Abstract
The E2A transcription factors are required for normal T lymphopoiesis and to prevent T-lymphocyte progenitor transformation. Ectopic expression of E2A proteins in E2A-deficient lymphomas results in growth arrest and apoptosis, indicating that these cells remain responsive to the targets of E2A. Here we identify the transcriptional repressor growth factor independent 1B (Gfi1b) as a target of E2A that promotes growth arrest and apoptosis in lymphomas. Gfi1b expression in primary T-lymphocyte progenitors is dependent on E2A and excess Gfi1b prevents the outgrowth of T lymphocyte progenitors in vitro. Gfi1b represses expression of Gata3, a transcription factor whose appropriate regulation is required for survival of lymphomas and T-lymphocyte progenitors. We also show that ectopic expression of Gata3 in lymphomas promotes expression of Gfi1b, indicating that these proteins may function in an autoregulatory loop that maintains appropriate levels of Gata3. Therefore, we propose that E2A proteins prevent lymphoma cell expansion, at least in part through regulation of Gfi1b and modulation of Gata3 expression.
Introduction
T lymphopoiesis is characterized by the differential expression of multiple cell surface receptors that reflect the changes in gene expression programs occurring within these cells.1,2 The majority of thymocytes are double positive (DP) for the coreceptors CD4 and CD8 and give rise to single-positive (SP) cells capable of responding to foreign pathogens. DP cells develop from double-negative (DN) cells, the most immature of which lack IL-2 receptor-α (CD25) but express CD44 and CD117 and are not restricted to T-lymphocyte development.3,4 These early thymic progenitors (ETPs/DN1) acquire CD25 (DN2) as they become more restricted to T-cell differentiation and down-regulate CD117 and CD44 as they initiate T-cell receptor (Tcr) gene rearrangement (DN3). DN3 cells producing a functional TCRβ chain form a pre-TCR by association with pre-Tα and pass to the DN4 stage (CD25−) through a process referred to as β-selection.5 Cells that successfully pass β-selection proliferate extensively and progress to become DP thymocytes that rearrange TCRα
Appropriate control of these dynamic gene expression programs is essential to ensure development of sufficient numbers of T lymphocytes to function in immune responses while avoiding self-reactivity and transformation. This fact is exemplified by the phenotype of mice lacking, or overexpressing, transcription factors that function in the regulation of these programs.6 For example, Gata3 is required for development of the earliest T-lineage progenitors but ectopic expression of Gata3 blocks T lymphopoiesis at the DN3 stage.6–8 In addition, mice lacking the transcription factors encoded by the E2A gene show reduced thymus cellularity and defective generation of DN2, DN3, and DP cells.9 In addition, E2A−/− mice develop T-cell lymphoma with a DN3-DP phenotype suggesting additional roles for these proteins as tumor suppressors.10 Inhibition of E2A activity is a common feature of human T-lineage acute lymphoblastic leukemia suggesting that E2A proteins function similarly in human T-lymphocyte progenitors.11 Ectopic expression of E2A in E2A−/− lymphomas results in growth arrest and apoptosis indicating that E2A proteins are able to induce their target genes in these cells.12,13 However, only a few E2A target genes have been identified including Ptcra and Hes1 and these targets do not explain potential growth/survival repressive functions of E2A.14–16
The E2A gene encodes 2 basic helix-loop-helix (bHLH) transcription factors, E12 and E47, that share redundant functions.17 Insights into the functions of these proteins, their target genes, and mechanism of regulation have come from studies in Drosophila, where daughterless (da), the homologue of E2A, plays essential roles in sex determination and neurogenesis, in addition to other processes.18–20 During neurogenesis da, in association with proneural bHLH proteins, functions to specify the proneural field through regulation of enhancer of split E (spl) proteins and senseless (sens), a transcription factor related to mammalian Gfi1 and Gfi1b.21,22 Sens, Gfi1, and Gfi1b bind DNA through carboxy-terminal zinc fingers and both Gfi1 and Gfi1b contain an amino-terminal Snail and Gfi1 (SNAG) domain that functions in transcriptional repression.23
Gfi1 and Gfi1b are expressed in T-lymphocyte progenitors and Gfi1 is required for normal progression beyond the ETP/DN1 stage.2,24 In contrast, a function for Gfi1b in T lymphopoiesis has not been described because mice lacking Gfi1b die during embryogenesis.25 Ectopic Gfi1 expression leads to IL-2–independent growth of mature T-cell lymphomas and the Gfi1 gene is a common integration site in murine leukemia virus-induced transformation indicating that Gfi1 can promote proliferation or survival of mature T lymphocytes.26,27 However, Gfi1 also restrains proliferation of hematopoietic stem cells (HSCs) indicating that the functions of Gfi1 proteins are cell context dependent.28,29
We have identified Gfi1b in a screen for E2A target genes in E2A−/− T-cell lymphomas. We show that Gfi1b mRNA is induced by E47 in these lymphomas without the need for additional E2A-dependent genes and that E47 binds to E-box sequences in the first intron of the Gfi1b gene. Moreover, we show that expression of Gfi1b in DN3 thymocytes requires E2A. We also demonstrate that ectopic expression of Gfi1b, like E2A, inhibits expansion and survival of T-cell lymphomas and suppresses expression of Gata3, a transcription factor required for lymphoma cell survival. Interestingly, Gata3 promotes expression of Gfi1b in E2A−/− lymphomas indicating that Gata3 may limit its own expression through regulation of Gfi1b. We propose that loss of E2A activity during T-lymphocyte progenitor transformation results in decreased Gfi1b and may stabilize Gata3, thereby promoting lymphoma cell survival.
Materials and methods
Cell lines and culture conditions
The E2A−/− lymphomas 0531 and 1.F9 were cultured in RPMI supplemented with 10% FBS (Invitrogen, Carlsbad, CA), penicillin, streptomycin, and glutamine and were maintained at 37°C in a humidified incubator with 5% CO2. E2A−/− lymphomas express CD4 and CD8, pre-TCR, activated Notch1; they are not cytokine dependent and have not been shown to differentiate under any conditions in vitro.10,32 HSCs were isolated from E13 fetal liver by cell sorting and cultured on a confluent layer of OP9-DL1 cells as described previously.30
Flow cytometry
Flow cytometry was performed on a FACSCalibur using CellQuest software and analyzed using FlowJo (Tree Star, Ashland, OR). Cell sorting was performed on a FACSAria or MoFlo using fluorescence-activated cell sorting (FACS) Diva software. Active caspase-3 staining was performed as recommended by the manufacturer (BD Biosciences, La Jolla, CA). Cell cycle analysis was performed as described previously.31 E47+/+ and E47−/− thymocyte populations were isolated after depletion of lineage-positive cells using a cocktail of biotinylated antibodies (CD4, CD8, TCRβ, B220, CD19, CD11b, CD11c, NK1.1, DX5, Ter119), followed by incubation with streptavidin-conjugated magnetic beads and passage over a magnetic column (Miltenyi Biotec, Auburn, CA). Enriched cell suspensions were stained with streptavidin-FITC, CD117-APC, and CD25-PE, followed by cell sorting. E13.5 fetal liver HSCs (Lin−CD117+CD27+) were isolated by magnetic bead depletion of Ter119+ and Gr1+ cells followed by sorting after staining with CD27-PE and CD117-APC. E2A+/+ and E2A−/− mice were on a C57Bl/6 background and were housed at the University of Chicago in accordance with the guidelines of the Institutional Animal Care and Use Committee.
