Abstract
Deficiency of glycosylphosphatidylinositol (GPI)–anchored molecules on blood cells accounts for most features of paroxysmal nocturnal hemoglobinuria (PNH) but not for the expansion of PNH (GPI−) clone(s). A plausible model is that PNH clones expand by escaping negative selection exerted by autoreactive T cells against normal (GPI+) hematopoiesis. By a systematic analysis of T-cell receptor beta (TCR-β) clonotypes of the CD8+ CD57+ T-cell population, frequently deranged in PNH, we show recurrent clonotypes in PNH patients but not in healthy controls: 11 of 16 patients shared at least 1 of 5 clonotypes, and a set of closely related clonotypes was present in 9 patients. The presence of T-cell clones bearing a set of highly homologous TCR-β molecules in most patients with hemolytic PNH is consistent with an immune process driven by the same (or similar) antigen(s)—probably a nonpeptide antigen, because patients sharing clonotypes do not all share identical HLA alleles. These data confirm that CD8+ CD57+ T cells play a role in PNH pathogenesis and provide strong new support to the hypothesis that the expansion of the GPI− blood cell population in PNH is due to selective damage to normal hematopoiesis mediated by an autoimmune attack against a nonpeptide antigen(s) that could be the GPI anchor itself.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired clonal disorder of the hematopoietic stem cell (HSC)1 characterized by 3 clinical hallmarks: intravascular hemolysis, tendency to venous thrombosis, and variable degrees of bone marrow failure.2–4 The primary molecular lesion responsible for PNH is a somatic mutation of the X-linked PIGA gene in HSCs,5,6 resulting in either complete or partial deficiency of all glycosylphosphatidylinositol (GPI)–linked proteins from the cell membrane of the progeny of the mutated HSC (GPI−).7,8 The deficiency of the GPI-linked proteins (including the complement-regulating proteins CD59 and CD55) from the surface of blood cells explains the intravascular hemolysis9 and probably underlies the increased tendency to venous thrombosis.10 However, a PIGA gene mutation per se does not explain the bone marrow failure and the expansion of the GPI− clone. In fact, very rare GPI− blood cells are present in healthy subjects,11 but only in PNH patients do the GPI− cells expand and contribute to hematopoiesis to various degrees, side by side with normal (GPI+) hematopoiesis.12,13 Clinical observations,14,15 in vitro hematopoietic colony studies,16,17 and data from PNH mouse models18–20 indicate that GPI− HSCs do not have an absolute growth advantage. The close relationship of PNH to idiopathic aplastic anemia (IAA) has suggested that autoreactive T cells against HSCs believed to be responsible for IAA may be at work also in PNH. Specifically, it has been hypothesized that in PNH autoreactive T cells destroy selectively GPI+ (normal) HSCs, whereas GPI− (PNH) HSCs can escape T-cell–mediated damage, thus being able to survive and expand.21 In recent years considerable evidence has accumulated in favor of an autoimmune mechanism for the expansion of the GPI− (PNH) clone. Indeed, the analysis of the T-cell receptor (TCR) repertoire has revealed an increased frequency of expanded T-cell clones in both PNH22,23 and IAA.23,24 The identity of the putative autoreactive T cells and of their molecular targets remains unknown. However, in PNH patients the expansion of large granular T lymphocytes (T-LGLs) with CD3+ CD8+ CD57+ immunophenotype is relatively common25,26 (L.L. and R.N., unpublished data, December 2004). Most of these T cells express inhibitory receptor superfamily (IRS) molecules that in PNH patients belong mainly to activating IRS isoforms, whereas in healthy controls they belong almost entirely to inhibitory IRS isoforms.27 This set of facts has suggested to us that the cells responsible for bone marrow damage in PNH may belong to the CD3+ CD8+ CD57+ cell population.
Here we show that CD8+ CD57+ T cells in PNH patients are oligoclonal and may include predominant clones. Moreover, by systematic sequence analysis of the complementary-determining region 3 (CDR3) of the T-cell receptor beta (TCR-β) chain, we show that identical or quasi-identical CDR3 sequences are present in CD8+ CD57+ T cells from most patients with hemolytic PNH. These data support the notion of an autoimmune process that is active in the pathogenesis of PNH, and they suggest that a restricted range of antigens may be the target of this process.
Patients, materials, and methods
Subjects
Peripheral blood samples from 20 patients with hemolytic PNH (median age, 41 years; range, 25 to 65 years) and from 31 healthy individuals (median age, 39 years; range, 25 to 67 years) were collected after informed consent was obtained according to institutional procedure and to the Declaration of Helsinki. Approval was obtained from the Istituto Nazionale per la Ricerca sul Cancro ([IST] Genoa, Italy) institutional review board for these studies. We included patients who had a large PNH population (more than 50% of GPI− granulocytes) and florid hemoglobinuria. All patients but one had primary classical hemolytic PNH and no severe cytopenias (Table 1). No patient was receiving any immunosuppressive drug at time of the study. Some patients have been studied more than once at 3-month intervals.
