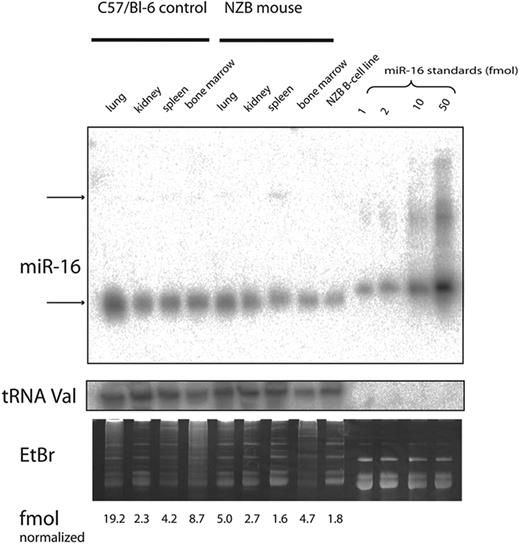
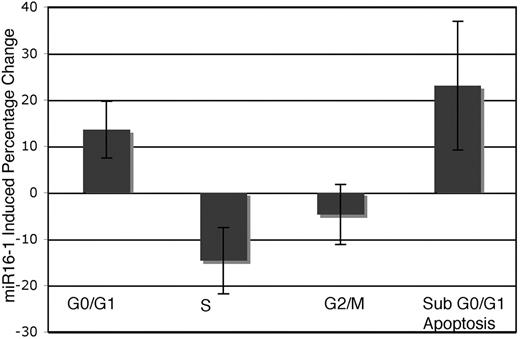
The level of expression of miR-16 was investigated by real-time PCR and Northern analysis of RNA from several tissue sources derived from NZB and compared with the expression in the control C57Bl/6 or DBA/2 strains (Table 3; Figure 5). NZB tissue sources of RNA showed a decrease in miR-16 in the spleen; however, the NZB kidney was not decreased in miR-16 expression compared with the control strain expression (Table 3). In addition, the NZB-derived malignant B-cell line, LNC, had an even greater decreased expression of miR-16 compared with C57Bl6 spleen (Figure 5). To study the effects of reconstitution of miR-16, electroporation was used to deliver either miR-16 or control mimic into the NZB line, LNC. Cell-cycle and sub-G0/G1 accumulation were used to monitor the induction of apoptosis by miR-16 (Figure 6). There was an increase in apoptosis following treatment with miR-16 and a decrease in cells in the S phase. A nonNZB-derived B-cell line, NJ11107, did not demonstrate any effects following introduction of exogenous miR-16 (data not shown). As a control, both cell lines were transfected with pMax-EGFP vector, and similar transfection efficiency was found (approximately 50%).
MicroRNA miR-16 expression
| . | NZB . | Control . | NZB/control ratio . |
|---|---|---|---|
| Real-time PCR* | |||
| Spleen | 0.97 ± 0.79 | −0.08 ± 0.21†† | 0.48 |
| Kidney | −0.08 ± 0.01 | 0.21 ± 2.08 | 1.24 |
| Northern NZB† | |||
| Spleen | 2.23 ± 1.19 | 4.36 ± 0.25 | 0.51 |
| Kidney | 3.8 ± 2.26 | 2.88 ± 0.58 | 1.32 |
| . | NZB . | Control . | NZB/control ratio . |
|---|---|---|---|
| Real-time PCR* | |||
| Spleen | 0.97 ± 0.79 | −0.08 ± 0.21†† | 0.48 |
| Kidney | −0.08 ± 0.01 | 0.21 ± 2.08 | 1.24 |
| Northern NZB† | |||
| Spleen | 2.23 ± 1.19 | 4.36 ± 0.25 | 0.51 |
| Kidney | 3.8 ± 2.26 | 2.88 ± 0.58 | 1.32 |
Real-time PCR detection of miR-16 in mouse tissue. The expression of miR-16 was compared with GAPDH control and the delta (Δ) CT determined (average ± SE) for NZB and control. The NZB/control ratio is the relative quantification determined using the ΔΔ CT method with a primer efficiency of 90% as reported by the manufacturer (Applied Biosystems), and compares the expression in NZB to the control strain, DBA/2 (mice were 5 weeks of age, n = 4).
Expression of miR-16 in mouse tissue was detected via Northern blot analysis and data are reported for NZB and control in fmol (average ± SE) normalized to the median of tRNA signals. The results are from 2 experiments with the control strain C57Bi/6. The NZB/control ratio is the ratio of the normalized miR-16 in NZB divided by the normalized miR-16 in C57Bi/6.
Significant difference between NZB and control, student t test P < .05.
Northern analysis. The expression of miR-16 was analyzed by Northern blots with RNA from 4 different tissues obtained from C57Bl/6 or NZB mice and the LNC cell line, a malignant B-1 cell line derived from NZB. The expression of mature miR16 (top gel, lower band) was quantitated and normalized to the expression of tRNA (middle gel). The gel was visualized following staining with ethidium bromide (lower gel). The top arrow (top gel) indicates the precursor form of miR-16.