RNA purification and microarray analysis
RNA was isolated using TRIzol reagent (Invitrogen, San Diego, CA). Microarray analysis was as described previously.32
Northern blot
Northern blots were generated and probed as described previously.32 Probes were prepared from polymerase chain reaction (PCR) fragments generated using the following primers:
Gfi1b forward: 5′-AGCACAGAGTCTCCCTTGGA-3′; Gfi1b reverse: 5′-CAAAGGTTTTGCCACAGACA-3′; Gfi1 forward: 5′-TCCGAGGGTCCAAACATCG-3′; Gfi1 reverse: 5′-TTGAAAGGCAGCGTGTAGGG-3′.
Retroviral infection
The retroviral vectors MigR1, pCS-E47-ER, pCS, Banshee, and Banshee-Gata3-siRNA were described previously.32–34 MigR1-Gfi1b and MigR1-Gfi1bΔSNAG were generated by PCR Gfi1b from wild-type thymus cDNA using primers: GFI1b-START: 5′-CCTCGAGAAAAATGCCACGGTCCTTT-3′ or ΔSNAG: 5′-AGATCTGAAAAATGCAGGGTGATGAGC-3′, with GFI1b-STOP: 5′-CCTTAAGTCTCACTTGAGATTGTGTTC-3′. MigR1-N290S was generated using the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA) with GFI1b-A999G: 5′-GCCAGAGCTCCAGCCTCATCACCCAC-3′, GFI1b-ANTISENSE A999G: 5′-GTGGGTGATGAGGCTGGAGCTCTGGC-3′.
Retroviral supernatants were produced in Phoenix cells and lymphomas were infected by spin inoculation.32
Western blot
Western blots were prepared using standard procedures and probed with antibodies against mouse Gfi1b (D-19) or Gata3 (HG3031) (Santa Cruz Biotechnology, Santa Cruz, CA) and horseradish peroxidase-coupled secondary reagents were used to develop the blots using enhanced chemiluminescence (ECL) plus (GE Healthcare, Piscataway, NJ).
Electrophoretic mobility shift assay
Whole-cell extracts were prepared as described previously.35 In vitro–translated Gfi1b proteins were generated from pSP64 vectors using TNT Quick coupled transcription/translation systems (Promega, Madison, WI). Double-stranded DNA probes were labeled and electrophoretic mobility shift assay (EMSA) was performed as described.35 The anti-E47 antibody (32.1) is from PharMingen (BD Biosciences). The sequences of the oligonucleotides used were: Gfi1bE-box1: 5′-TCCTTGGTTGCAGGTGGGGA-3′; Gfi1bE-box2: 5′-GTGCACGCCACACCTGCGGGGG-3′.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed after fixation of cells with 1% formaldehyde at 37°C for 10 minutes and neutralization with glycine at a final concentration of 0.125 M using the Upstate Biotechnology kit (Lake Placid, NY). Protein-DNA complexes were precipitated using polyclonal anti-E2A (N-649; Santa Cruz Biotechnology) or anti-aH3 (Upstate Biotechnology). After reversing the cross-links, DNA was purified using a PCR purification kit (Qiagen, Valencia, CA) and eluted in Tris-HCl, pH8.0. Primers used for DNA amplification of the Gfi1b locus were: Gfi1b-ChIP-Ebox1 forward: 5′-CTCACTGCAGAGGGACAACA-3′; Gfi1b-ChIP-Ebox1 reverse: 5′CCTGCCTCTGTCCTGACTTC-3′; Gfi1b-ChIP-Ebox2 forward: 5′-GCATGGGATAGTGTGCCTTT-3′; and Gfi1b-ChIP-Ebox2 reverse: 5′-TCATCAAGACAGTGCCAAGC-3′.
PCR amplification was performed for 38 cycles prior to ethidium bromide-agarose gel analysis.
Quantitative real-time PCR
Total RNA was reverse transcribed with Superscript II and oligo dT18. The cDNA was amplified in an iCycler in a 50-μL reaction containing the iQ SYBR Green Supermix from Bio-Rad (Hercules, CA). Expression was calculated for each gene relative to HPRT using the ΔCT method. The primers used were: Gfi1b Q forward: 5′-CCTGTGATGTCTGTGGCAAAACC-3′; Gfi1b Q reverse: 5′-AGGGTGGATGAACGCTTGAAGG-3′; Gfi1 Q forward: 5′-AAGACCCTTTGCGTGCGAGATG-3′; Gfi1 Q reverse: 5′-ATGTGTGGACAGCGTGGATGAC-3′; Gata3 Q forward: 5′-AGGCAAGATGAGAAA GAGTGCCTC-3′; Gata3 Q reverse: 5′-CTCGACTTACATCCGAACCCGGTA-3; HPRT Q forward: 5′-ACCTCTCGAAGTGTTGGATA-3′; HPRT Q reverse: 5′-CAACAACAAACTTGTCTGGA-3′.
Results
Identification of E2A inducible genes in E2A−/− T-cell lymphomas
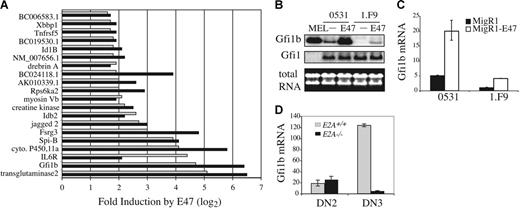
Ectopic expression of E2A proteins in E2A−/− lymphomas results in growth arrest and apoptosis indicating that E2A can induce target genes that inhibit expansion of these cells.12 To identify E2A-regulated genes we performed a microarray analysis comparing RNA isolated from 2 E2A−/− lymphomas infected with a bi-cistronic retrovirus producing GFP (S003) or E47 plus GFP (S003-E47). Approximately 107 genes were found to be differentially expressed by more than 2-fold in the presence or absence of E47 in both lymphomas. Of these 107 genes, 20 were increased by E47 by more than 3-fold including 5 transcriptional regulators, growth factor independent 1B (Gfi1b), SpiB, Idb2, Id1B, and X-box binding protein (Xbbp1), 4 proteins involved in signal transduction, interleukin 6 receptor (IL-6R), Jagged2, ribosomal S6 kinase (Rps6ka2), and the tumor necrosis factor receptor superfamily member 5 (Tnfsf5), and 5 sequences of unknown function (Figure 1A). Therefore, E2A proteins can influence transcriptional programs and signal transduction in T-cell lymphomas.
Induction of Gfi1b by E47 in E2A−/− T-cell lymphomas. (A) Fold induction of the indicated genes 24 hours after infection of E2A−/− lymphomas 0531 (▪) and 1.F9 (⊡) with GFP-producing (S003) or E47 plus GFP-producing (S003-E47) retrovirus. Expression levels were determined using Affymetrix 430 2.0 gene arrays and the fold increase in gene expression in E47 plus GFP-expressing cells as compared to GFP-expressing cells is presented (log2 scale) for genes that showed a greater than 3-fold induction in both cell lines. (B) Northern blot analysis of RNA isolated from sorted GFP+ lymphomas 24 hours after infection with MigR1 (−) or MigR1-E47 (E47) retrovirus. MEL cells were included as a positive control. Total RNA is shown on the bottom and the blot was probed sequentially with Gfi1b and Gfi1 cDNA. (C) Quantification of Gfi1b mRNA by QPCR in 0531 or 1.F9 cells infected with MigR1 (▪) or MigR1-E47 (□). The level of Gfi1b mRNA was determined relative to HPRT mRNA by the ΔCT method and relative expression was determined using the formula 2−ΔΔCT. The standard error of triplicate measurements is shown. (D) QPCR analysis of Gfi1b mRNA in DN2 (Lin−CD117+CD25+) and DN3 (Lin−CD117−CD25+) cells isolated from E2A+/+ (⊡) or E2A−/− (▪) thymocytes.
Induction of Gfi1b by E47 in E2A−/− T-cell lymphomas. (A) Fold induction of the indicated genes 24 hours after infection of E2A−/− lymphomas 0531 (▪) and 1.F9 (⊡) with GFP-producing (S003) or E47 plus GFP-producing (S003-E47) retrovirus. Expression levels were determined using Affymetrix 430 2.0 gene arrays and the fold increase in gene expression in E47 plus GFP-expressing cells as compared to GFP-expressing cells is presented (log2 scale) for genes that showed a greater than 3-fold induction in both cell lines. (B) Northern blot analysis of RNA isolated from sorted GFP+ lymphomas 24 hours after infection with MigR1 (−) or MigR1-E47 (E47) retrovirus. MEL cells were included as a positive control. Total RNA is shown on the bottom and the blot was probed sequentially with Gfi1b and Gfi1 cDNA. (C) Quantification of Gfi1b mRNA by QPCR in 0531 or 1.F9 cells infected with MigR1 (▪) or MigR1-E47 (□). The level of Gfi1b mRNA was determined relative to HPRT mRNA by the ΔCT method and relative expression was determined using the formula 2−ΔΔCT. The standard error of triplicate measurements is shown. (D) QPCR analysis of Gfi1b mRNA in DN2 (Lin−CD117+CD25+) and DN3 (Lin−CD117−CD25+) cells isolated from E2A+/+ (⊡) or E2A−/− (▪) thymocytes.
Gfi1b mRNA is regulated by E2A in lymphomas and DN3 thymocytes
We were particularly interested in the finding that multiple transcriptional regulators were induced by E47 in these lymphomas since E2A regulates B lymphopoiesis, in part, through regulation of essential transcription factors.37 Gfi1b was of interest because it is highly induced by E47 and because the related drosophila gene, sens, is a known target of E-proteins in drosophila sensory organ progenitors.38 Therefore, we examined the ability of E47 to regulate Gfi1b mRNA in T-cell lymphomas. Consistent with our microarray data we observed an increase in Gfi1b mRNA by Northern blot analysis in both lymphomas 24 hours after infection with E47 virus as compared with control virus (Figure 1B). In contrast, mRNA encoding the related transcription factor Gfi1 was not induced by E47 (Figure 1B). Quantitative real-time (Q) PCR revealed that E47 induced Gfi1b mRNA by approximately 4-fold in both lymphomas although the baseline level of Gfi1b was different between the 2 lines (Figure 1C). Therefore, Gfi1b mRNA is regulated downstream of E47 in E2A−/− lymphomas.
As shown by others, Gfi1b mRNA is expressed in DN2 stage thymocytes, increased at the DN3 stage, and extinguished at the DN4 stage1,39,40 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). QPCR analysis of Gfi1b mRNA in E2A+/+ and E2A−/− DN2 and DN3 thymocytes revealed that E2A is not required for the low level of Gfi1b mRNA in DN2 thymocytes, but E2A is required for Gfi1b expression at the DN3 stage (Figure 1D). We also detected a 2- to 5-fold increase in Gfi1b mRNA after infection of primary T-cell progenitors cultured on OP9-DL1 stromal cells with E47-producing retrovirus (data not shown). Taken together, our data indicate that Gfi1b mRNA is induced by E47 in T-cell lymphomas and that Gfi1b expression in DN3 thymocytes is E2A dependent.
Gfi1b is a target of E2A
To determine whether Gfi1b is a direct target of E2A we infected the lymphomas with virus producing a tamoxifen-inducible form of E47 (E47-ER) and examined gene expression 4 hours after addition of 4-hydroxytamoxifen (4-OHT) in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX). 4-OHT releases E47-ER from endogenous chaperonins allowing induction of E47 activity without the need for new protein synthesis. In the presence of CHX de novo protein synthesis cannot occur and only mRNA induced by E2A, without the need for additional E2A targets, will be increased in response to 4-OHT. Using this approach we found a 6- and 4-fold increase in Gfi1b mRNA in the 0531 and 1.F9 lymphomas, respectively, after induction of E47-ER (Figure 2A). In contrast, Gfi1b was not induced in control virus-infected (pCS) cells or by E47-ER in the absence of 4-OHT (Figure 2A). Therefore, Gfi1b is induced by E47 without the need for additional protein synthesis in lymphomas.
Gfi1b is directly regulated by E47. (A) 0531 (top graph) and 1.F9 (bottom graph) cells were infected with pCS retrovirus (▪) or pCS-E47ER (□) and treated with 4-OHT (1 μM) for 4 hours in the presence or absence of CHX (4 μM) starting 24 hours after infection. Gfi1b and HPRT mRNA were quantified by QPCR. The standard error of triplicate measurements is shown. (B) Schematic of the Gfi1b gene showing the approximate location of the conserved E-boxes (▾) within the first intron. (C) EMSA using labeled Gfi1b E-box1, E-box2, or μE5 oligos and protein extracts prepared from 0531 cells infected with S003 or S003-E47 retrovirus 24 hours after infection. Prior to addition of labeled probe the extracts were treated with either ddH20 (−), 100 × excess of competitor oligo (E-box1 or E-box2), or anti-E47 antibody for 15 minutes. (D) ChIP assay performed on 0531 cells infected with MigR1 or MigR1-E47 using polyclonal anti-E2A antibody (E2A), anti–acetyl-histone H3 (aH3), or no antibody (−).
Gfi1b is directly regulated by E47. (A) 0531 (top graph) and 1.F9 (bottom graph) cells were infected with pCS retrovirus (▪) or pCS-E47ER (□) and treated with 4-OHT (1 μM) for 4 hours in the presence or absence of CHX (4 μM) starting 24 hours after infection. Gfi1b and HPRT mRNA were quantified by QPCR. The standard error of triplicate measurements is shown. (B) Schematic of the Gfi1b gene showing the approximate location of the conserved E-boxes (▾) within the first intron. (C) EMSA using labeled Gfi1b E-box1, E-box2, or μE5 oligos and protein extracts prepared from 0531 cells infected with S003 or S003-E47 retrovirus 24 hours after infection. Prior to addition of labeled probe the extracts were treated with either ddH20 (−), 100 × excess of competitor oligo (E-box1 or E-box2), or anti-E47 antibody for 15 minutes. (D) ChIP assay performed on 0531 cells infected with MigR1 or MigR1-E47 using polyclonal anti-E2A antibody (E2A), anti–acetyl-histone H3 (aH3), or no antibody (−).
To identify E2A-binding sites in the Gfi1b gene we compared the mouse and human genomic sequences and identified 2 conserved E-boxes within the first intron (Figure 2B). EMSA revealed that E47 is able to bind both of these E-boxes, whereas only a low level of binding was observed with extracts prepared from lymphomas lacking E2A (Figure 2C). This low level of binding is likely due to E2-2 or HEB in these cells, which bind the same E-box sequences as E2A.41,42 To determine whether these E-boxes are bound by E47 in lymphomas, we performed ChIP using an antibody directed against the amino-terminus of E2A. Lymphomas were infected with control or E47 virus and extracts were prepared from formaldehyde-treated cells. PCR analysis of DNA precipitated with anti-E2A antibody revealed an association of E47 with both E-boxes (Figure 2D). However, the 3′ E-box (E-box2) was precipitated more robustly than the 5′ E-box (E-box1). A low level of precipitation of both E-boxes was observed in E2A−/− lymphomas, which may be due to cross-reactivity of this antibody with E2-2 and HEB, as observed by Western blot analysis (Figure 2D and data not shown). As a positive control we were able to precipitate DNA flanking the Ptcra enhancer, a known E2A target, with anti-E2A antibody at higher levels in E47-expressing than in E2A−/− lymphomas and little DNA was precipitated in the absence of antibody (data not shown).14 We were also able to precipitate both E-boxes with an anti–acetyl-histone H3 antibody indicating that histone H3 in this region of the Gfi1b gene is acetylated in E2A−/− lymphomas (Figure 2D). Taken together, the data provide strong evidence that Gfi1b is regulated by E47 through direct interaction of E47 with the Gfi1b gene.
Expression of Gfi1b inhibits proliferation and survival of E2A−/− lymphomas
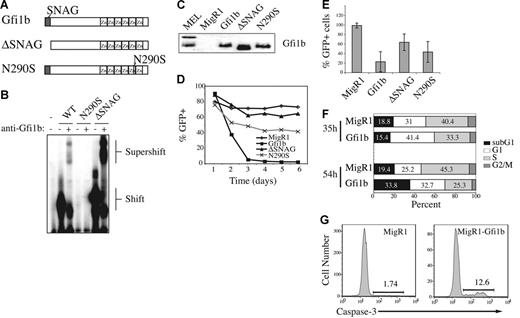
Lck-Gfi1b transgenic mice have reduced numbers of DP thymocytes and an altered CD4/CD8 cell ratio.39 Therefore, we hypothesized that Gfi1b may be an E2A target gene that inhibits expansion of T lineage cells or lymphomas. To test this hypothesis we examined the consequences of Gfi1b expression in E2A−/− lymphomas, which have a hybrid DN3-DP phenotype. We created retrovirus producing Gfi1b or a mutant Gfi1b that lacks the SNAG repression domain (ΔSNAG; Figure 3A). We also created retrovirus producing Gfi1b harboring a N290S mutation, which is similar to a mutation found in Gfi1 in severe congenital neutropenia (SCN) that prevents DNA binding (Figure 3A).43 Using in vitro-translated proteins and EMSA we confirmed that Gfi1b and ΔSNAG bind to the consensus Gfi1b-binding site, whereas the N290S protein fails to bind this sequence (Figure 3B). We also demonstrated that each retrovirus produces protein after infection of E2A−/− lymphomas (Figure 3C).
Ectopic expression of Gfi1b inhibits proliferation and survival of T-cell lymphoma. (A) Schematic of Gfi1b, ΔSNAG, and N290S. (B) EMSA using in vitro-translated Gfi1b, ΔSNAG, and N290S and labeled oligos containing consensus Gfi1/1b-binding sequence (B30). Anti-Gfi1b antibody was used to confirm the presence of Gfi1b proteins. The complex shifted by Gfi1b protein and supershifted with anti-Gfi1b antibody is shown. (C) Western blot analysis of 0531 cell extracts 24 hours after infection with the indicated virus. The blot was probed with anti-Gfi1b antibody. Two Gfi1b isoforms are detected in MEL cells. (D) Time course of GFP expression in 0531 cells infected with MigR1(♦), MigR1-Gfi1b (▪), MigR1-ΔSNAG (▴), or MigR1-N290S (×) as determined by flow cytometry. Identical results were obtained with 2 other E2A−/− lymphoma lines. (E) Average relative percent GFP+ cells 5 days after infection combining data from 3 experiments. Relative percent GFP+ was determined by dividing the percent GFP+ on day 5 by the percent GFP+ on day 1 (× 100). The decrease in GFP+ cells in Gfi1b virus-infected cultures is greater than in all other experimental groups (P < .05, paired Student t test). (F) 0531 cells were infected with MigR1 or MigR1-Gfi1b retrovirus and BrDU was added to cultures for 30 minutes 35 or 54 hours after infection. The GFP+ cells were isolated and stained with anti–BrDU-FITC and propidium iodide and examined by flow cytometry.31 G1 = 2N DNA, BrDU−; S = BrDU+; G2+M = 4N DNA, BrDU−; apoptotic cells = < 2N DNA. (G) 0531 cells infected with MigR1 or MigR1-Gfi1b retrovirus were stained 48 hours after infection with an antibody that detects active caspase-3. Caspase-3 staining on GFP+ cells is shown. Similar results were observed with 1.F9 cells.
Ectopic expression of Gfi1b inhibits proliferation and survival of T-cell lymphoma. (A) Schematic of Gfi1b, ΔSNAG, and N290S. (B) EMSA using in vitro-translated Gfi1b, ΔSNAG, and N290S and labeled oligos containing consensus Gfi1/1b-binding sequence (B30). Anti-Gfi1b antibody was used to confirm the presence of Gfi1b proteins. The complex shifted by Gfi1b protein and supershifted with anti-Gfi1b antibody is shown. (C) Western blot analysis of 0531 cell extracts 24 hours after infection with the indicated virus. The blot was probed with anti-Gfi1b antibody. Two Gfi1b isoforms are detected in MEL cells. (D) Time course of GFP expression in 0531 cells infected with MigR1(♦), MigR1-Gfi1b (▪), MigR1-ΔSNAG (▴), or MigR1-N290S (×) as determined by flow cytometry. Identical results were obtained with 2 other E2A−/− lymphoma lines. (E) Average relative percent GFP+ cells 5 days after infection combining data from 3 experiments. Relative percent GFP+ was determined by dividing the percent GFP+ on day 5 by the percent GFP+ on day 1 (× 100). The decrease in GFP+ cells in Gfi1b virus-infected cultures is greater than in all other experimental groups (P < .05, paired Student t test). (F) 0531 cells were infected with MigR1 or MigR1-Gfi1b retrovirus and BrDU was added to cultures for 30 minutes 35 or 54 hours after infection. The GFP+ cells were isolated and stained with anti–BrDU-FITC and propidium iodide and examined by flow cytometry.31 G1 = 2N DNA, BrDU−; S = BrDU+; G2+M = 4N DNA, BrDU−; apoptotic cells = < 2N DNA. (G) 0531 cells infected with MigR1 or MigR1-Gfi1b retrovirus were stained 48 hours after infection with an antibody that detects active caspase-3. Caspase-3 staining on GFP+ cells is shown. Similar results were observed with 1.F9 cells.
Infection of lymphomas with Gfi1b-retrovirus revealed that Gfi1b-expressing cells (GFP+) are rapidly lost from culture (Figure 3D-E). In contrast, GFP+ cells were not lost from MigR1 virus–infected cultures and were lost at a significantly reduced rate from cultures infected with ΔSNAG-producing virus (Figure 3D-E). Cells expressing N290S were also lost from culture more slowly than Gfi1b-expressing cells, although the N290S protein did have some effect on the cells, possibly due to competition for corepressor complexes (Figure 3D-E). Gfi1b-expressing lymphomas incorporated less BrDU than MigR1-infected lymphomas 35 and 54 hours after infection, indicating that there are fewer cells in the S phase of the cell cycle (Figure 3F). Staining with propidium iodide in conjunction with BrDU revealed more cells in G1 (BrDU−, 2N DNA content) and, at later time points, more cells in the sub-G1 stage (< 2N DNA content = apoptotic cells) in the presence of Gfi1b (Figure 3F). Gfi1b-expressing lymphomas also showed increased expression of activate caspase-3, the executioner of apoptosis, when compared to control virus-infected cells (Figure 3G). These data demonstrate that ectopic expression of Gfi1b in E2A−/− lymphomas leads to growth arrest and apoptosis and these effects are dependent on the SNAG domain.
We next examined the consequences of ectopic expression of Gfi1b in primary T-lymphocyte progenitors cultured in vitro.30 We observed a decline in the percent of GFP+ cells over time in populations infected with Gfi1b virus but not control virus similar to that observed in lymphomas (Figure 4A). The majority of this decline was due to loss of cells with the highest levels of GFP, representing cells with the highest expression of Gfi1b. Surprisingly, we found an enrichment of DN3 cells among the remaining GFPlo cells indicating that Gfi1b may be less detrimental for DN3 thymocytes than for cells at other stages of T-cell development (Figure 4B). Consistent with that observation, by day 9 afterinfection there were more Gfi1b-expressing DN3 cells than control virus-expressing DN3 cells (Figure 4C). Therefore, ectopic expression of Gfi1b is detrimental to primary T-cell progenitors but lower levels of Gfi1b preferentially support DN3-like cells in vitro.
Differential effects of ectopic Gfi1b expression in primary T-lymphocyte progenitors cultured in vitro. (A) Fetal liver-derived HSCs (Lin−CD27+CD117+) were cultured on OP9-DL1 for 6 days prior to infection with MigR1 (◊) or MigR1-Gfi1b (▪). The percent of cells expressing GFP 1, 3, 5, 7, and 9 days after infection was determined by flow cytometry. One of 2 independent experiments is shown. (B) Expression of CD44 and CD25 on GFP+Lin− cells 5 days after infection with MigR1 or MigR1-Gfi1b retrovirus. The percent of cells in each quadrant is indicated. Results are representative of 4 experiments. (C) Total number of DN3 cells (Lin−CD44−CD25+) in cultures of wild-type HSCs infected with MigR1 (⊡) or MigR1-Gfi1b (▪) virus on day 6 of culture and examined at the indicated time after infection. The initial infection efficiency was similar for both retroviruses.
Differential effects of ectopic Gfi1b expression in primary T-lymphocyte progenitors cultured in vitro. (A) Fetal liver-derived HSCs (Lin−CD27+CD117+) were cultured on OP9-DL1 for 6 days prior to infection with MigR1 (◊) or MigR1-Gfi1b (▪). The percent of cells expressing GFP 1, 3, 5, 7, and 9 days after infection was determined by flow cytometry. One of 2 independent experiments is shown. (B) Expression of CD44 and CD25 on GFP+Lin− cells 5 days after infection with MigR1 or MigR1-Gfi1b retrovirus. The percent of cells in each quadrant is indicated. Results are representative of 4 experiments. (C) Total number of DN3 cells (Lin−CD44−CD25+) in cultures of wild-type HSCs infected with MigR1 (⊡) or MigR1-Gfi1b (▪) virus on day 6 of culture and examined at the indicated time after infection. The initial infection efficiency was similar for both retroviruses.
Identification of Gfi1b repressible genes in T-cell lymphomas
Our data indicate that ectopic expression of Gfi1b, like E2A, inhibits proliferation and survival of E2A−/− lymphomas through a SNAG domain-dependent mechanism. To begin to address how Gfi1b functions we isolated RNA from lymphomas infected with Gfi1b or control virus and examined gene expression by microarray analysis. We identified 19 genes whose mRNA was decreased more than 2-fold by Gfi1b in both lymphomas (Figure 5A). These genes include the transcription factors Gfi1, Gata3, and Lmo4. Interestingly, Gata3 is also repressed after expression of E47 in these lymphomas (data not shown). In addition, the E-protein antagonist Idb3 (Id3), chemokine receptor Cmkbr10, and the RNA-binding protein ELAV/HuD were decreased by Gfi1b in E2A−/− lymphomas (Figure 5A). Therefore, Gfi1b represses many genes in lymphomas including the essential T-lineage transcription factors Gfi1 and Gata3.
Gfi1b suppresses Gfi1 and Gata3 expression in a SNAG domain-dependent manner. (A) Fold repression of the indicated genes after infection of 0531 (▪) or 1.F9 (⊡) with MigR1-Gfi1b as compared to MigR1 virus, as determined by Affymetrix 430 2.0 gene arrays. Gfi1b-mediated repression was confirmed by QPCR for (B) Gfi1 or (C) Gata3 mRNA after infection of 0531 or 1.F9 cells with MigR1, MigR1-Gfi1b, or MigR1-ΔSNAG. (D) Western blot analysis of Gfi1 and Gata3 protein in 1.F9 and 0531 cells expressing MigR1 (−) or MigR1-Gfi1b retrovirus. (E) QPCR analysis of Gfi1b and Gata3 mRNA isolated from primary E2A−/− lymphomas (labeled A-D). Data are presented as the difference in CT value between HPRT and Gfi1b ( ) or Gata3 (▪). The standard error of triplicate measurements is shown for panels B, C, and E.
) or Gata3 (▪). The standard error of triplicate measurements is shown for panels B, C, and E.
Gfi1b suppresses Gfi1 and Gata3 expression in a SNAG domain-dependent manner. (A) Fold repression of the indicated genes after infection of 0531 (▪) or 1.F9 (⊡) with MigR1-Gfi1b as compared to MigR1 virus, as determined by Affymetrix 430 2.0 gene arrays. Gfi1b-mediated repression was confirmed by QPCR for (B) Gfi1 or (C) Gata3 mRNA after infection of 0531 or 1.F9 cells with MigR1, MigR1-Gfi1b, or MigR1-ΔSNAG. (D) Western blot analysis of Gfi1 and Gata3 protein in 1.F9 and 0531 cells expressing MigR1 (−) or MigR1-Gfi1b retrovirus. (E) QPCR analysis of Gfi1b and Gata3 mRNA isolated from primary E2A−/− lymphomas (labeled A-D). Data are presented as the difference in CT value between HPRT and Gfi1b ( ) or Gata3 (▪). The standard error of triplicate measurements is shown for panels B, C, and E.
) or Gata3 (▪). The standard error of triplicate measurements is shown for panels B, C, and E.
Gfi1 has been shown previously to be a target of Gfi1b.36,44 However, regulation of Gata3 downstream of Gfi1b has not been reported. We confirmed, by QPCR, that Gfi1 and Gata3 mRNA are repressed 24 hours after expression of Gfi1b in E2A−/− lymphomas (Figure 5B-C). Moreover, repression of these mRNAs requires the SNAG domain (Figure 5B-C). We also found a decrease in Gfi1 and Gata3 protein in E2A−/− lymphomas expressing Gfi1b but not control virus (Figure 5D). Analysis of multiple primary E2A−/− lymphomas revealed that Gfi1b mRNA is present at very low levels, whereas Gata3 mRNA is more highly expressed (Figure 5E). Our data indicate that Gata3 mRNA is directly, or indirectly, repressed by Gfi1b expression in E2A−/− lymphomas.
Comparison of the mouse and human GATA3 genes revealed 2 conserved potential Gfi1b-binding sites; however, these sequences failed to bind in vitro-translated Gfi1b in EMSA (Figure S2). Therefore, Gfi1b may regulate Gata3 mRNA indirectly or directly through binding sites located in distant regulatory regions.
Gata3 is required for survival of T-cell lymphomas
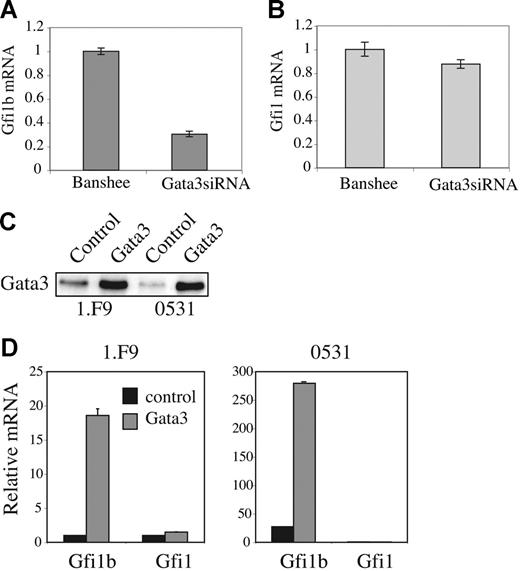
Gata3 plays an essential role in T lymphopoiesis; however, it is not known whether Gata3 is essential for survival of immature T-cell lymphomas. To determine whether repression of Gata3 downstream of E2A and Gfi1b might reduce viability of these lymphomas we examined the effects of reducing Gata3 mRNA in these cells. The Banshee retroviral vector was used to produce Gata3siRNA, or irrelevant siRNA (NSsiRNA), as well as GFP in E2A−/− lymphomas. Gata3siRNA decreased Gata3 mRNA in 0531 cells by approximately 3-fold 48 hours after infection (Figure 6A). 34 Moreover, Gata3siRNA, but not NSsiRNA, decreased Gata3 protein compared with control virus-infected cells (Figure 6B). Notably, the percent of cells expressing GFP declined over time in cultures infected with Gata3siRNA virus, although the decline was slower than that observed with ectopic expression of E47 or Gfi1b (Figure 6C-D and data not shown). In contrast, the percent of GFP+ cells did not decline in cultures infected with NSsiRNA or control virus (Figure 6C-D). Gata3siRNA-expressing lymphomas also showed increased expression of active caspase-3 when compared to control virus-infected cells (Figure 6E). Therefore, decreased expression of Gata3 leads to decreased viability of E2A−/− lymphomas.
Gata3 is required for optimal survival of E2A−/− lymphomas. (A) Gata3 mRNA expression in Gata3siRNA-expressing lymphomas determined by real-time QPCR. (B) Western blot analysis of Gata3 expression in GFP+ 0531 cells infected with Banshee (control), irrelevant siRNA (NSsiRNA), or Gata3siRNA retrovirus. (C) Time course of relative percent GFP+ cells after infection of 0531 cells (△, ▴) or 1.F9 cells (○, •) with Banshee (○, △) or Gata3siRNA (•, ▴) retrovirus (left graph), or 0531 cells with Banshee (⊡) and NSsiRNA (♦) retrovirus (right graph). One representative experiment is shown. (D) Relative GFP+ cells 5 days after infection of 0531 cells with Banshee, Gata3siRNA, or NSsiRNA retrovirus. The average and standard error of 3 independent experiments is presented. (E) Percent of GFP+ cells staining with anti–caspase-3 antibody 24 hours after infection of 1.F9 or 0531 cells with Banshee (□) or Gata3siRNA (▪) retrovirus. Results are the average with standard error of 3 replicate experiments.