Clinical and hematologic features of investigated patients
| PNH patient . | Sex/age, y . | Time since diagnosis, y . | Hemoglobin level, g/L . | WBC, × 109/L . | ANC, × 109/L . | Lymphocyte count, × 109/L . | Platelet count, × 109/L . | Reticulocytes, % . | Size of PNH cell population,* % . | Thrombosis . | Blood transfusion . | Other treatment . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RBC . | PMN . | ||||||||||||
| 1 | M/53 | 1 | 102 | 4.4 | 3.2 | 0.9 | 249 | 6.0 | 21 | 81 | No | Occasional | Supportive |
| 2 | M/45 | 2 | 94 | 2.9 | 1.6 | 1.1 | 97 | 4.8 | 20 | 55 | No | Never | Supportive |
| 3 | M/34 | 2 | 101 | 4.7 | 2.5 | 1.7 | 178 | 10.2 | 50 | 80 | No | Occasional | Supportive† |
| 4 | F/54 | 10 | 81 | 3.3 | 2.4 | 0.7 | 57 | 7.1 | 36 | 98 | Hepatoportal | Never | Supportive |
| 5 | F/54 | 10 | 77 | 3.8 | 1.8 | 1.5 | 193 | 6.4 | 30 | 93 | No‡ | Never | Supportive |
| 6 | M/33 | 5 | 123 | 2.1 | 0.9 | 0.9 | 87 | 2.2 | 22 | 69 | No | Never | Supportive |
| 7 | F/25 | 8 | 67 | 2.5 | 1.1 | 1.2 | 83 | 14.5 | 23 | 82 | No | Regular | Supportive§ |
| 8 | F/53 | 7 | 121 | 3.6 | 1.6 | 1.7 | 64 | 2.1 | 51 | 96 | Budd-Chiari | Occasional | Supportive |
| 9 | F/25 | 1 | 109 | 4.5 | 2.4 | 1.8 | 323 | 9.8 | 26 | 80 | No‡ | Never | Supportive |
| 10 | F/45 | 5 | 86 | 8.7 | 7.5 | 1.0 | 143 | 17.0 | 75 | 99 | No | Occasional | Supportive |
| 11 | M/33 | 5 | 101 | 5.6 | 3.3 | 1.8 | 262 | 6.8 | 37 | 97 | No | Occasional | Supportive§ |
| 12 | F/55 | 23 | 84 | 3.9 | 2.5 | 1.0 | 185 | 10.1 | 54 | 90 | No | Regular | Supportive |
| 13 | F/32 | 3 | 78 | 4.0 | 2.2 | 1.3 | 193 | 2.4 | 91 | 69 | No | Occasional | Supportive§ |
| 14 | M/41 | 2 | 82 | 4.6 | 2.7 | 1.5 | 20 | 2.5 | 24 | 93 | No | Regular | Supportive |
| 15 | F/46 | 1 | 68 | 3.7 | 1.8 | 1.6 | 172 | 4.5 | 59 | 91 | No | Never | Supportive |
| 16 | M/25 | 5 | 79 | 5.0 | 3.6 | 1.0 | 164 | 14.1 | 16 | 84 | No | Regular | Supportive |
| 17 | M/38 | 6 | 78 | 7.0 | 6.3 | 0.6 | 327 | 9.5 | 83 | 99 | No‡ | Regular | Supportive§ |
| 18 | M/36 | 1 | 82 | 5.3 | 3.8 | 1.2 | 252 | 8.5 | 83 | 97 | No | Occasional | Supportive |
| 19 | M/65 | 6 | 112 | 7.7 | 6.7 | 0.8 | 203 | 6.7 | 35 | 67 | No | Occasional | Supportive |
| 20 | F/46 | 3 | 98 | 4.3 | 3.4 | 0.5 | 240 | 19.7 | 28 | 89 | No | Regular | Supportive |
| PNH patient . | Sex/age, y . | Time since diagnosis, y . | Hemoglobin level, g/L . | WBC, × 109/L . | ANC, × 109/L . | Lymphocyte count, × 109/L . | Platelet count, × 109/L . | Reticulocytes, % . | Size of PNH cell population,* % . | Thrombosis . | Blood transfusion . | Other treatment . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RBC . | PMN . | ||||||||||||
| 1 | M/53 | 1 | 102 | 4.4 | 3.2 | 0.9 | 249 | 6.0 | 21 | 81 | No | Occasional | Supportive |
| 2 | M/45 | 2 | 94 | 2.9 | 1.6 | 1.1 | 97 | 4.8 | 20 | 55 | No | Never | Supportive |
| 3 | M/34 | 2 | 101 | 4.7 | 2.5 | 1.7 | 178 | 10.2 | 50 | 80 | No | Occasional | Supportive† |
| 4 | F/54 | 10 | 81 | 3.3 | 2.4 | 0.7 | 57 | 7.1 | 36 | 98 | Hepatoportal | Never | Supportive |
| 5 | F/54 | 10 | 77 | 3.8 | 1.8 | 1.5 | 193 | 6.4 | 30 | 93 | No‡ | Never | Supportive |
| 6 | M/33 | 5 | 123 | 2.1 | 0.9 | 0.9 | 87 | 2.2 | 22 | 69 | No | Never | Supportive |
| 7 | F/25 | 8 | 67 | 2.5 | 1.1 | 1.2 | 83 | 14.5 | 23 | 82 | No | Regular | Supportive§ |
| 8 | F/53 | 7 | 121 | 3.6 | 1.6 | 1.7 | 64 | 2.1 | 51 | 96 | Budd-Chiari | Occasional | Supportive |
| 9 | F/25 | 1 | 109 | 4.5 | 2.4 | 1.8 | 323 | 9.8 | 26 | 80 | No‡ | Never | Supportive |
| 10 | F/45 | 5 | 86 | 8.7 | 7.5 | 1.0 | 143 | 17.0 | 75 | 99 | No | Occasional | Supportive |
| 11 | M/33 | 5 | 101 | 5.6 | 3.3 | 1.8 | 262 | 6.8 | 37 | 97 | No | Occasional | Supportive§ |
| 12 | F/55 | 23 | 84 | 3.9 | 2.5 | 1.0 | 185 | 10.1 | 54 | 90 | No | Regular | Supportive |
| 13 | F/32 | 3 | 78 | 4.0 | 2.2 | 1.3 | 193 | 2.4 | 91 | 69 | No | Occasional | Supportive§ |
| 14 | M/41 | 2 | 82 | 4.6 | 2.7 | 1.5 | 20 | 2.5 | 24 | 93 | No | Regular | Supportive |
| 15 | F/46 | 1 | 68 | 3.7 | 1.8 | 1.6 | 172 | 4.5 | 59 | 91 | No | Never | Supportive |
| 16 | M/25 | 5 | 79 | 5.0 | 3.6 | 1.0 | 164 | 14.1 | 16 | 84 | No | Regular | Supportive |
| 17 | M/38 | 6 | 78 | 7.0 | 6.3 | 0.6 | 327 | 9.5 | 83 | 99 | No‡ | Regular | Supportive§ |
| 18 | M/36 | 1 | 82 | 5.3 | 3.8 | 1.2 | 252 | 8.5 | 83 | 97 | No | Occasional | Supportive |
| 19 | M/65 | 6 | 112 | 7.7 | 6.7 | 0.8 | 203 | 6.7 | 35 | 67 | No | Occasional | Supportive |
| 20 | F/46 | 3 | 98 | 4.3 | 3.4 | 0.5 | 240 | 19.7 | 28 | 89 | No | Regular | Supportive |
All patients experienced recurrent episodes of hemoglobinuria.
WBC indicates white blood count; ANC, absolute neutrophil count; RBC, red blood cells; PMN, granulocytes.
The percentage of PNH PMN reflects the relative size of the PNH cell population more accurately than the percentage of PNH RBC because the latter will be grossly underestimated as a consequence of selective hemolysis; this may be further complicated by blood transfusion.
Previous prednisone treatment.
Recurrent attacks of severe abdominal pain but no documented thrombosis.
Previous antithymocyte globulin treatment.