Northern analysis. The expression of miR-16 was analyzed by Northern blots with RNA from 4 different tissues obtained from C57Bl/6 or NZB mice and the LNC cell line, a malignant B-1 cell line derived from NZB. The expression of mature miR16 (top gel, lower band) was quantitated and normalized to the expression of tRNA (middle gel). The gel was visualized following staining with ethidium bromide (lower gel). The top arrow (top gel) indicates the precursor form of miR-16.
Functional analysis of miR-16. The NZB malignant B-cell line, LNC, was transfected with either the miR-16 or control mimics. The cells were harvested 24 hours after transfection, and the cell-cycle stages were determined by flow cytometric techniques. The columns represent the mean percentage change in the NZB B-cell line treated with miR16 versus control mimic treatment (n = 4). Error bars indicate mean ± SEM.
Functional analysis of miR-16. The NZB malignant B-cell line, LNC, was transfected with either the miR-16 or control mimics. The cells were harvested 24 hours after transfection, and the cell-cycle stages were determined by flow cytometric techniques. The columns represent the mean percentage change in the NZB B-cell line treated with miR16 versus control mimic treatment (n = 4). Error bars indicate mean ± SEM.
Discussion
In the present report, genetic loci linked to the development of B-cell malignancy was determined by using NZB, a strain that develops a highly penetrant phenotype of B-cell lymphoma and leukemia.18,26 We have described the phenotypic characterization and genomic analysis of NZB LPD in crosses of NZB and DBA/2 mouse strains.27 The F1BC mice were derived from F1 crosses of NZB × DBA/2, and the F1 were backcrossed to the NZB parent. Aged F1BC mice were characterized for anemia, serum IgM globulin levels, spleen histopathology, spleen cell DNA content, and surface-marker expression, and a genomic scan was conducted using microsatellite polymorphism typing. Splenic histopathology revealed LPD in 36.6%, while splenic aneuploidy was found to be present in 43.2% of the F1BC colony. Most of the F1BC mice that developed histopathologic evidence of LPD were classified as MZLs expressing surface IgM, elevated levels of CD5, and dim B220. This B-cell malignancy is linked to the NZB alleles at D14Mit160, D18Mit4, and D19Mit6. In addition, sequence analysis of the mir-16-1 locus located near D14Miit160 demonstrated a nucleotide change in NZB that was not present in all other mouse strains analyzed, including the nearest neighbor strain, NZW. Since this nucleotide is noncoding and located in the 3′ flanking region of mir-16-1, we examined expression of miR-16 in NZB tissues. Using RT-PCR and Northern analysis, there is reduced expression of miR-16 in NZB spleen and in an in vitro NZB B-cell line. Add-back experiments involving miR-16 resulted in increased apoptosis and decreased cell growth. The finding of NZB MZL linkage to D14Mit160 is a new finding, and the fact that its human homolog is located at 13q14 further strengthens the rationale for using NZB as a model for human CLL.
LPD and aneuploidy occurred in approximately one-third of the F1BC mice, and was not unexpected.15 We were surprised with the finding of MZL. Based on LPD in NZB as a model for human CD5+ CLL, one would have expected the spleen to show evidence of a small lymphocytic lymphoma. Although hyperdiploidy has been previously described in the NZB spleen,14 human CLL does not typically show evidence of aneuploidy as determined by the flow cytometric analysis of PI-stained cells (G.E.M., unpublished observation, 1991-2000). Chromosomal abnormalities, however, are well known in CLL.6 It would now seem that MZL histopathology, aneuploidy, and surface IgM, CD5, and B200 expression are linked to a region near D14Mit160 (D14Mit160 is approximately 11 Mb from mir-16-1).
As noted in the Introduction, the 13q14 site is frequently involved in human CLL, and 13q14 deletions also occur in other closely related human LPD, such as mantle cell lymphoma and multiple myeloma.28 The process of leukemogenesis or lymphomagenesis is a multistep process, as evidenced by the fact that not all tumors in the F1BC were MZL, but were instead other B-cell malignancies. In addition, individual mice homozygous for all 3 loci not always have histopathologic evidence of MZL. Conversely, mice could be homozygous for 2 of the loci and still have malignant disease. Therefore, there must exist additional genetic loci controlling the expression of LPD in this murine model. In our familial CLL studies, we find a variety of other lymphoid and hematologic malignancies.3 Of greater interest, we find a significant increase of 13q14 deletions in familial CLL,8 and thus the NZB may be a model not only for sporadic CLL but for familial CLL. This highlights the fact that additional factors influence the type of B-cell malignancy that develops.