Gata3 is required for optimal survival of E2A−/− lymphomas. (A) Gata3 mRNA expression in Gata3siRNA-expressing lymphomas determined by real-time QPCR. (B) Western blot analysis of Gata3 expression in GFP+ 0531 cells infected with Banshee (control), irrelevant siRNA (NSsiRNA), or Gata3siRNA retrovirus. (C) Time course of relative percent GFP+ cells after infection of 0531 cells (△, ▴) or 1.F9 cells (○, •) with Banshee (○, △) or Gata3siRNA (•, ▴) retrovirus (left graph), or 0531 cells with Banshee (⊡) and NSsiRNA (♦) retrovirus (right graph). One representative experiment is shown. (D) Relative GFP+ cells 5 days after infection of 0531 cells with Banshee, Gata3siRNA, or NSsiRNA retrovirus. The average and standard error of 3 independent experiments is presented. (E) Percent of GFP+ cells staining with anti–caspase-3 antibody 24 hours after infection of 1.F9 or 0531 cells with Banshee (□) or Gata3siRNA (▪) retrovirus. Results are the average with standard error of 3 replicate experiments.
Gata3 promotes Gfi1b mRNA expression in E2A−/− lymphomas
Previous studies have implicated Gata proteins, particularly Gata1, in the regulation of Gfi1b in erythroleukemia cells.45 Moreover, transient transfection of a human Gfi1b promoter-luciferase reporter with an expression vector for Gata3 resulted in a 4-fold induction of luciferase activity in 3T3 fibroblasts.45 Therefore, we questioned whether Gata3 might influence Gfi1b expression in E2A−/− lymphomas. QPCR analysis of Gata3siRNA-expressing cells revealed that Gata3, like E2A, regulates Gfi1b expression (Figure 7A). Gfi1b mRNA was decreased approximately 3-fold 48 hours after infection of lymphomas with Gata3siRNA virus (Figure 7A). In contrast, Gfi1 mRNA was not altered by Gata3siRNA (Figure 7B). To further test the ability of Gata3 to promote Gfi1b mRNA expression we used a Gata3-producing retrovirus to ectopically express Gata3 protein (Figure 7C). Gata3 induced Gfi1b mRNA by 12- or 6-fold over control virus-infected cells in 1.F9 or 0531 lymphomas, respectively (Figure 7D). Therefore, whereas Gfi1b can repress Gata3, Gata3 may promote Gfi1b expression, either directly or indirectly, thereby limiting Gata3 expression. Notably, we were unable to detect binding of Gata3 to the GATA site previously identified in the Gfi1b promoter by EMSA (Figure S3). In addition, 2 conserved nonconsensus GATA sequences in the Gfi1b promoter failed to bind Gata3 in an EMSA. Therefore, Gata3 may bind with poor affinity to these sites or Gata3 may regulate Gfi1b expression through distant sites or other mechanisms.46–47 Regardless, our data demonstrate a regulatory loop between Gata3 and Gfi1b in E2A−/− lymphomas and suggest that loss of E2A during lymphocyte progenitor transformation may lead to relaxed regulation of Gata3.
Gata3 promotes Gfi1b expression in E2A−/− lymphomas. QPCR analysis of (A) Gfi1b and (B) Gfi1 mRNA in 0531 cells 48 hours after infection with Banshee or Gata3siRNA retrovirus. (C) Western blot analysis of Gata3 expression in 1.F9 and 0531 cells 48 hours after infected with control (S003) or pXMI-Gata3 retrovirus. (D) QPCR analysis of Gfi1b and Gfi1 mRNA in 1.F9 (left graph) and 0531 (right graph) 48 hours after infection with control (▪) or Gata3-producing retrovirus ⊡. The standard error of triplicate measurements is shown for panels A, B, and D.
Gata3 promotes Gfi1b expression in E2A−/− lymphomas. QPCR analysis of (A) Gfi1b and (B) Gfi1 mRNA in 0531 cells 48 hours after infection with Banshee or Gata3siRNA retrovirus. (C) Western blot analysis of Gata3 expression in 1.F9 and 0531 cells 48 hours after infected with control (S003) or pXMI-Gata3 retrovirus. (D) QPCR analysis of Gfi1b and Gfi1 mRNA in 1.F9 (left graph) and 0531 (right graph) 48 hours after infection with control (▪) or Gata3-producing retrovirus ⊡. The standard error of triplicate measurements is shown for panels A, B, and D.
Discussion
We have identified the transcription factor Gfi1b as a target of E2A in T-cell lymphomas. E2A proteins also play a role in Gfi1b expression in primary T-lymphocyte progenitors since the up-regulation of Gfi1b mRNA observed in wild-type DN3 thymocytes is not observed in DN3 cells from E2A−/− mice. Ectopic expression of Gfi1b, like E2A, induces growth arrest and apoptosis in E2A−/− lymphomas and these functions are dependent on the SNAG repression domain. Moreover, we show that Gfi1b, as well as E2A, represses expression of the essential transcription factor Gata3. Gata3, when ectopically expressed in E2A−/− lymphomas, is also able to promote expression of Gfi1b suggesting that these proteins can function in a regulatory loop. We hypothesize that loss of E2A during the process of T-lymphocyte progenitor transformation may lead to relaxed regulation of Gata3 through loss of Gfi1b and contribute to aberrant survival of immature T-cell lymphomas.
Our observation that Gfi1b is an E2A target gene is consistent with the known role of da in regulation of sens in Drosophila sensory organ development.21,38 We have identified binding sites for E2A in the Gfi1b gene that lie within the first intron, upstream of the start site for translation. This region of Gfi1b has not been investigated previously as a regulatory region for Gfi1b gene expression, although DNAse I hypersensitivity sites have been localized near the 5′ E-box (E-box1) in mouse splenocytes.44 Gfi1b promoter-luciferase reporters have been characterized that contain up to 1 kb of sequence upstream of the Gfi1b transcription start site and 16 to 30 bp of 5′ untranslated sequence.45,46 Although these reporters have been very useful in identifying regulators of Gfi1b, it is likely that additional regulatory regions exist, for example, enhancers or the intronic region containing hypersensitivity sites and the E-boxes identified here. In Drosophila, da and proneural bHLH proteins regulate sens through a 3′ enhancer and it remains possible that E2A proteins also function in enhancers of Gfi1b that have yet to be identified.38 Given the restricted expression of Gfi1b during early T-lymphocyte development, and its possible utility in suppressing expansion of lymphomas, it will be of interest to further elucidate the mechanisms controlling Gfi1b expression.