Immunostaining and flow sorting of lymphocytes
Peripheral blood mononuclear cells (PBMNCs) were isolated by density gradient centrifugation (Lympholyte; Cedarlane Laboratories, Burlington, ON, Canada). PBMNCs were washed twice in ice-cold PBS, resuspended, and stained with fluorescein isothiocyanate (FITC)–CD57 and phycoerythrin (PE)–CD8 (Becton Dickinson, San Jose, CA) mouse antihuman monoclonal antibodies for 30 minutes on ice. After 2 washes with ice-cold PBS, the surface expression of CD8 and CD57 was examined by using a FACScan (Becton Dickinson). Live cells were gated on the basis of forward and side scatter. Background staining was determined by using appropriate isotypic controls (mouse PE-IgG2a and FITC-IgG1; Becton Dickinson). Because CD8+(bright) cells are mostly T cells (whereas CD8+(dim) cells are mostly natural killer cells),28 the percentage of the CD8+ CD57+ T-cell population among PBMNCs has been determined by counting the CD8+(bright) CD57+ cells. The absolute numbers of CD8+(bright) CD57+ T cells in peripheral blood have been calculated on the basis of their percentage and of the PBMNC counts.
The CD8+(bright) CD57+ T-cell population, stained as described in the previous paragraph, was purified by flow sorting using either FACScan and FACS-Aria (Becton Dickinson). The purity of the sorted CD8+(bright) CD57+ population was higher than 80%.
Size analysis of TCR-β gene complementary-determining region 3 (CDR3)
Total DNA was extracted from PBMNCs and from sorted CD8+ CD57+ T cells by using the QIAmp DNA Mini Kit (Qiagen, Valencia, CA) and dissolved in diethylpyrocarbonate (DEPC) water to a final concentration of 2 ng/μL.
To analyze the TCR-β CDR3 region we have used a 4-reaction multiplex polymerase chain reaction (PCR) designed to detect all possible TCR-β gene rearrangements using a set of degenerated Vβ forward primers and 13 Jβ reverse primers. For the Vβ (TRBV) and Jβ (TRBJ) genes, nomenclature was according to The International Immunogenetics Information System (IMGT).29,30 “Reactions 1 and 2” detect rearrangements using Vβ genes TRBV 2 to 5, 7, 9, 10 to 12, 14, 16, 18, 21, 23, 26. “Reactions 3 and 4” detect rearrangements using Vβ genes TRBV 1, 3, 6, 8, 10, 12, 13, 15, 17, 19, 20, 24 to 30. “Reactions 1 and 3” detect rearrangements using JB genes 1.1 to 1.6, 2.2, 2.6, 2.7, while “reactions 2 and 4” detect those using JB genes 2.1, 2.3, 2.4, and 2.5. Primer sequences were generously supplied by M. Ulivi (TibMolBio, Genoa, Italy). The PCR reaction (total volume, 30 μL) contained 10 nM dNTP, 1 unit of hot-start AmpliTaq Gold DNA polymerase with its buffer with 1.5 mM MgCl2 (Applera Europe, Nieuwerkerk aan den Ijssel, The Netherlands), 20 ng genomic DNA, and the appropriate primer mixture at a final concentration of 20 pM. PCR amplification was carried out by using the I-cycler (Bio-Rad, Hercules, CA) under the following conditions: after activation of DNA polymerase for 10 minutes at 95°C, the cycling conditions were 9 cycles (60 seconds at 94°C, 30 seconds at 62°C, and 10 seconds at 72°C), 24 cycles (40 seconds at 94°C, 30 seconds at 61°C, and 15 seconds at 72°C), 12 cycles (40 seconds at 94°C, 60 seconds at 60°C, and 20 seconds at 72°C), and a final extension of 10 minutes at 72°C. Ten microliters of each of the 4 amplified products, after denaturation (5 minutes at 95°C in 20 mM formamide followed by 10 minutes on ice), have been resolved by electrophoresis at 50 V/cm for 4 hours on a 7.5% polyacrylamide denaturing gel and detected by silver staining. After staining the gel, the products of each reaction are displayed as a ladder of bands in which each band differs by a multiple of 3 bp from the others.
Cloning and sequencing of CDR3 region
The remaining 20 μL of each of the 4 PCR products was run on 2% agarose gel and purified by using QIAEX II Gel Extraction Kit (Qiagen) according to manufacturer's instructions. The purified product was ligated into a pCR2.1-TOPO vector and transfected into TOP 10 Escherichia coli (TOPO TA cloning kit; Invitrogen, Carlsbad, CA). After kanamycin selection, at least 25 white bacterial colonies for each of the 4 PCR products were selected and expanded. Plasmid DNA was isolated by boiling and PCR amplified with M13 primers; the PCR products, after purification, were directly sequenced by using ABI PRISM BigDye Terminator v3.0 Cycle Sequencing Ready Reaction Kit (Applera Europe) and run on ABI 3130 Genetic Analyzer (Applera Europe). An average of 81 sequences have been obtained from each of the PNH patients and the healthy controls.
The CDR3 sequences have been analyzed with Chromas Lite software (version 2.0; Technelysium, Helensvale, Australia) and the amino acid translation determined. The frequency of recurrent sequences was calculated on the total number of sequences obtained from the cloning of each of the 4 PCR reactions. To identify individual CDR3s with identical or quasi-identical sequence in different patients and healthy controls, all CDR3 sequences have been aligned by using ClustalX software.31
Clonotype-specific PCR
Clonotype-specific PCR was performed on sorted CD8+ CD57+ T cells from PNH patients (n = 16) and from healthy individuals (n = 25) by using a seminested PCR strategy. For each of 3 clonotypes (S1, S4, S5; Table 2) we have designed 2 seminested sense primers specific for the appropriate Vβ family and 1 clonotype-specific antisense primer that includes a portion of the nDn region and the junction with Jβ. This design of the clonotypic primers has 2 advantages: (1) it enables the amplification of both identical and quasi-identical CDR3, and (2) the sequence of the clonotype is not completely forced by the clonotypic primer; thus, the sequencing allows the positive confirmation of the identity of the amplified fragment.
The primers were as follows. (1) Clonotype S1: sense 1, 5′-CAGGTGCTGGAGTCTCCCAG-3′; sense 2, 5′-AACCCTTTATTGGTACCGACA-3′. Clonotype-specific antisense: 5′-GGACCCGAGCAGTACTTCG-3′. (2) Clonotype S4 and S5: sense 1, 5′-GGCCCCAAAGCTGCTGTTCCAC-3′; sense 2, 5′-AGCAGACACCCCTGATAACTTCC-3′. Clonotype-specific antisense for S4: 5′-GCCCGAAGTACTGGGTCTCCC-3′. Clonotype-specific antisense for S5: 5′-GGAGAGACGCAGTTCTTC-3′.
The first step of the clonotype-specific PCR reaction (total volume, 30 μL) contained 10 nM dNTP, 1 unit of AmpliTaq DNA polymerase with its buffer with 1.5 mM MgCl2 (Applera Europe), 20 ng genomic DNA, and 0.4 μM of each primer (sense 1 and clonotype-specific antisense; see above). PCR amplification was carried out by using the I-cycler (Bio-Rad) under the following conditions: 5 minutes of initial denaturation at 95°C followed by 35 cycles (30 seconds at 94°C, 30 seconds at 57°C, and 45 seconds at 72°C) and a final extension of 7 minutes at 72°C. Five microliters of this PCR product were purified from primers by treatment with exonuclease I and shrimp alkaline phosphatase (ExoSap; USB, Cleveland, OH) and subjected to the second PCR step using the appropriate primers (sense 2 and clonotype-specific antisense; see previous paragraph) and similar PCR conditions (see above). Amplified products were separated by electrophoresis on 2% agarose gel; the identity of the amplified products after the appropriate purification was always verified by sequencing either directly or after cloning.
Statistical analysis
Data are expressed as mean ± SD. The Student t test and nonparametric Mann-Whitney have been used when appropriate. Differences were regarded as statistically significant at P < .05.
Results
Analysis of CD8+ CD57+ T-cell population
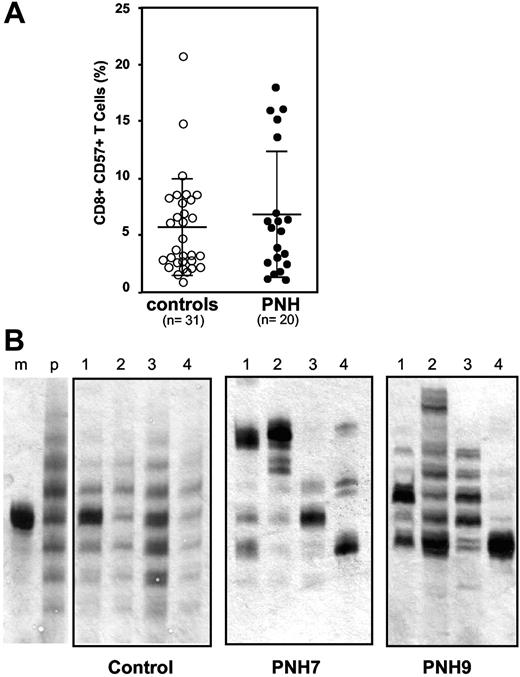
T-LGL expansion having the immunophenotype CD8+ CD57+ has been previously observed in a significant proportion of PNH patients.25,26 To further investigate the possible role of CD8+ CD57+ T cells in the pathogenesis of PNH, we first measured the size of this population in 20 hemolytic PNH patients with large PNH populations and in 31 age-matched healthy controls. The percentage of CD8+ CD57+ T cells was similar in the 2 groups: 6.9% ± 5.6% (range, 1.4% to 18.1%) versus 5.7% ± 4.3% (range, 1.0% to 21.2%) (Student t test, P = .38; Figure 1A). However, this finding does not rule out the possibility that in PNH patients this population might include abnormal and/or expanded clones. To investigate this possibility, because each CDR3 defines a TCR clonotype, we have analyzed the size distribution of the TCR-β CDR3 of the sorted CD8+ CD57+ T cells by a 4-reaction multiplex PCR designed to detect all possible TCRB gene rearrangements. This analysis revealed a gaussian distribution of the TCR-β CDR3 size in healthy controls (n = 20; a typical result is shown in the left panel of Figure 1B). In contrast, in 14 of 14 PNH patients CDR3 size analysis showed a nongaussian distribution of CDR3 length: in addition, we often observed individual heavy bands of PCR products (Figure 2). Repeat analysis carried out in 7 patients has yielded similar patterns after 3 to 6 months (data not shown). Thus, the TCR-β repertoire is skewed in CD8+ CD57+ T cells from PNH patients, suggesting that this T-cell population is oligoclonal and includes dominant clones.
Analysis of circulating CD8+ CD57+ T cells in PNH patients and in healthy controls. (A) Quantitation of CD8+ CD57+ T cells. Percentage of CD8+ CD57+ T cells within peripheral blood mononuclear cells (PBMNCs). Each circle represents 1 control subject or 1 PNH patient. Average and standard deviation are shown for the patient group and for the control group (P = .38). (B) Size distribution of the CDR3 region of the TCRB genes within the CD8+ CD57+ cell population of individual subjects. Each 4-lane panel displays the analysis, by electrophoresis on a denaturing polyacrylamide gel, of the products of 4 multiplex PCRs (lanes 1 to 4) designed to detect all possible TCRB gene rearrangements that can take place in T cells. After silver staining, each lane shows a ladder of bands differing from each other by 3 bp or a multiple of 3 bp. The 2 left lanes are for reference: lane m shows the product of the amplification, obtained with the appropriate primers, of the TCR-β CDR3 from a monoclonal population of T cells (from a patient with a T-cell leukemia); lane p shows the product of the amplification, with 1 of the 4 sets of primers, of the TCR-β CDR3 from a polyclonal T-cell population (from a healthy person). The panel labeled “control” shows, in a healthy subject, a gaussian distribution of CDR3 sizes in each of the 4 lanes. The next 2 panels illustrate the results in 2 PNH patients (Table 1). In patient PNH 7, one sees nongaussian distributions of CDR3 sizes in all 4 lanes, with 1 band predominating in each of the 4 lanes. In patient PNH 9, there is only 1 heavy band in lane 4, and in the other 3 lanes the distributions are also markedly skewed.
Analysis of circulating CD8+ CD57+ T cells in PNH patients and in healthy controls. (A) Quantitation of CD8+ CD57+ T cells. Percentage of CD8+ CD57+ T cells within peripheral blood mononuclear cells (PBMNCs). Each circle represents 1 control subject or 1 PNH patient. Average and standard deviation are shown for the patient group and for the control group (P = .38). (B) Size distribution of the CDR3 region of the TCRB genes within the CD8+ CD57+ cell population of individual subjects. Each 4-lane panel displays the analysis, by electrophoresis on a denaturing polyacrylamide gel, of the products of 4 multiplex PCRs (lanes 1 to 4) designed to detect all possible TCRB gene rearrangements that can take place in T cells. After silver staining, each lane shows a ladder of bands differing from each other by 3 bp or a multiple of 3 bp. The 2 left lanes are for reference: lane m shows the product of the amplification, obtained with the appropriate primers, of the TCR-β CDR3 from a monoclonal population of T cells (from a patient with a T-cell leukemia); lane p shows the product of the amplification, with 1 of the 4 sets of primers, of the TCR-β CDR3 from a polyclonal T-cell population (from a healthy person). The panel labeled “control” shows, in a healthy subject, a gaussian distribution of CDR3 sizes in each of the 4 lanes. The next 2 panels illustrate the results in 2 PNH patients (Table 1). In patient PNH 7, one sees nongaussian distributions of CDR3 sizes in all 4 lanes, with 1 band predominating in each of the 4 lanes. In patient PNH 9, there is only 1 heavy band in lane 4, and in the other 3 lanes the distributions are also markedly skewed.
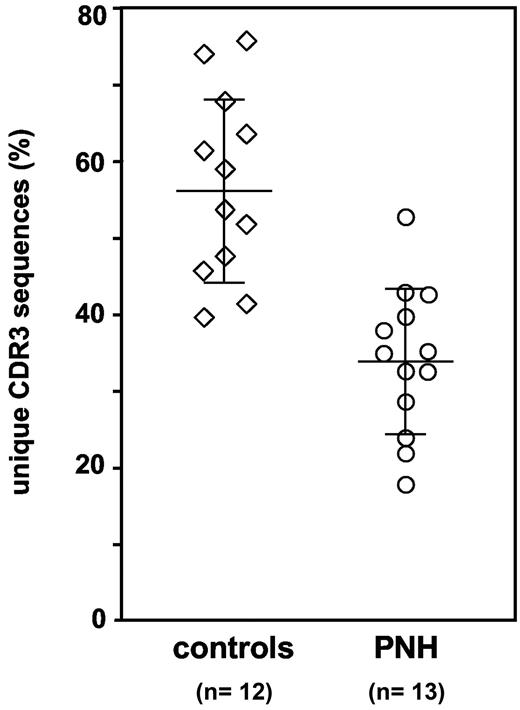
Decreased clonal diversity in CD8+ CD57+ T-cell populations from PNH patients. Percentage of different CDR3 sequences, and thus of different clones, in the CD8+ CD57+ T cells. Each symbol represents 1 healthy control subject or 1 PNH patient. Average and standard deviation are shown for the patient group (34.0% ± 9.6%) and for the control group (56.7% ± 12.1%); Mann-Whitney U test, P < .001.
Decreased clonal diversity in CD8+ CD57+ T-cell populations from PNH patients. Percentage of different CDR3 sequences, and thus of different clones, in the CD8+ CD57+ T cells. Each symbol represents 1 healthy control subject or 1 PNH patient. Average and standard deviation are shown for the patient group (34.0% ± 9.6%) and for the control group (56.7% ± 12.1%); Mann-Whitney U test, P < .001.
Clonal diversity of CD8+ CD57+ T-cell population
To confirm that in PNH patients the CD8+ CD57+ T-cell populations are oligoclonal, we have performed a systematic analysis of their TCR-β CDR3 sequences by cloning and sequencing products from each of the 4 PCR reactions that had been designed to amplify all possible TCRB gene rearrangements.
From each of 14 PNH patients and 12 healthy controls we have obtained an average of 81 sequences. Based on the fact that each CDR3 sequence identifies a T-cell clone, a measure of the clonal diversity of the CD8+ CD57+ T-cell population in each individual was obtained by dividing the number of different sequences observed by the total number of sequenced clones.
In the 12 healthy controls we have found that 56.7% ± 12.1% of sequences were different (range, 39.5% to 75.6%): this established the extent of physiological clonal diversity within the CD8+ CD57+ T-cell population. In the 13 evaluable PNH patients (1 patient has been excluded from this analysis because the yield of clones was too low) we have found that only 34.0% ± 9.6% of sequences were different (range, 17.8% to 52.7%) (Mann-Whitney U test compared with controls, P < .001; Figure 2). In addition, in 12 of 13 patients at least 1 sequence represented more than 40% of all sequences obtained. Thus, the analysis of this unbiased dataset of TCR-β CDR3 sequences has confirmed that the TCR repertoire of this T-cell population is less diverse in PNH patients than in controls.
Different PNH patients share certain TCR-β CDR3 sequences
Because each TCR-β CDR3 sequence identifies a T-cell clone, it is called a clonotype. Given the large diversity of the physiologic TCR repertoire, the detection of identical or quasi-identical clonotypes in more than 1 PNH patient would be consistent with an immune response driven by the same antigen. To test whether indeed the same clonotypes are shared by different individuals, we have aligned all the TCR-β CDR3 sequences (after translation into amino acid sequences) that we observed in both PNH patients and healthy controls.
None of the healthy controls shared identical or quasi-identical clonotypes with others controls (n = 12) or with PNH patients (n = 14). In contrast, we have identified 5 TCR-β CDR3 sequences shared by at least 2 PNH patients (Table 2). Each of the sequences S1 (TRBV-7.9 and TRBJ-2.7), S2 (TRBV-15 and TRBJ-1.2), and S3 (TRBV-30 and TRBJ-1.1) have been found in 2 patients (Table 2). Three patients (patients 5, 7, and 9) shared the TCR-β CDR3 S4, produced by a rearrangement involving TRBV-15 and TRBJ-2.1. In each of the same 3 patients we have also found a sequence (S5) that differs from S4 by only 2 of 7 amino acids in the nDn region: this sequence has the same TCR Vβ gene (TRBV-15) but a different TCR Jβ gene (TRBJ-2.5). In 1 of these 3 patients we found an additional CDR3 (S5a) that differs by 5 nucleotides and by only 1 amino acidic residue. In addition, in a fourth patient (patient 2) we found a CDR3 (S5b) that differs from S5 by 4 nucleotides and by 1 amino acidic residue (a threonine replaces a valine).
Shared TCR-β CDR3 sequences found in PNH patients by systematic cloning and sequencing
| Sequence ID . | Patients . | TRBV . | TCR-β CDR3 . | TRBJ . | ||
|---|---|---|---|---|---|---|
| Vβ . | nDn . | Jβ . | ||||
| S1 | 7, 9 | 7.9 | CASS | LVGGPEQ | YFGP | 2.7 |
| S2 | 6, 16 | 15 | CATS | RGRTQGLDYG | YTFG | 1.2 |
| S3 | 8, 19 | 30 | CA | WEQVIA | FFGQ | 1.1 |
| S4 | 5, 7, 9 | 15 | CATS | GIAGETQ | FFGP | 2.1 |
| S5 | 5, 7, 9 | 15 | CATS | RVAGETQ | YFGP | 2.5 |
| S5a | 9 | 15 | CATS | RIGGETQ | YFGP | 2.5 |
| S5b | 2 | 15 | CATS | RTAGETQ | YFGP | 2.5 |
| Sequence ID . | Patients . | TRBV . | TCR-β CDR3 . | TRBJ . | ||
|---|---|---|---|---|---|---|
| Vβ . | nDn . | Jβ . | ||||
| S1 | 7, 9 | 7.9 | CASS | LVGGPEQ | YFGP | 2.7 |
| S2 | 6, 16 | 15 | CATS | RGRTQGLDYG | YTFG | 1.2 |
| S3 | 8, 19 | 30 | CA | WEQVIA | FFGQ | 1.1 |
| S4 | 5, 7, 9 | 15 | CATS | GIAGETQ | FFGP | 2.1 |
| S5 | 5, 7, 9 | 15 | CATS | RVAGETQ | YFGP | 2.5 |
| S5a | 9 | 15 | CATS | RIGGETQ | YFGP | 2.5 |
| S5b | 2 | 15 | CATS | RTAGETQ | YFGP | 2.5 |
Identical and similar TCR-β CDR3 sequences identified in different PNH patients by cloning and sequencing of the TCR-β repertoire of CD8+ CD57+ T cells. Nomenclature of Vβ (TRBV) and Jβ (TRBJ) genes is according to the International Immunogenetics Information System.30 Patients are numbered as in Table 1.
TCR-β CDR3 sequences from some PNH patients are found in others by clonotype-specific PCR amplification
To find out whether previously identified TCR-β CDR3 clonotypes are present in additional patients, we have designed clonotype-specific PCR methodology. For the 3 clonotypes S1, S4, and S5 (Table 2) we have designed 2 seminested sense primers specific for the appropriate Vβ gene and a clonotype-specific antisense primer that includes a portion of the nDn region and the junction with the appropriate Jβ gene. These primers do not overlap the entire V-nDn-J junction and therefore have 2 advantages: (1) they enable the amplification of either identical or quasi-identical CDR3, and (2) the primer does not force amplification of the entire clonotypic sequence; thus, we can obtain positive confirmation of the identity of the amplified fragment by sequencing.
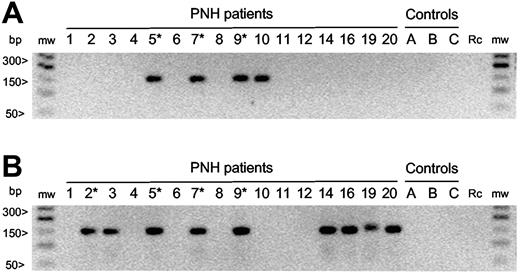
By this approach we have searched for clonotypes S1, S4, and S5 in sorted CD8+ CD57+ T cells from 16 PNH patients and 25 healthy controls. First, we have confirmed the presence of these clonotypes in those PNH patients in which they had been originally found by cloning and sequencing. In addition, by clonotypic PCR we have detected the presence of all these sequences in the same patients during follow-up. None of these clonotypes have been detected in the 25 healthy controls. Clonotype S1 has not been found in PNH patients other than that in which it had been originally found (data not shown). Clonotype S4 has been found in a fourth patient (10; Figure 3A), and clonotype S5 has been found in 4 additional patients (Figure 3B). The identity of all fragments amplified by clonotype-specific PCR has been confirmed by sequencing. Thus, we found that the clonotype detected by S4-specific clonotypic PCR in patient 10 was identical to that found in the original 3 patients, whereas the S5-specific clonotypic PCR has identified a set of identical or quasi-identical clonotypes (Table 3). Because the clonotype S4 and the S5-like clonotypes have a high degree of homology, 9 of 16 PNH patients share identical or quasi-identical TCR-β CDR3.
Clonotype-specific PCR amplification. Seminested PCR was carried out with clonotype-specific primers that, because they do not overlap the entire V-nDn-J junctions, enable the amplification of either identical or quasi-identical CDR3 sequences. Numbers identify PNH patients as from Table 1. Capital letters indicate healthy controls; *, patients in whom the clonotype had been previously identified by systematic cloning and sequencing; mw, molecular weight markers; rc, reagent control. (A) Clonotype-specific PCR for S4 clonotype. A specific S4 clonotype band is present in the 3 patients in whom it had been previously found by systematic cloning and sequencing (PNH 5, 7, 9; Table 2) and in 1 additional patient (PNH 10). The sequencing of the amplified products confirmed the identity of the sequence. (B) Clonotype-specific PCR for S5-like clonotype. A specific S5-like clonotype band is present in the 4 patients in whom it had been previously found by systematic cloning and sequencing (PNH 2, 5, 7, 9; Table 2) and in 4 additional patients (PNH 3, 14, 16, 20). The sequencing of the amplified products demonstrates the presence of both S5-identical and S5 quasi-identical sequences (Table 3). In addition, the PCR amplification in patient 19 yielded a longer PCR product whose sequence (CATSRGTSGRETQYFGP) revealed a CDR3 that uses the same TRBV-15 and TRBJ-2.5 of S5-like clonotypes but with a 6 nucleotide insertion that modifies significantly the nDn region. Therefore, this sequence has not been counted as belonging to the S5-like clonotype group.
Clonotype-specific PCR amplification. Seminested PCR was carried out with clonotype-specific primers that, because they do not overlap the entire V-nDn-J junctions, enable the amplification of either identical or quasi-identical CDR3 sequences. Numbers identify PNH patients as from Table 1. Capital letters indicate healthy controls; *, patients in whom the clonotype had been previously identified by systematic cloning and sequencing; mw, molecular weight markers; rc, reagent control. (A) Clonotype-specific PCR for S4 clonotype. A specific S4 clonotype band is present in the 3 patients in whom it had been previously found by systematic cloning and sequencing (PNH 5, 7, 9; Table 2) and in 1 additional patient (PNH 10). The sequencing of the amplified products confirmed the identity of the sequence. (B) Clonotype-specific PCR for S5-like clonotype. A specific S5-like clonotype band is present in the 4 patients in whom it had been previously found by systematic cloning and sequencing (PNH 2, 5, 7, 9; Table 2) and in 4 additional patients (PNH 3, 14, 16, 20). The sequencing of the amplified products demonstrates the presence of both S5-identical and S5 quasi-identical sequences (Table 3). In addition, the PCR amplification in patient 19 yielded a longer PCR product whose sequence (CATSRGTSGRETQYFGP) revealed a CDR3 that uses the same TRBV-15 and TRBJ-2.5 of S5-like clonotypes but with a 6 nucleotide insertion that modifies significantly the nDn region. Therefore, this sequence has not been counted as belonging to the S5-like clonotype group.
Closely related TCR-β CDR3 sequences found within individual patients and in different patients with PNH
| Sequence ID . | Patients . | TRBV . | TCR-β CDR3 . | TRBJ . | ||
|---|---|---|---|---|---|---|
| Vβ . | nDn . | Jβ . | ||||
| S4 | 5,* 7,* 9,* 10† | 15 | CATS | GIA GETQ | FFGP | 2.1 |
| S5 | 5,* 7,* 9,* 14† | 15 | CATS | RVA GETQ | YFGP | 2.5 |
| S5a | 9,* 20† | 15 | CATS | RTG GETQ | YFGP | 2.5 |
| S5b | 2* | 15 | CATS | RTA GETQ | YFGP | 2.5 |
| S5c | 5,† 16† | 15 | CATS | RIA GETQ | YFGP | 2.5 |
| S5d | 3† | 15 | CATS | RVG GETQ | YFGP | 2.5 |
| S5e | 5† | 15 | CATS | REI GETQ | YFGP | 2.5 |
| S5f | 5† | 15 | CATS | RDLAGETQ | YFGP | 2.5 |
| Sequence ID . | Patients . | TRBV . | TCR-β CDR3 . | TRBJ . | ||
|---|---|---|---|---|---|---|
| Vβ . | nDn . | Jβ . | ||||
| S4 | 5,* 7,* 9,* 10† | 15 | CATS | GIA GETQ | FFGP | 2.1 |
| S5 | 5,* 7,* 9,* 14† | 15 | CATS | RVA GETQ | YFGP | 2.5 |
| S5a | 9,* 20† | 15 | CATS | RTG GETQ | YFGP | 2.5 |
| S5b | 2* | 15 | CATS | RTA GETQ | YFGP | 2.5 |
| S5c | 5,† 16† | 15 | CATS | RIA GETQ | YFGP | 2.5 |
| S5d | 3† | 15 | CATS | RVG GETQ | YFGP | 2.5 |
| S5e | 5† | 15 | CATS | REI GETQ | YFGP | 2.5 |
| S5f | 5† | 15 | CATS | RDLAGETQ | YFGP | 2.5 |
Identical and quasi-identical TCR-β CDR3 sequences identified in different PNH patients by (1) “cloning and sequencing”* or (2) by clonotypic-specific PCR.† Nomenclature of Vβ (TRBV) and Jβ (TRBJ) genes is according to the International Immunogenetics Information System.30 Patients are numbered as in Table 1, and sequence ID is as identified in Table 2.
Identical and quasi-identical TCR-β CDR3 sequences identified in different PNH patients by “cloning and sequencing.”
Identical and quasi-identical TCR-β CDR3 sequences identified in different PNH patients by clonotypic-specific PCR.
Discussion
The presence of a large population of blood cells deficient in GPI-linked molecules, which we call PNH cells, is the main determinant of the most characteristic features of PNH—namely, intravascular hemolysis and thrombosis. Rare PNH blood cells harboring PIG-A mutations are present in healthy people, and this does not result from an increased rate of somatic mutations.32 Therefore, the major outstanding question in the pathogenesis of PNH is what determines the expansion of a PNH clone.
In principle, one can envisage 2 mechanisms. On one hand, (1) expansion may result from an acquired somatic mutation, other than the PIG-A mutation, that confers to the clone a growth advantage. On the other hand, (2) expansion may be the consequence of a selective immune attack against normal (GPI+) HSCs to which PNH (GPI−) HSCs are invulnerable. The 2 possibilities are not mutually exclusive.
The first mechanism has been recently exemplified33 by 2 patients with PNH in whom an acquired rearrangement of chromosome 12 produces ectopic expression of the HMG2 gene, which might favor growth. It will be interesting to determine whether HMG2 is up-regulated, by some other mechanism, in PNH patients who do not have a chromosome 12 abnormality. However, this finding does not justify the failure in the bone marrow of normal hematopoiesis (documented as pre-existent in 1 of the 2 patients), which would still require a separate explanation.
Here we are concerned with the second possibility. A large body of evidence links PNH to IAA,34–36 and because the latter is thought to be a cell-mediated autoimmune disease,36,37 the same may be true of the former.21,38 Only recently, however, have we had clues that have helped to circumscribe the nature of suspect autoreactive T cells. In PNH patients the TCR-β repertoire is skewed22 and the degree of skewness is even greater among CD8+ T cells.23 The expansion of CD3+ CD8+ CD57+ LGLs can be considerable25 and relatively frequent26 (L.L. and R.N., unpublished data, December 2004), and most of these cells express inhibitory receptor superfamily (IRS) molecules that, in contrast to what applies to healthy subjects, belong to the group of activating IRS isoforms.27
Based on these data we have systematically analyzed CD8+ CD57+ T cells in patients suffering from full-blown hemolytic PNH. This subset of lymphocytes, although similar in size to that found in healthy controls (Figure 1A), showed consistently a nongaussian distribution of TCRB gene CDR3 size (Figure 1B), suggesting that it is oligoclonal and that it may include abnormal and/or expanded T-cell clones. In addition, an analysis of the TCR-β CDR3 sequences (clonotypes) carried out by shotgun cloning and sequencing of the entire TCRB gene repertoire has shown that in PNH patients the proportion of different clonotypes, and thus of T-cell clones, is reduced (34.0% versus 56.7%; Mann-Whitney U test, P < .001; Figure 2), further confirming the reduced clonal diversity of the CD8+ CD57+ T-cell population.
An autoimmune attack against GPI+ HSCs implies the presence of autoreactive T cells able to recognize a molecule present on GPI+ HSCs but not on GPI− HSCs. There must be a limited range of such molecules; therefore, there may be a limited range of CDR3 sequences that recognize them and, significantly, we have found recurrently identical or quasi-identical sequences in different PNH patients but not in healthy controls: 5 clonotypes shared by at least each of 2 PNH patients (Table 2). Interestingly, 3 of these shared clonotypes (present in 10 of 16 patients; Tables 2 and 3) use the TRBV15 gene (Vβ-24 in the Arden nomenclature29 ), which belongs to 1 of the 4 TCR-Vβ families we have previously found to be more frequently skewed in a separate series of 19 PNH patients.22
These findings are in good agreement with previous work by Plasilova et al,39 who have analyzed CD8+ T cells belonging to TCR-Vβ families that appeared to be expanded by using Vβ family-specific antibodies. Nine TCR-β CDR3 sequences were obtained and, by using clonotypic PCR in 19 PNH patients, it was found that 2 of these were shared by 2 patients, 1 by 3 patients, and 1 by 4 patients as well as 1 of 8 control subjects. On the other hand, we have found a higher frequency of PNH patients sharing some clonotypes (11 of 16; Tables 2 and 3) and, even by using a highly sensitive methodology, we have not detected any of these clonotypes in healthy controls (n = 25). There may be several reasons to account for both of these discrepancies. First, we have included only patients with florid hemolytic PNH. Second, we have analyzed selectively CD8+ CD57+ T lymphocytes because of independent existing evidence suggesting that they may be involved in the pathogenesis of PNH (see “Introduction”). Third, because potently autoreactive T-cell clones need not be large, we have used an unbiased cloning and sequencing strategy able to pick up CDR3 sequences regardless of expansion of the respective clones. By our approach we have found in our patients clonotypic sequences that were remarkably related in several ways. First, 3 patients shared each of 2 sequences (obtained by shotgun cloning), and these 2 (S4 and S5) were in turn quasi-identical (Table 2). Second, another quasi-identical sequence was found in a fourth patient (Table 2), and sequences identical or quasi-identical to S4 and S5 were found by clonotypic PCR in 5 additional patients (Table 3). Thus, 9 of 16 patients share the same group of highly homologous clonotypes. Given that the estimated diversity of TCR CDR3 sequences is of the order of millions, these findings cannot be due to chance. On the other hand, we have not found significant homology of any of these sequences with those previously reported in PNH39 or in other related diseases (IAA40 and T-LGL41 ). Most importantly, by using a very sensitive technique (seminested clonotypic PCR followed by sequencing), we have never found any of the above sequences in any of 25 healthy controls.
Given the close relationship between PNH and IAA, our findings are in apparent contrast with the report that in 54 patients with IAA sequence analysis of the TCR-β CDR3 of expanded TCR-Vβ families has yielded no identical clonotypes.40 However, this discrepancy is not at all in conflict with the notion that autoreactive T cells may be important in the pathogenesis of both diseases. Indeed, specific T-cell clones may be responsible for damage to normal HSCs in both PNH and IAA. However, in IAA a variety of different autoantigens may be involved, whereas in PNH the range of potential target antigens is much more restricted, because they must be present on normal HSCs but not on PNH HSCs, thus enabling them to survive the autoimmune attack and to expand.
The finding that within the CD8+ CD57+ T-cell population identical or quasi-identical clonotypes are regularly present in PNH patients, and that a group of highly homologous clonotypes (S4 and S5-like) is shared by more than half of hemolytic PNH patients, is consistent with an immune process driven by one and the same antigen (or a set of very closely related antigens). This notion is corroborated further by the finding in few patients (patients 5, 7, and 9; Table 3) of different clones (up to 5 in number) bearing closely related clonotypes–for example, clonotypes S4, S5, S5c, S5e, and S5f in patient 5. These facts, together with the persistence of these shared clonotypes during follow-up, support strongly the hypothesis that an immune response directed against such antigen(s) may play a central role in the pathogenesis of PNH.
Unfortunately, the identity of the putative antigenic molecule that is the target of these clonotypes remains as yet unknown. Because PNH patients that share identical or quasi-identical clonotypes do not all share the same HLA-A or HLA-B alleles (Table 4), and because recognition by TCR of peptide antigens is major histocompatibility complex (MHC) restricted, we infer that the antigen in question is not a peptide. On the other hand, T lymphocytes may respond to nonpeptide antigens, and lipids are presented to T cells by the CD1 molecules, a class of nonpolymorphic, β2-microglobulin–associated, MHC I–like proteins.42,43 A member of the CD1 family, CD1d, presents glycolipids to specific T-cell subsets44,45 ; and the GPI molecule is one of its possible ligands.46,47 In fact, it was suggested some years ago that in PNH the target of autoimmune T cells might be the GPI molecule itself, presented in the context of CD1d48 ; and it is now known that a subset of immature HSCs expresses CD1d.49
HLA-A and HLA-B of PNH patients that share related TCR-β CDR3 sequences
| PNH patients . | HLA-A . | HLA-B . |
|---|---|---|
| 2 | 03, 11 | 07, 35 |
| 3 | 01, 26 | 14, 38 |
| 5 | 01, 11 | 15, 35 |
| 6 | 02, 03 | 44, 51 |
| 7 | 02, 32 | 07, 51 |
| 8 | ND | ND |
| 9 | 11, 24 | 39, 57 |
| 10 | 24, 33 | 14, 49 |
| 14 | 01, 19 | 12, 53 |
| 16 | 29, 32 | 08, 40 |
| 20 | 03, 24 | 07, 51 |
| PNH patients . | HLA-A . | HLA-B . |
|---|---|---|
| 2 | 03, 11 | 07, 35 |
| 3 | 01, 26 | 14, 38 |
| 5 | 01, 11 | 15, 35 |
| 6 | 02, 03 | 44, 51 |
| 7 | 02, 32 | 07, 51 |
| 8 | ND | ND |
| 9 | 11, 24 | 39, 57 |
| 10 | 24, 33 | 14, 49 |
| 14 | 01, 19 | 12, 53 |
| 16 | 29, 32 | 08, 40 |
| 20 | 03, 24 | 07, 51 |
Patients are numbered as in Table 1.
ND indicates not done.
In conclusion, in most PNH patients we have demonstrated a CD8+ CD57+ T-cell population bearing a set of highly homologous TCR-β molecules that is likely to recognize the same nonpeptide antigen. These data provide strong new support to the hypothesis that the expansion of the GPI− blood cell population in PNH is due to selective damage to GPI+ HSCs mediated by an autoimmune attack against the GPI anchor or a closely related molecule.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by a grant from Associazione Italiana Ricerca sul Cancro (L.L., R.N.) and grants from the Fondo Investimenti Ricerca di Base–Ministero dell'Università e della Ricerca (MIUR) (L.L., R.N., S.Z.).
We thank D. Reverberi for helpful technical suggestions. We are grateful to D. Rapezzi and O. Racchi for helping with patients. We are grateful to M. De Angioletti and P. Piccioli for helpful suggestions, stimulating discussions, and support.
Authorship
Contribution: L.G. participated in designing and performing the research, analyzing data, and writing the paper; S.L., M.S., and F.L. participated in performing the research; G.C. and S.Z performed CDR3 size analysis; and L.L and R.N. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosario Notaro, Laboratory of Genetics and Gene Transfer, Core Research Laboratory, Istituto Toscano Tumori (ITT-CRL), viale Pieraccini 6 (Cubo), 50139 Florence, Italy; e-mail: notaror@yahoo.it.