There are a number of candidate genes on chromosomes 14, 18, and 19 located near the site of SSLP-identified linkage. However, because of the synteny of mouse chromosome 14 to human 13q14, this loci was further studied. The significance of the other 2 loci on mouse chromosomes 18 and 19 remains to be determined. In the present study, identification of loci linked to the development of LPD in the NZB mouse model has been consistent with suggested tumor suppressor loci in human CLL. In particular, the linkage of lymphoproliferative disease with mouse chromosome 14 (human 13q14) is not unexpected, since up to 50% of human cases of CLL have a loss of human 13q14.3.28 Two genes in this region have been shown to be highly conserved between mouse and humans, DLEU2 and RFP2 (also called LEU5).29 Despite the linkage to this particular region of DNA in CLL, no somatic mutations in this region have been detected in CLL patients,30 and none were found in the NZB strain of mouse (data not shown). In addition to the presence of DLEU2 and RFP2 in the human 13q14 and the region of synteny in the mouse chromosome 14, there are 2 microRNAs genes found in the intronic region of DLEU2 in both murine and human, mir-15a and mir-16-1.
miRNAs have been found to play a role in oncogenesis,31 and have differential expression in tissues.32 Alterations in microRNAs in human B-CLL consist of both deletions and mutations.33 Specifically, the most frequently deleted genomic region contains the mir-15a and mir-16-1 genes. Recent SNP array studies also found that mir15a/16-1 genes were the targets of the recurrent 13q14 deletions and strengthen the conclusion that these 2 microRNAs are critical genes for CLL pathogenesis.34 Mutations in this region have been reported (reviewed in Calin et al33 ). In the present report, we describe a mutation in NZB in the immediate 3′ flanking region of mir-16-1. Analysis of levels of mature miR-16 indicated decreased expression in the NZB spleen as well as in the NZB-derived malignant B-cell line, LNC. The flanking region, containing the mutation, may be important for proper tertiary structure of the stem-loop in the pre-mir.35 This is consistent with the finding reported by others of decreased miR-16 in some patients with CLL.33 One possible outcome of decreased miR-16 is a failure to properly reduce target gene expression. One of the target genes for miR-16 (based on the presence of sequences complementary to miR-16 in the target gene 3′ UTR) is bcl-2.36 Due to the presence of the mutation, decreased miR-16 may result in increased bcl-2 expression. In both the murine model of CLL and some CLL patients with the disease, increased bcl-2 may lead to a failure to undergo apoptosis, which may play a role in disease. To test this possibility, the NZB cell line LNC with the mir-16-1 point mutation was treated with exogenous miR-16; following treatment, induction of apoptosis was observed. This was not found in other non-NZB B-cell lines. This suggests that miR-16 is important in the induction of apoptosis in the murine NZB CLL line. Of note, the mature miR-16 is also encoded by a second transcription unit, mir-15b–mir-16-2, located on chromosome 3.37 It remains unclear if this transcription unit might partially compensate for the loss of expression of the mir-16-1 gene.
The genetic linkage of autoimmune disease in NZB has been studied extensively as a model for SLE,38,39 while the loci linked to leukemia and lymphoma have not been determined. Based on our findings, the loci linked to LPD in NZB mice were not linked to previously identified loci linked to autoimmunity. In our present study, we found linkage to chromosomes 14, 18, and 19 in NZB. In other mouse CLL studies, transgenic mice derived by using the human TCL1 sequence (mouse homolog on chromosome 12) have been produced that develop a variety of B-cell neoplasias, including CLL, follicular lymphomas, Burkitt lymphoma, and diffuse large-cell lymphomas; however, no linkage was found to this gene locus in the present study.40,41 It is of interest that a locus involving CD5 B cells in NZB has also been mapped to chromosome 4, but this was not linked to LPD in the present study,42 while a locus on chromosome 13 has been found to be linked to marginal zone hyperplasia in NZB mice.43
The results from this present study describe 3 loci in NZB mice linked to the development of B-cell lymphoproliferative disease. The significance of 2 of these loci located on mouse chromosomes 18 and 19 remains to be determined. The third locus was found to be the site of a point mutation in a microRNA in NZB mice. Alterations in miRNA miR-16 levels may be an important factor in the development of both human CLL and the NZB mouse model since overexpression of miR-16 in an NZB cell line induces apoptosis. The NZB mouse, because it spontaneously develops B lymphoproliferative disease in an environment characterized by anti–self-reactivity, can shed insight not only into disease mechanisms but also in the development of therapy targeted at the loci linked to the development of CLL.
An Inside Blood analysis of this article appears at the front of this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Supported in part by a NIH grant to E.S.R. (CA/AI 71478-07). The support for P.L. was through the Dr Mildrede Scheel Stiftung fur Krebsforschung of the Deutsche Krebshilfe (German Cancer Aid).
E.S. and B.J.S. are PhD candidates at GSBS/NJMS, and this work is submitted in partial fulfillment of the requirement for PhD.
National Institutes of Health
Authorship
Contributions: E.S.R. and G.E.M. contributed to scientific design, performed experiments, contributed reagents, and wrote the paper; E.S., B.J.S., V.M., F.A., Y.-C.L., T.F., P.L., S.R., and R.A.M. performed research and analyzed data; V.E.Z. performed statistical analysis; and K.H. and J.R.T. analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerald E. Marti, Chief, Flow and Image Cytometry Section/Cell and Tissue Therapy Branch, Division of Cell and Gene Therapies/Office of Cellular, Tissues and Gene Therapies, CBER FDA NIH, Bldg 29B, Rm 2NN08, 8800 Rockville Pike, Bethesda, MD 20892; e-mail: gemarti@helix.nih.gov.