Our data indicate that Gata3 also can regulate Gfi1b expression in E2A−/− T-cell lymphomas. This observation is consistent with previous studies demonstrating a role for Gata proteins in Gfi1b expression in a erythroleukemia cell line.45–47 These previous studies indicated a complex mechanism by which Gata1 may regulate Gfi1b promoter activity including direct binding to a GATA site as well as via interactions with other DNA-binding proteins including NF-Y and Gfi1b.45–46 We did not detect binding of Gata3 to the GATA site previously identified in this promoter by EMSA; however, it remains possible that Gata3 functions at distant regulatory regions of Gfi1b or through interactions with other DNA-binding factors. In addition to determining the mechanism by which Gata3 regulates Gfi1b it will be of interest to determine whether E2A and Gata3 cooperatively regulate Gfi1b in DN3 thymocytes and whether loss of E2A alters the threshold of Gata3 required for induction of Gfi1b in lymphomas or primary T-lymphocyte progenitors.
We have shown that E2A−/− lymphomas express very low levels of Gfi1b mRNA and that ectopic expression of Gfi1b results in growth arrest and apoptosis. Therefore, reduced Gfi1b expression in the absence of E2A may contribute to survival and transformation of T lymphocyte progenitors. However, E2A−/− lymphomas have a phenotype with characteristics of DN3, DN4, and DP thymocytes, and they have extremely high levels of the Notch1 transcription factor,32 making it difficult to determine whether gene regulation in these cells occurs by a similar mechanism as in primary T-lymphocyte progenitors. Indeed, each stage of T-cell differentiation is associated with a unique complement of transcriptional regulators and the factors regulating any individual gene may be constantly changing during T-lymphocyte differentiation.1,2 We have tested the consequences of retroviral-mediated Gfi1b expression in primary T-lymphocyte progenitors cultured in vitro and, consistent with our findings in lymphomas, cells with the highest levels of Gfi1b were lost from culture. However, DN3-like cells with low levels of Gfi1b were selectively retained. The observed accumulation of DN3-like cells may be the result of increased survival of Gfi1b-expressing DN3 cells or, alternatively, these cells may fail to differentiate in the presence of Gfi1b. Gfi1b is normally expressed in DN3 cells and down-regulated in DN4 cells in vivo and DN4-like cells expressing retroviral Gfi1b were difficult to detect in vitro (data not shown). Therefore, growth arrest and apoptosis of lymphomas may be a consequence of mis-expression as well as ectopic expression of Gfi1b. This conclusion is consistent with the observed decline in DP and SP thymocytes in lck-Gfi1b transgenic mice, although the effect of Gfi1b on DN thymocytes was not examined in these animals.39
Our observation that Gfi1b can repress Gfi1 is consistent with previous studies in lymphoid cells.36,44 To our knowledge, however, suppression of Gata3 by Gfi1b, either directly or indirectly, has not been reported. It is likely that both Gfi1b and Gfi1 can repress Gata3 in lymphomas since these proteins have been shown to be functionally redundant in hematopoietic cells.48 Nonetheless, Gfi1 and Gfi1b can have different affinities for Gfi1-binding sites, indicating that they may have overlapping but not identical sets of target genes.44 Different functions for Gfi1 and Gfi1b are suggested by the phenotype of lck-Gfi1 and lck-Gfi1b transgenic mice since Gfi1b inhibits TCR signaling, whereas Gfi1 augments signaling and ectopic expression of Gfi1 can overcome the inhibitory effects of Gfi1b.39 In addition, Gfi1 and Gfi1b have different patterns and levels of expression, which may result in differences in their ability to activate target genes. Recent studies have revealed that the concentration of sens can determine whether it functions as a transcriptional repressor or activator indicating that concentration may have a crucial effect on function.38,49 Interestingly, Gfi1b activates transcription in some contexts demonstrating that gene regulation by this family of factors may be highly complex and cell context dependent.50
Our data show that Gfi1b is a direct target of E2A proteins that can inhibit the growth of established lymphomas. Whether deregulated Gfi1b expression in E2A−/− thymocytes plays a role in transformation remains to be determined. Regardless, induction of Gfi1b by E47 in E2A−/− lymphomas may not be the only cause of the growth and survival inhibitory effects of E2A in these cells, even though it is sufficient to mimic these effects. We identified approximately 107 genes that are altered in E2A−/− lymphomas by E47 and many of these proteins may contribute to E2A-induced growth arrest and apoptosis. Interestingly, a similar microarray analysis undertaken by Schwartz et al33 revealed some of the same genes that we identified in our screen, although our results differ from this other report in a number of ways. For example, we did not find a significant regulation of Gfi1 by E47 and we confirmed this by Northern blot analysis. Other genes identified by Schwartz et al,33 such as RORγt, were not expressed at statistically significant levels in the presence or absence of E2A in either lymphoma in our study (data not shown). The reason for these discrepancies remains to be determined but may be related to differences in culture conditions, experimental design, or the microarrays used in each study. In addition, Schwartz et al33 examined gene expression in a single lymphoma, 1.F9, and we found multiple genes that were induced or repressed by E47 in 1.F9 that were not regulated by E47 in 0531 and therefore were not included in our list of potential E2A targets.
Our study reveals a mechanism for suppressing expansion of immature T-cell lymphomas in response to E2A protein and demonstrates the existence of a regulatory network that can modulate the expression of Gata3 in these cells. Future studies should be directed at accessing the role of Gfi1b or Gfi1 or both in suppressing T-cell progenitor transformation and the necessity for this transcriptional network in normal and malignant T-cell development.
Authorship
Contribution: W.X. designed and performed experiments and helped to write the manuscript, and B.L.K. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara L. Kee, Department of Pathology, The University of Chicago, 5841 S Maryland Ave, MC1089, Chicago, IL 60637; e-mail: bkee@bsd.uchicago.edu
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported in part by an award to the University of Chicago's Division of Biological Sciences under the Research Resources Program for Medical Schools of the Howard Hughes Medical Institute, a New Investigator Award from the Leukemia Research Foundation, the Concern Foundation, and the Cancer Research Foundation.
We thank Lee Grimes, Tarik Moroy, Iannis Aifantis, Piers Nash, John Crispino, and Guido Franzoso for helpful discussions; Ryan Duggan and David LeClerc for cell sorting; Jim DiSanto for the Gata3 retroviral vector; and Jose Alberola-Ila for the Banshee and Gata3siRNA retroviral vectors. We also thank members of the Kee laboratory for discussions and comments on the manuscript.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal