Abstract
Patients with advanced stages of chronic myeloid leukemia (CML) often manifest imatinib mesylate resistance associated with point mutations in BCR-ABL. AMN107 is a new higher-potency inhibitor of BCR-ABL. To identify mutations in BCR-ABL that could result in resistance to AMN107, a cDNA library of BCR-ABL mutants was introduced into Ba/F3 cells followed by selection in AMN107 (0.125-0.5 μM). A total of 86 individual, drug-resistant colonies were recovered, and the SH3, SH2, and kinase domains of BCR-ABL were sequenced. A total of 46 colonies had single point mutations in BCR-ABL, with a total of 17 different mutations, all within the kinase domain. The other 40 colonies had multiple point mutations and were not analyzed further. Each of the 17 single point mutants were reconstructed by site-directed mutagenesis of native BCR-ABL and found to be approximately 2.5- to 800-fold more resistant to AMN107 than native BCR-ABL. The mutations included 6 known imatinib mesylate–resistant mutations, including T315I, which showed complete resistance to AMN107. Interestingly, most AMN107-resistant mutants were also resistant to imatinib mesylate. These results may predict some of the resistance mutations that will be detected in clinical trials with this kinase inhibitor.
Introduction
Chronic myelogenous leukemia (CML) is caused by the fusion oncogene BCR-ABL arising from a reciprocal t(9;22) chromosome translocation in hematopoietic stem cells.1,2 This fusion gene encodes a chimeric BCR-ABL protein, in which the tyrosine kinase activity of Abl is constitutively activated. BCR-ABL–mediated unregulated phosphorylation of intracellular proteins results in growth-factor–independent proliferation, reduced differentiation, and enhanced survival of hematopoietic stem cells. A 210-kDa BCR-ABL protein (p210) is expressed in 95% of patients with CML, and a smaller 190 kDa BCR-ABL in approximately 33% of patients with acute lymphoblastic leukemia (ALL).3,4 However, approximately one-third of patients with ALL also express the p210 form of the protein.
Imatinib mesylate (Gleevec, STI571; Novartis Pharma AG, Basel, Switzerland) is a drug that targets the tyrosine kinase activity of BCR-ABL5 and is very effective in the treatment of CML.6 However, in genetically unstable, advanced-stage CML, highly specific protein tyrosine kinase inhibition can lead to a problem in that the target protein can develop escape mutations that lead to drug resistance. In fact, many patients with advanced stages of CML ultimately manifest imatinib mesylate resistance.7–9 Similar tyrosine kinase inhibitor drug resistance has also been reported for imatinib mesylate in gastrointestinal stromal tumor (GIST) and for gefitinib (AstraZeneca, London, United Kingdom) in non–small-cell lung cancer.10 In CML, although relapse is occasionally linked with amplification of the BCR-ABL gene,7,11,13–15 it is more often associated with mutations in BCR-ABL.11,12 Point mutations within the BCR-ABL kinase domain interfere with drug binding while retaining the capability to bind ATP and catalyze protein phosphorylation, and constitute the major mechanism of resistance detected in patients.11,14 Thus, there is a need for more potent BCR-ABL tyrosine kinase inhibitors, which minimize the number of residual BCR-ABL+ cells capable of developing resistance and which maintain activity against imatinib mesylate-resistant BCR-ABL mutants.
AMN107 is a new, highly selective Abl kinase inhibitor, which is substantially more potent than imatinib mesylate as an inhibitor of the BCR-ABL tyrosine kinase activity in cellular systems.12,16 AMN107 has also demonstrated efficacy in experimental models of BCR-ABL+ leukemias in mice.12 This compound is currently undergoing phase 2 clinical trials and has demonstrated activity in patients with advanced CML resistant to imatinib mesylate. In phase 1 studies, responses were achieved in patients possessing G250E, M351T, E355G, Y253F, F311L, F317L, F359V, H396P, H396R, and E459Q.17 However, it can be predicted that BCR-ABL mutants could also arise that would confer resistance to AMN107. The identification of all of these possible mutations is important for analysis of clinical trial data, and could contribute to the future design of next generation Abl inhibitors. Here, following the general strategies of Azam et al18 and Cools et al,19 we report a cell culture–based system to identify the spectrum of mutations in BCR-ABL that confer resistance to AMN107.
Materials and methods
General strategy
In order to generate libraries of mutagenized BCR-ABL cDNAs, the target cDNA was cloned into a retroviral vector. The library of mutations was generated in the target gene by propagating the vector through a bacterial strain deficient in 3 major DNA repair pathways, namely mutS (error-prone mismatch repair), mutD (deficient in 3′- to 5′-exonuclease of DNA polymerase III), and mutT (unable to hydrolyze 8-oxodGTP) mutations. Murine Ba/F3 cells were then transfected with the mutated vector, and AMN107-resistant colonies were isolated under different concentrations of AMN107 to select for resistance. Genomic DNA was recovered and sequenced. Possible resistance mutations were then recreated by site-directed mutagenesis, and stable cell lines expressing the mutant forms of BCR-ABL were generated. Subsequently, the resistance phenotype of the mutations was characterized by proliferation assays in the presence of AMN107.
Retroviral construct and mutagenesis
A full-length BCR-ABL cDNA was cloned into pMSCV-IRES-GFP retroviral vector,20 generating MSCV-IRES-GFP-B/A. XL-1 Red competent Escherichia coli cells (Stratagene, La Jolla, CA) deficient in DNA repair mechanisms were used for transformation, according to the recommended Stratagene protocol guide. The basics steps of mutagenesis, such as bacterial transformation and DNA isolation, were performed following Azam et al18 and Cools et al.19
Retroviral transduction and infection of Ba/F3 cells
Retrovirus was generated by transfecting HEK 293T cells (5 × 105 cells per plate, cultured in DMEM medium containing 10% FCS, pen/strep, and 2 mM l-glutamine) with either the mutagenized BCR-ABL cDNA library or a native BCR-ABL cDNA in the presence of FuGENE-6 (Roche, Indianapolis, IN) and the retroviral packaging construct EcoPAC (Cell Genesys, Redwood City, CA).19 The cDNA and the packaging construct were added in a 1:1 ratio. The viral supernatant was collected 24, 48, 72, and 96 hours after transfection, and concentrated by centrifugation at 36 026 g for 3 hours at 4°C. The concentrated pellet was resuspended in 10% HBSS buffer in PBS (200 μL for 25 mL of supernatant) and used for infection of Ba/F3 cells.
Ba/F3 cells, a murine pro-B cell line, need IL-3 for growth and viability. Expression of BCR-ABL results in IL-3 (growth-factor) independence.12 BaF3 cells were grown in RPMI medium containing 10% FCS, pen/strep, 2 mM l-glutamine, and 15% WEHI-3B–conditioned medium (a source of IL-3). With a range of 105 to 108 IU/mL of retroviral supernatant, we infected 1 × 106 BaF3 cells in the presence of 8 μg/mL polybrene (Sigma-Aldrich, St Louis, MO). Following an overnight incubation at 37°C, 5% CO2, selection for BCR-ABL–expressing cells was initiated by the removal of IL-3 from the medium. The cells were incubated in IL-3–free medium for 4 to 5 days before exposure to AMN107.
Selection of Ba/F3 cells resistant to AMN107
A total of 6 different retroviral libraries of mutagenized BCR-ABL were prepared, and each was separately used to infect Ba/F3 cells (1 × 106 cells) in 6 separate experiments. One day after infection, IL-3 was removed from the medium, and the cells were cultured in flasks for 4 to 5 days. At this point, the remaining cells were divided into 3 equal aliquots and exposed to AMN107 at a concentration of 0.125, 0.25, or 0.5 μM, respectively, and cultured in flasks for an additional 3 days. At this point, viable cells were counted and transferred to 96-well plates at an average of 1 cell per well. Ultimately, 86 wells had outgrowth of clones of Ba/F3 cells, out of approximately 14 400 wells plated.
Sequencing and alignment
Expanded colonies were harvested, and genomic DNA was isolated using the DNeasy kit (Qiagen, Valencia, CA). Specific subregions of BCR-ABL cDNA were amplified by high-fidelity polymerase chain reaction (PCR) from genomic DNA on a Stratagene Autocycler (Robocycler Gradient 40) using the following primer-pairs: (1) SH3-SH2-upper, 5′-CTCCAGACTGTCCACAGCATTCC-3′, and SH3-SH2-lower, 5′-CTGTCATCAACCTGCTCAGGC-3′; 2785 nt to 3916 nt (Bcr-Abl–spanning region to amino acid residue G380); (2) SH2-kinase-upper, 5′-CGTGAGAGTGAGAGCAGTCC-3′, and SH2-kinase-lower, 5′-GCAGCTCTCCTGGAGGTCCTCG-3′; 3202 nt to 4364 nt (amino acid residue R152 to A539); and (3) BCR-ABL–spanning region-upper and Bcr-Abl–spanning region-lower, 1890 nt to 3200 nt (Bcr domain to V151). The PCR products were sequenced and sequencing results were analyzed by DNASTAR (DNASTAR, Madison, WI) and Vector NTI (Invitrogen, Carlsbad, CA).
Generation of BCR-ABL mutants by site-directed mutagenesis and generation of stable Ba/F3 cell lines with these mutations
Site-directed mutagenesis was performed on the pMSCV-IRES- B/A-GFP plasmid using the QuiKchange XL Mutagenesis Kit (Strategene). Specific oligonucleotide primers were designed to create the point mutations found in the screen. In all cases, individual point mutants were confirmed by sequence analysis. Stable Ba/F3 lines were generated by using retroviral infection as described with the appropriate mutated plasmid. At 48 hours after infection, infected Ba/F3 cells were washed with PBS and sorted for GFP+ cells in a Beckman cell sorter (Beckman Coulter, Miami, FL). Sorted cells were further allowed to expand in RPMI 1640 plus 10% FCS medium (without the IL-3) to provide the stable mutant Ba/F3 cell lines.
Cell proliferation studies
The effect of the inhibitor on cell viability (and proliferation) was studied by treating the mutant Ba/F3 cells with increasing concentrations of AMN107 (dissolved in ultrapure DMSO) for 72 hours (in triplicate). Exponentially growing Ba/F3 (5 × 104/mL) were plated in each well of a 24-well dish with 2 mL of RPMI medium 1640/10% FCS containing the appropriate drug as indicated.
Control experiments were also performed by simultaneously treating the cells with an equivalent amount of DMSO vehicle. Viable cells were counted in a hemacytometer using trypan blue exclusion (Trypan Blue solution; Sigma-Aldrich, St Louis, MO). In some experiments, cell proliferation was measured using an MTS assay (using the Cell Titer 96 AQueous One Solution Cell Proliferation Assay kit; Promega, Madison WI). The drug concentration resulting in 50% cell viability was scored as the cellular IC50.
Structural modeling
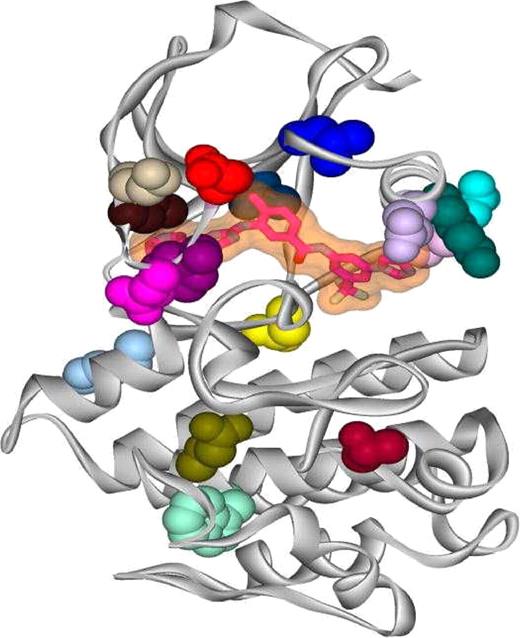
The potential structural changes associated with all of the isolated mutations were analyzed using the molecular graphics program O (DatOno, Uppsala, Sweden).21 Using the crystal structure of AMN107 in complex with Abl kinase,12 each of the side-chain mutations were modeled, and structural effects were estimated based on comparisons with other Abl kinase structures, such as the wild-type kinase domain in complex with imatinib mesylate,22,23 the assembled inactive state (SH3-SH2-kinase domains) in complex with PD166326,24 and an active conformation of the Abl kinase domain in complex with a staurosporin analog (S.W.C.-J. et al, unpublished data). The locations of all the single site mutations described in the work reported here are shown in Figure 2.
Results
Random mutagenesis screen for Bcr-Abl mutations
We conducted a saturation mutagenesis screening for AMN107-resistant BCR-ABL subclones, using methods previously described by Azam et al18 and Cools et al19 for imatinib mesylate against BCR-ABL and PKC412 against Flt3, respectively. The approach uses a DNA repair–deficient E coli strain to produce random mutagenesis of a BCR-ABL retroviral plasmid, infection of Ba/F3 cells, and selection for Ba/F3 clones conferring varying degrees of drug resistance. We chose 0.125, 0.25, and 0.5 μM concentrations of AMN107 (“Materials and methods”). The resistant colonies grew up after a median of 17 days (range, 10-25 days). In a series of screens with 6 independently derived mutagenized libraries, we isolated 86 individual colonies from our selection, ultimately resulting in the identification of 17 point mutations that confer resistance to AMN107 (Table 1). As stated earlier, the colonies with multiple point mutations were not analyzed further (see the supplemental information available on the Blood website; click the Supplemental Materials link at the top of the online article).
Properties of different point mutations
| AMN107-resistant BCR-ABL mutant . | Concentration at recovery, nM . | No. of colonies . | Percentage of total* . | IC50 of AMN107, nM . | IC50 of imatinib mesylate, nM . |
|---|---|---|---|---|---|
| Wild-type | — | — | — | < 10 | 450 |
| K247N | 125 | 1 | 1.16 | 90 | ≈850 |
| L248V | 250 | 1 | 1.16 | 675 | ≈10 000 |
| Q252H | 125 | 2 | 2.32 | 40 | ≈1 300 |
| Y253C | 250 | 8 | 9.30 | 600 | ≈10 000 |
| Y253H | 500 | 7 | 8.14 | 1300 | ≈10 000 |
| E255K | 250 | 3 | 3.49 | 200 | ≈10 000 |
| L273F | 125 | 1 | 1.16 | 51 | 750 |
| E282K | 250 | 2 | 2.32 | 150 | ≈1 550 |
| K285N | 250 | 2 | 2.32 | ≈500 | ≈3 500 |
| V289L | 125 | 1 | 1.16 | 25 | 1 000 |
| E292K | 125 | 1 | 1.16 | 90 | ≈1 500 |
| N297T | 125 | 1 | 1.16 | 30 | 650 |
| T315I | 500 | 11 | 12.79 | ≈8000 | ≈10 000 |
| H375P | 125 | 1 | 1.16 | 50 | 1 100 |
| T406I | 125 | 1 | 1.16 | 80 | ≈1 400 |
| W430L | 125 | 2 | 2.32 | 90 | 800 |
| E431G | 125 | 1 | 1.16 | 35 | ≈650 |
| AMN107-resistant BCR-ABL mutant . | Concentration at recovery, nM . | No. of colonies . | Percentage of total* . | IC50 of AMN107, nM . | IC50 of imatinib mesylate, nM . |
|---|---|---|---|---|---|
| Wild-type | — | — | — | < 10 | 450 |
| K247N | 125 | 1 | 1.16 | 90 | ≈850 |
| L248V | 250 | 1 | 1.16 | 675 | ≈10 000 |
| Q252H | 125 | 2 | 2.32 | 40 | ≈1 300 |
| Y253C | 250 | 8 | 9.30 | 600 | ≈10 000 |
| Y253H | 500 | 7 | 8.14 | 1300 | ≈10 000 |
| E255K | 250 | 3 | 3.49 | 200 | ≈10 000 |
| L273F | 125 | 1 | 1.16 | 51 | 750 |
| E282K | 250 | 2 | 2.32 | 150 | ≈1 550 |
| K285N | 250 | 2 | 2.32 | ≈500 | ≈3 500 |
| V289L | 125 | 1 | 1.16 | 25 | 1 000 |
| E292K | 125 | 1 | 1.16 | 90 | ≈1 500 |
| N297T | 125 | 1 | 1.16 | 30 | 650 |
| T315I | 500 | 11 | 12.79 | ≈8000 | ≈10 000 |
| H375P | 125 | 1 | 1.16 | 50 | 1 100 |
| T406I | 125 | 1 | 1.16 | 80 | ≈1 400 |
| W430L | 125 | 2 | 2.32 | 90 | 800 |
| E431G | 125 | 1 | 1.16 | 35 | ≈650 |
Six mutations, namely L248V, Q252H, Y253C, Y253H, E255K, and T315I, identified here from the screen, have previously been reported in patients with CML receiving imatinib mesylate, and are known to be associated with imatinib mesylate resistance. All double-mutant clones were recovered at 500 nM concentration.
— indicates no recovery.
(no. of individual colonies/total no. of colonies obtained) × 100.
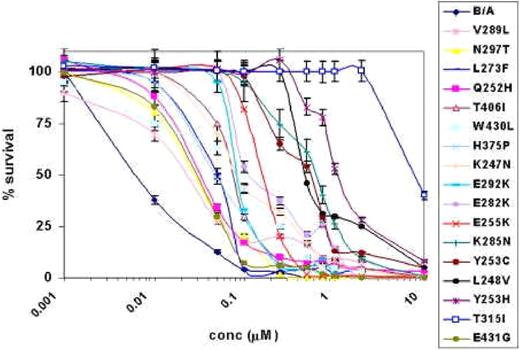
AMN107 treatment of BCR-ABL point mutant and wild-type BCR-ABL–expressing Ba/F3 cell lines. This figure shows the results of AMN107 treatment of different BCR-ABL point mutants and wild-type Bcr-Abl–expressing Ba/F3 cell lines using a wide range of concentrations for 72 hours. The dose response curves obtained from the MTS assay are presented in a semilog plot. Error bars indicate the standard deviation of the mean for each dose.
AMN107 treatment of BCR-ABL point mutant and wild-type BCR-ABL–expressing Ba/F3 cell lines. This figure shows the results of AMN107 treatment of different BCR-ABL point mutants and wild-type Bcr-Abl–expressing Ba/F3 cell lines using a wide range of concentrations for 72 hours. The dose response curves obtained from the MTS assay are presented in a semilog plot. Error bars indicate the standard deviation of the mean for each dose.
Resistant colonies have mutations in the kinase domain of BCR-ABL
All mutations were found to lie within the kinase domain of Abl. A total of 4 mutations (L248V, Q252H, Y253H/C, and E255K) located in the ATP-binding site (P-loop) have previously been identified in patients with imatinib mesylate resistance.18,25
One of the mutants (T315I) is located at a key AMN107 contact site12 and has also been identified as an important contact residue for imatinib mesylate binding in the Abl kinase domain.26 This particular residue appears with a frequency higher than that of all other isolates. In fact, this mutation has also a high frequency of occurrence (approximately 20%) in patients with CML.11 X-ray crystallographic studies show that like imatinib mesylate, AMN107 binds to an inactive conformation of the Abl kinase domain, with the Thr315 residue participating in a hydrogen bond with the aniline NH.12,18 Mutated residues were also identified in the C-helix lining the AMN107 binding site (K285N, V289L), or close to the C-helix but not in contact with AMN107 (L273F, E292K) (Figure 2).12 We also isolated a mutation (H375P) located in a loop spatially close to the hinge region and others (T406I, W430L, E431G) located beyond the activation loop in the C-terminal lobe of the protein kinase domain that appeared under low selection pressure of AMN107. In the screen, no mutations were found within the SH2 domain (residues 127-217). This domain is involved in the BCR-ABL transformation, and as shown earlier by Pendergast et al27 and Azam et al.18 Several mutations in the SH2 domain of Abl were detected by Azam et al18 in their study with imatinib mesylate, out of which S187P was found to confer moderate resistance to imatinib mesylate. SH2 domain mutations may not confer sufficient resistance against AMN107 for detection in an in vitro assay such as described here.
AMN107 bound to the kinase domain of Abl. The ribbon diagram depicts the kinase domain of Abl (shown in gray) with AMN107 (shown as a transparent surface with its skeleton highlighted) bound within the ATP pocket, and with different point mutants highlighted to show their locations within the kinase domain. The side chains of the mutated residues are color-coded according to the color scheme used in Figure 1, such that E255K is red, Y253H/C is purple, and so on.
AMN107 bound to the kinase domain of Abl. The ribbon diagram depicts the kinase domain of Abl (shown in gray) with AMN107 (shown as a transparent surface with its skeleton highlighted) bound within the ATP pocket, and with different point mutants highlighted to show their locations within the kinase domain. The side chains of the mutated residues are color-coded according to the color scheme used in Figure 1, such that E255K is red, Y253H/C is purple, and so on.
Validation of AMN107 resistance of the isolated mutants
We validated the AMN107 resistance of the mutants recovered from the screening by generating individual BCR-ABL retroviral constructs containing the point mutation of interest. The constructs were then introduced into Ba/F3 cells, which were used to evaluate the effects of AMN107 on the proliferation (expressed as IC50 values) of each of these mutant cell lines (Table 1). Viable cell counts normalized to the control (solvent-DMSO) were preformed after 72 hours of growth in triplicate. T315I conferred the greatest degree of resistance to AMN107 with an IC50 value approximately 800-fold greater than that against wild-type BCR-ABL.
The next most resistant mutant was Y253H, also found in imatinib mesylate-resistant patients, which showed an IC50 value approximately 130-fold over that of native BCR-ABL. The Q252H, E255K, K285N, and Y253C mutants showed moderate resistance, with IC50 values ranging from approximately 5-fold to 60-fold of that of the wild-type BCR-ABL.
Comparison of AMN107 and imatinib mesylate resistance
The proliferation of the mutant cell lines in the presence of imatinib mesylate was also done for comparison, and the IC50 values were well within the range of previously reported values.
Discussion
We describe here the use of a random mutagenesis-based screening method for the prediction of Abl mutations that cause resistance to the small-molecule kinase inhibitor AMN107. This method has previously been shown to be of value in studying resistance to imatinib mesylate and PKC412.18,19 AMN107 is a newly developed selective inhibitor of Abl that inhibits the proliferation of human CML cell lines, such as K562 and Ku-812F, as well as cell lines (32D or Ba/F3) transfected to express the BCR-ABL protein, 10 to 50 times more effectively than imatinib mesylate.12 X-ray crystallographic studies have shown that both imatinib mesylate and AMN107 bind to the “DFG-out” inactive conformation of Abl kinase, with the greater potency of AMN107 over imatinib mesylate as a BCR-ABL inhibitor being attributed to a better topologic fit of AMN107 to the protein.11,12 (For the structure, see Weisberg et al12 ).
The T315I mutant is highly insensitive to inhibition by AMN107. This is consistent with the loss of a hydrogen bond and introduction of a steric clash at a vital inhibitor-contact site, as in the case of imatinib mesylate.26 The side chain of Y253 in the P-loop, when changed to histidine, provides the second most resistant mutant. The cysteine mutant at this position would cause less electrostatic differences compared with the histidine replacement, which may explain why Y253C is less resistant to AMN107. It has been inferred from modeling studies that P-loop mutations like Y253C/H and E255K/V destabilize the inactive conformation of the protein, which is necessary for the binding of AMN107 and imatinib mesylate.11 It is possible that this contributes to AMN107 resistance in these mutants, although it is not formally proven. For L248V, resistance is probably due to a change in the shape of the binding pocket and destabilization of the inactive conformation of the P-loop. Disruption of the conformation of the P-loop is also a probable mechanism of K247N, Q252H, and E255K resistance. Although the K285N mutation has not been isolated from imatinib mesylate-treated patients, it has been detected in a cell based screening for imatinib mesylate resistance by von Bubnoff et al.28 This lysine side chain contributes to the hydrophobicity of the AMN107-binding surface surrounding the imidazole substituent, and substitution of lysine with asparagine, to change both the shape and the hydrophobicity of the AMN107-binding surface, could be an explanation for the decrease in sensitivity of the K285N mutant toward AMN107. Other mutants, such as L273F, N297T, T406I, and W430L, which show relatively weak resistance to AMN107, are located further away from the binding site. Valine 289 forms part of the hydrophobic surface against which AMN107 packs, and the leucine substitution would alter the shape of the binding site. Mutants such as H375P and E431G would cause small protein conformational changes, but would not be expected to disturb the binding of AMN107 very much. E292K faces the solvent and would not be expected to cause any resistance to the binding of AMN107, unless it has a role in regulatory interactions with other proteins or the regulatory domains of Abl. E282K probably destabilizes the positioning of the C-helix by losing a salt bridge with R386 from the activation loop.
Dasatanib (BMS354825) is another new agent developed for the treatment of CML, which, in addition to inhibiting BCR-ABL, also inhibits the src-family kinases Src, Lyn, Yes, and Lck.29 In a random mutagenesis study with dasatinib, Shah and coworkers29 isolated a total of 10 resistance mutants of BCR-ABL involving 6 residues: L248R, Q252H, E255K, V299L, T315I/A, and F317L/V/I/S. With the exception of the V299L mutant, all of these are also found in mutagenesis studies with imatinib mesylate and AMN107. (Although F317 was not isolated in this study, the effect of AMN107 against F317L expressing Ba/F3 cell line has previously been shown by Weisberg et al.12 )
In 2 recent reports, one based on the characterization of mutations in BCR-ABL that confer resistance to dasatinib,29 and another to PD166326,28 respectively, a cell-based screening strategy has been used. The latter report, in contrast to the method described by Azam et al18 as followed by us and others,29 used a complete cell-culture–based method that allows selection of resistant cells and can also detect mechanisms of inhibitor resistance other than mutations of the BCR-ABL gene. The retroviral-based mutagenesis method relies upon random mutations in the BCR-ABL gene, followed by the expression of the mutated protein under selection pressure. For the kinase domain mutations, depending upon the nature of the drug as well as the mutant, the dose-response curves will shift toward higher inhibitor concentrations, marginally for the “weak” mutants and significantly for the “strong” ones, as has been observed in the present study. However, in a cell-culture–based system, an increase of phosphorylated BCR-ABL in resistant sublines can be observed without mutations in the gene.28 If a drug has more than one target, the acquired resistance of a subline could well be due to the effect of the drug on multiple targets.
It is also important to consider the limitations of random mutagenesis screen. Although 6 mutations (L248V, Q252H, Y253H, Y253C, E255K, and T315I) that were identified from the screen have been previously reported in patients receiving imatinib mesylate, we did not recover other clinically identified mutants that confer imatinib mesylate resistance (eg, E255V, Y253F, or T315S/G). Thus, this mutagenesis strategy is unlikely to be able to detect all possible resistance mutations. One possible reason could be that the number of clones isolated from this type of screen may not be sufficient enough to detect all point mutants. Moreover, as this study reflects a series of random events, there is always a possibility that we might have missed some new/novel mutations.
Peng et al30 reported earlier that the mean plasma trough concentration in patients receiving imatinib mesylate is 0.57 μg/mL (approximately 1 μM) 24 hours after administration of 350 mg of the drug. AMN107 is substantially more potent than imatinib mesylate, and recent trials demonstrated activity in patients with advanced CML resistant to imatinib mesylate.31 In this study, the mean serum peak concentrations of AMN107 among patients receiving 400 mg of AMN107 twice daily was 3.6 μM at steady state.31 The mean serum trough level at the steady-state level was 1.0 μM at 400 mg daily, 1.7 μM at 400 mg twice daily, and 2.3 μM at 600 mg twice daily. With the exception of T315I, these concentrations of AMN107 would be expected to at least partially inhibit proliferation of leukemic cells with essentially all of the other mutations detected in this study. This suggests that the clinical resistance to AMN107 might be predominantly associated with the emergence of T315I.
In contrast with previous studies dealing with imatinib mesylate resistance, the results of the screen with AMN107 suggest that mutations of the P-loop, C-helix, SH2 contact, and A-loop might occur less frequently, or would be manageable with moderate dose increases of AMN107. As clinical studies of this compound in CML expand to include larger numbers of patients, the mutagenesis screen may anticipate mechanisms of clinical resistance. It will be of interest to determine whether the BCR-ABL mutations showing the highest degrees of resistance in our screen/validation would also account for most cases of clinical resistance. Since these mutations are also resistant to imatinib mesylate treatment, a different kind of combination therapy would be necessary under those circumstances. Although the molecular basis for drug resistance has been characterized only for a handful of diseases, the mechanism remains quite consistent for all of them: selection for tumor subclones bearing secondary kinase domain mutations that block drug action. Any further insight into the mechanism of acquired resistance of BCR-ABL to small-molecule inhibitors will help in designing an advanced therapeutic strategy for the treatment of patients with CML.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Supported by NIH grant CA66996 and by research funding from Novartis Pharma AB to J.D.G.
Authorship
Conflict-of-interest statement: S.W.C-J, P.W.M., and J.M. are employees of Novartis, whose product was used in the current study. J.D.G. receives research support from Novartis. A.R. has no conflicts.
Correspondence: James D. Griffin, Department of Medical Oncology, Dana Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: james_griffin@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal