Abstract
Four of 9 PAX transcription factor genes have been associated with chromosomal translocations in human tumors, although their oncogenic potential has not yet been demonstrated in transgenic mouse models. The B-lymphoidPAX5 gene participates in the generation of the t(9;14)(p13;q32) translocation in germinal center B cells, which leads to deregulated PAX5 expression under the control of the immunoglobulin heavy-chain (IgH) locus in a subset of B-cell non-Hodgkin lymphomas. Here we reconstructed a human t(9;14) translocation in a knock-in mouse by inserting a PAX5 minigene into the IgH locus. The IgHP5ki allele, which corresponds to a germline rather than somatic mutation, is activated in multipotent hematopoietic progenitors and is subsequently expressed in dendritic cells (DCs) and in natural killer (NK), T, and B cells. Ectopic Pax5 expression interferes with normal T-cell development and causes immature T-lymphoblastic lymphomas in IgHP5ki/+ and IgHP5ki/P5ki mice. Aggressive T-cell lymphomas develop even faster in IkPax5/+ mice expressing Pax5 from the Ikaros locus. Pax5 expression in thymocytes activates B-cell–specific genes and represses T-lymphoid genes, suggesting that Pax5 contributes to lymphomagenesis by deregulating the T-cell gene-expression program. These data identify Pax5 as a potent oncogene and demonstrate that the T-lymphoid lineage is particularly sensitive to the oncogenic action of Pax5.
Introduction
B-cell commitment critically depends on the transcription factor Pax5 (BSAP), which is required for B-cell development beyond an early pro–B-cell stage in the bone marrow.1 Pax5−/− pro-B cells are uncommitted lymphoid progenitors with a broad developmental potential and are able to differentiate into mature B cells only once their multilineage potential is suppressed by restoration of Pax5 expression.2 Conversely, the loss of Pax5 expression by conditional gene inactivation converts committed pro-B cells with a restricted B-cell potential into lymphoid progenitors with a broad developmental potential.3 Moreover, ectopic Pax5 expression in hematopoietic stem cells (HSCs) and their progeny strongly skews the developmental options of lymphoid progenitors by promoting B-cell development at the expense of T lymphopoiesis.4 Consistent with its B-lineage commitment function, Pax5 is exclusively expressed within the hematopoietic system from the pro-B to the mature B-cell stage and is essential for maintaining the identity and function of B lymphocytes.5 At the molecular level, Pax5 fulfills a dual role in gene regulation. It activates a multitude of B-cell–specific genes, some of which code for essential components of (pre)BCR signaling.6 At the same time, Pax5 represses lineage-inappropriate genes with important functions in receptor signaling, cell adhesion, migration, and transcriptional regulation in other hematopoietic lineages.2,7 Hence, Pax5 controls the B-cell phenotype by shutting down inappropriate signaling systems and by simultaneously facilitating B-lymphoid signal transduction from the (pre)BCR.
Tumors of hematopoietic origin are frequently associated with specific chromosomal translocations, which result in the activation of proto-oncogenes controlling differentiation, proliferation, or cell survival. In B-cell non-Hodgkin lymphomas (B-NHLs), these proto-oncogenes are often deregulated by translocation adjacent to regulatory elements of the immunoglobulin heavy-chain (IgH) locus on human chromosome 14q32.8 The PAX5 gene on chromosome 9p13 is involved in the recurrent translocation t(9;14)(p13;q32), which juxtaposes the intact coding sequences of PAX5 in opposite orientation next to the constant gene region of the IgH locus.9-11 The molecular analysis of 5 breakpoints revealed that the t(9;14) translocation arises in germinal center B cells as a result of misguided IgH class switch recombination or somatic hypermutation. In 3 cases, the breakpoints were found in the switch Sμ or Sα region of the IgH locus and in exon 1B of PAX5, which resulted in replacement of the 2 alternative PAX5 promoters by antisense promoters present in the IgH switch region.10,-,12 The breakpoints of 2 additional translocations occurred 1.8 kb (KIS-1 lymphoma) or 2 kb upstream of the distal exon 1A of PAX5, which brought the 2 PAX5 promoters under the control of the juxtaposed intronic Eμ enhancer or 3′ locus control region of the IgH locus.9,10,13 The regulatory t(9;14)(p13;q32) translocation may participate in lymphomagenesis in 2 ways. Overexpression of the translocated PAX5 gene10,11 may deregulate the transcription of Pax5-dependent genes in B cells. The expression of PAX5 is normally down-regulated during terminal B-cell differentiation, whereas the IgH gene is transcriptionally most active in immunoglobulin-secreting plasma cells.7,14 Hence, the presence of IgH control elements forces the translocated PAX5 allele to remain active during terminal differentiation, which may contribute to lymphoma formation by interfering with plasma-cell generation or function.9-11 Consistent with this idea, the IgH-PAX5 translocation was considered to be a characteristic hallmark of low-grade lymphoplasmacytic lymphomas (LPLs).10,15 However, reassessment of the t(9;14) translocation frequency revealed a low incidence in LPL,16 but a higher prevalence in aggressive B-NHL.12,17
To provide direct evidence that PAX5 can act as an oncogene in the pathogenesis of NHL, we have reconstructed the KIS-1 t(9;14) translocation in a knock-in mouse by inserting a PAX5 minigene adjacent to the Eμ enhancer into the IgH locus (Figure 1A) We found that the IgHP5ki allele is first transcribed in multipotent hematopoietic progenitors and is then expressed in dendritic cells (DCs) and in natural killer (NK), T, and B cells. Forced Pax5 expression within the T-lymphoid lineage interferes with normal T-cell development and leads to the formation of immature T-lymphoblastic lymphomas in IgHP5ki mutant mice. Pax5 expression under the control of the Ikaros locus4 generates even more aggressive T-cell lymphomas in IkPax5/+ mice. Ectopic Pax5 expression in thymocytes activates B-cell–specific genes and represses key T-lymphoid genes, which may contribute to lymphomagenesis. These data identify Pax5 as a potent oncogene and demonstrate that the T-cell lineage is particularly sensitive to the oncogenic action of Pax5.
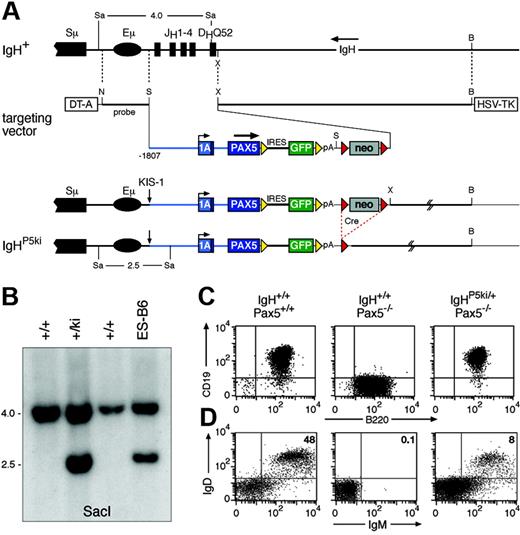
Reconstruction of the KIS-1 translocation by PAX5 insertion into the IgH locus. (A) Structure of the targeted IgH locus. The IgH sequences from the JH1 to the DHQ52 element were replaced by a human PAX5 minigene linked via an internal ribosomal entry sequence (IRES) to a GFP gene. The PAX5 minigene consisted of 1807 bp of 5′ flanking sequences, the entire exon 1A, a shortened intron 1, and exon 2 fused in-frame to PAX5 cDNA containing exons 3 to 10. The IRES-GFP gene was flanked by frt sites (yellow arrowheads) and the downstream neomycin (neo) resistance gene by loxP sites (red arrowheads). The lengths of SacI fragments indicative of correct targeting are shown in kilobases. B indicates BamHI; N, NotI: S, SalI; Sa, SacI; X, XhoI; DT-A, diphtheria toxin A; HSV-TK, herpes simplex virus thymidine kinase; and pA, polyadenylation site. (B) Southern blot analysis of SacI-digested DNA of the indicated mice and targeted ES cell line using a NotI-SalI probe shown in panel A. (C) Expression of functional Pax5 protein from the IgHP5ki allele. Bone marrow pro-B cells from mice of the indicated genotypes were cultured in vitro prior to fluorescence-activated cell sorting (FACS) analysis of CD19 and B220 expression. (D) Partial rescue of B-cell development in Pax5−/− mice by the IgHP5ki allele. Splenocytes from 2-month-old mice of the indicated genotypes were analyzed for IgM and IgD expression.
Reconstruction of the KIS-1 translocation by PAX5 insertion into the IgH locus. (A) Structure of the targeted IgH locus. The IgH sequences from the JH1 to the DHQ52 element were replaced by a human PAX5 minigene linked via an internal ribosomal entry sequence (IRES) to a GFP gene. The PAX5 minigene consisted of 1807 bp of 5′ flanking sequences, the entire exon 1A, a shortened intron 1, and exon 2 fused in-frame to PAX5 cDNA containing exons 3 to 10. The IRES-GFP gene was flanked by frt sites (yellow arrowheads) and the downstream neomycin (neo) resistance gene by loxP sites (red arrowheads). The lengths of SacI fragments indicative of correct targeting are shown in kilobases. B indicates BamHI; N, NotI: S, SalI; Sa, SacI; X, XhoI; DT-A, diphtheria toxin A; HSV-TK, herpes simplex virus thymidine kinase; and pA, polyadenylation site. (B) Southern blot analysis of SacI-digested DNA of the indicated mice and targeted ES cell line using a NotI-SalI probe shown in panel A. (C) Expression of functional Pax5 protein from the IgHP5ki allele. Bone marrow pro-B cells from mice of the indicated genotypes were cultured in vitro prior to fluorescence-activated cell sorting (FACS) analysis of CD19 and B220 expression. (D) Partial rescue of B-cell development in Pax5−/− mice by the IgHP5ki allele. Splenocytes from 2-month-old mice of the indicated genotypes were analyzed for IgM and IgD expression.
Materials and methods
Detailed methods are provided in Supplemental Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article.
IgHP5ki knock-in mouse
The IgHP5ki targeting vector was generated based on the IgH targeting vector pIVhL2neor,18 as described in detail in the Supplementary Materials. E14.1 ES cells were electroporated, and correctly targeted ES cells were injected into C57BL/6 blastocysts. The IgHP5ki allele was genotyped by amplifying a 780-bp polymerase chain reaction (PCR) fragment from the rabbit β-globin polyadenylation region with the primers 5′-GCGGAGCTCCTCAGGTGCAGGCTGCCTATC-3′ and 5′-CGCGTCGACGCTAACATCCTGAGGGACTGT-3′.
Mice
Competitive repopulation assay
C57BL/6 mice (homozygous Ly5.1/Ly5.1) were γ-irradiated with 12 Gy 24 hours prior to intravenous injection of 3 × 106IgHP5ki/+ bone marrow cells (Ly5.2/Ly5.2) and 3 × 106IgHμMT/+ bone marrow cells (Ly5.1/Ly5.2).
Antibodies and flow cytometry
Antibodies were obtained from PharMingen (San Diego, CA). Single-cell suspensions were stained with the respective antibodies and analyzed on a Calibur or Canto flow cytometer (Becton Dickinson, San Jose, CA). A wide forward/side scatter (FSC/SSC) gate was used for analysis of the different hematopoietic cell types.
In vivo transplantation of T-cell tumors
Single-cell suspensions were prepared from the enlarged thymus and spleen of IgHP5ki/+ tumor mice, and 106 thymocytes or splenocytes were injected into the tail vein of syngeneic wild-type mice.
Tumor cell lines and in vitro culture
Tumor cell lines were derived from IgHP5ki/+ and IgHP5ki/P5ki lymphomas and cultured like pro-B cells on γ-irradiated ST2 cells in IL-7–containing IMDM medium.1
Histologic analysis
Dissected tissues were fixed overnight at room temperature in 4% paraformaldehyde/PBS followed by paraffin sectioning and staining with hematoxylin and eosin.
TCRβ rearrangement analysis
DNA was prepared from single-cell suspensions of T-cell tumors prior to PCR amplification of the Dβ2-Jβ2 rearrangements of the TCRB locus.22
Semiquantitative reverse transcription–PCR analysis
Results
Reconstruction of the KIS-1 translocation by insertion of PAX5 into the IgH locus
To study the role of Pax5 in lymphomagenesis, we inserted a human PAX5 gene into the mouse IgH locus in the same configuration as described for the t(9;14)(p13;q32) translocation of the KIS-1 lymphoma.9,10,24 To this end, we generated a human PAX5 minigene containing 1807 bp of its 5′ flanking sequences and linked this gene via an internal ribosome entry sequence (IRES) to the green fluorescent protein (GFP) gene (Figure 1A). The PAX5-IRES-GFP expression cassette was inserted into the mouse IgH locus adjacent to the intronic Eμ enhancer in such a way that the DHQ52 element and all 4 JH segments were deleted, thus resulting in a nonfunctional IgH allele (Figure 1A). Targeted ES cells were subsequently used to generate IgHP5ki/+ mice (Figure 1B). The IgHP5ki allele was crossed into Pax5 mutant mice to investigate whether Pax5 expression from the IgH locus can rescue transcription of the Pax5 target gene CD19.1 Flow cytometric analysis revealed similar CD19 expression on cultured wild-type and IgHP5ki/+Pax5−/− pro-B cells in contrast to Pax5−/− pro-B cells, indicating that the IgHP5ki allele expresses a functional Pax5 protein (Figure 1C). The IgHP5ki allele was also able to rescue the early B-cell developmental block in Pax5−/− mice because mature IgM+IgD+ B cells could be detected in the spleens of IgHP5ki/+Pax5−/− mice (Figure 1D). However, these mice contained 6-fold fewer splenic B cells than wild-type mice (Figure 1D) due to a partial block of B-cell development at the pro–B- to pre–B-cell transition in the bone marrow (Figure S1). In summary, Pax5 expression from the IgHP5ki allele could restore B lymphopoiesis in Pax5−/− mice, although with low efficiency.
Expression of the IgHP5ki allele in lymphoid and dendritic cells
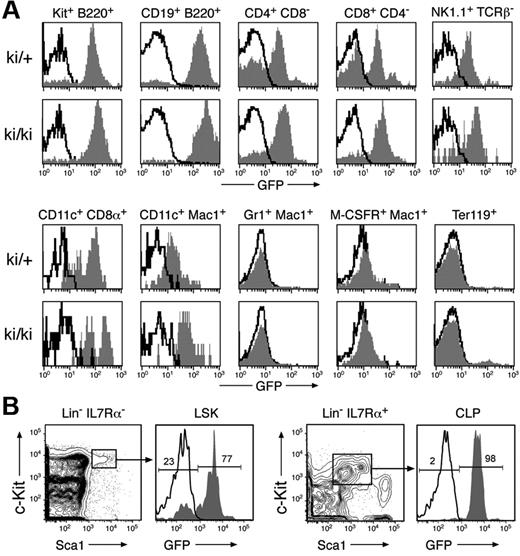
The IgH locus is transcriptionally active in T cells in addition to B cells, as thymocytes express germline Cμ transcripts.25 Moreover, the immunoglobulin Eμ enhancer can direct transgene expression in both B and T cells.26 To investigate the activity of the genetically modified IgH locus within the hematopoietic system, we analyzed GFP expression of the IgHP5ki allele in bone marrow and spleen cells of heterozygous and homozygous IgHP5ki mutant mice (Figure 2A) No GFP expression was detected in granulocytes (Gr1+Mac1+), monocyte/macrophages (M-CSFR+Mac1+), and erythroblasts (Ter119+). In contrast, the IgHP5ki allele was expressed in all lymphoid cell types, because GFP expression was observed in B lymphocytes (B220+CD19+), peripheral CD4+ and CD8+ T cells, NK cells (NK1.1+TCRβ−) as well as in CD11c+CD8α+ and CD11c+Mac1+ DCs (Figure 2A). As expected, GFP expression was higher in B cells compared to all other lymphoid cell types (Figure 2A). Surprisingly, GFP expression was strongly variegated in peripheral T lymphocytes because a large fraction of these cells failed to express GFP in heterozygous IgHP5ki/+ mice. Less variegation of GFP expression was, however, observed in T cells of homozygous IgHP5ki/P5ki mice (Figure 2A). We next studied the initiation of IgHP5ki expression in uncommitted bone marrow progenitors by analyzing GFP expression in multipotent progenitors (MPPs or LSKs; IL7Rα−Lin−Sca1hic-Kithi) and common lymphoid progenitors (CLPs; Lin−IL7Rα+Sca1loc-Kitlo). GFP expression was absent in 23% of the IgHP5ki/+ MPPs (Figure 2B), which included the LT-HSCs (Flt3−Lin−Sca-1hic-Kithi; data not shown). In contrast, all CLPs in the IgHP5ki/+ bone marrow expressed GFP at a high level (Figure 2B). Together these results indicate that the IgHP5ki allele is activated in a subset of MPPs and is subsequently expressed in DCs and all cell types of the lymphoid system.
Expression of the IgHP5ki allele in lymphoid and dendritic cells. (A) GFP expression in different hematopoietic cell types. Bone marrow (BM) cells and splenocytes (SPL) of 6-week-old IgHP5ki/+ (ki/+) and IgHP5ki/P5ki (ki/ki) mice were stained with the indicated antibodies, and GFP expression of the IgHP5ki knock-in alleles (gray area) was displayed for c-Kit+B220+ pro-B cells (BM), B220+CD19+ B lymphocytes (BM), CD4+ and CD8+ T cells (SPL), NK1.1+TCRβ− NK cells (SPL), CD11c+CD8α+ and CD11c+Mac1+ DCs (SPL), Gr1+Mac1+ granulocytes (BM), M-CSFR+Mac1+monocyte/macrophages (BM), and Ter119+ erythroblasts (BM). Black lines indicate the absence of GFP expression in the corresponding wild-type cells. (B) Expression of the IgHP5ki allele in uncommitted progenitors. GFP expression is displayed for the Lin−IL7Rα−Sca1hic-Kithi MPPs and Lin−IL7Rα+Sca1loc-Kitlo CLPs in IgHP5ki/+ (gray area) and wild-type (black line) BM. The percentage of GFP− and GFP+ cells is shown for MPPs and CLPs of the IgHP5ki/+ mouse.
Expression of the IgHP5ki allele in lymphoid and dendritic cells. (A) GFP expression in different hematopoietic cell types. Bone marrow (BM) cells and splenocytes (SPL) of 6-week-old IgHP5ki/+ (ki/+) and IgHP5ki/P5ki (ki/ki) mice were stained with the indicated antibodies, and GFP expression of the IgHP5ki knock-in alleles (gray area) was displayed for c-Kit+B220+ pro-B cells (BM), B220+CD19+ B lymphocytes (BM), CD4+ and CD8+ T cells (SPL), NK1.1+TCRβ− NK cells (SPL), CD11c+CD8α+ and CD11c+Mac1+ DCs (SPL), Gr1+Mac1+ granulocytes (BM), M-CSFR+Mac1+monocyte/macrophages (BM), and Ter119+ erythroblasts (BM). Black lines indicate the absence of GFP expression in the corresponding wild-type cells. (B) Expression of the IgHP5ki allele in uncommitted progenitors. GFP expression is displayed for the Lin−IL7Rα−Sca1hic-Kithi MPPs and Lin−IL7Rα+Sca1loc-Kitlo CLPs in IgHP5ki/+ (gray area) and wild-type (black line) BM. The percentage of GFP− and GFP+ cells is shown for MPPs and CLPs of the IgHP5ki/+ mouse.
Abnormal T-cell development in IgHP5ki mutant mice
We next investigated whether the early expression of the IgHP5ki allele in lymphoid progenitors affects the B- versus T-lineage choice similar to the pan-hematopoietic Pax5 expression of the IkPax5 allele.4 For this purpose, we reconstituted lethally irradiated C57BL/6 mice with a 1:1 mixture of syngeneic IgHP5ki/+ and control IgHμMT/+ bone marrow, which could be distinguished by expression of the allogenic markers Ly5.1 and Ly5.2 (Figure S2). Nine months after transplantation, B cells were preferentially derived from the IgHP5ki/+ HSCs (Figure S2A), whereas thymocytes exclusively developed from the IgHμMT/+ HSCs (Figure S2B). Hence, precocious Pax5 expression from the IgHP5ki allele prevents T lymphopoiesis at the expense of B-cell development as described for the IkPax5 allele.4
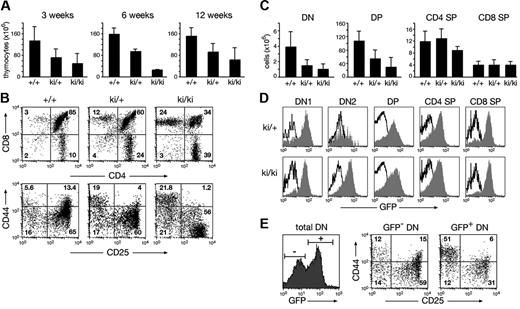
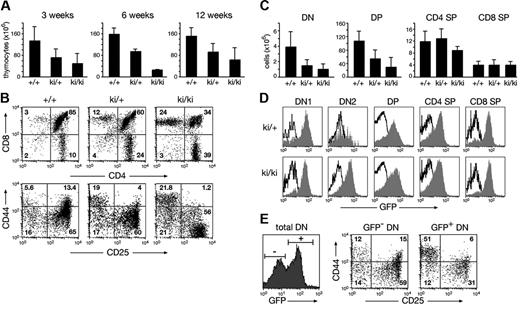
Under noncompetitive conditions, the pro-B cell compartment (c-Kit+B220+) was increased 2-fold in IgHP5ki/+ and IgHP5ki/P5ki mice relative to wild-type mice, whereas B-cell numbers were normalized at later developmental stages in IgHP5ki/+ mice (Figure S3A-C). The thymic cellularity of IgHP5ki/+ and IgHP5ki/P5ki mice was, however, reduced on average to 60% and 33% of the wild-type level, respectively (Figure 3A) Flow cytometric analysis revealed an abnormal distribution of the different thymocyte subsets in IgHP5ki/+ and IgHP5ki/P5ki mice (Figure 3B). The absolute number of the most immature CD4−CD8− double-negative (DN) and CD4+CD8+ double-positive (DP) thymocytes progressively decreased from heterozygous to homozygous mutant mice, whereas the total number of CD4+ and CD8+ single-positive (SP) thymocytes remained constant regardless of the genotype (Figure 3B-C). The early phase of T-cell development can be divided into 4 stages, progressing from CD44+CD25− (DN1) progenitors through the CD44+CD25+ (DN2) and CD44−CD25+ (DN3) stages to CD44−CD25− (DN4) cells.27 Analysis of the CD44/CD25 expression profile of Lin− DN thymocytes revealed a partial block at the earliest stage of pro-T cell development, as DN1 cells were 4- to 10-fold increased within the DN cell compartment of IgHP5ki/+ and IgHP5ki/P5ki mice compared to wild-type mice (Figure 3B; data not shown). Consistent with an early block in pro-T cell development, most DN1 cells of IgHP5ki/+ and IgHP5ki/P5ki mice expressed GFP at a high level (Figure 3D). The GFP expression profile of DN2 cells and thymocytes at later developmental stages was broader and thus more variegated, as a sizeable fraction of the cells failed to express GFP (Figure 3D). Analysis of the GFP− DN thymocytes revealed normal progression through the different stages of early T-cell development in IgHP5ki/+ mice (Figure 3E). In contrast, the GFP+ DN cells were preferentially blocked at the DN1 stage (Figure 3E). We conclude therefore that T-cell development is partially blocked at the earliest pro-T cell stage in IgHP5ki mutant mice and that expression of the IgHP5ki allele is variegated at later developmental stages.
Abnormal T-cell development in IgHP5ki mutant mice. (A) Decreased cellularity of the thymus in IgHP5ki/+ and IgHP5ki/P5ki mice. The average number of thymocytes (× 106) was determined at 3, 6, and 12 weeks with at least 3 mice analyzed per genotype and time point. Error bars represent standard deviations of the mean. (B) Aberrant T lymphopoiesis in IgHP5ki mutant mice. Total Thy1.2+ thymocytes of 3-week-old mice were examined for CD4 and CD8 expression and the Lin−Thy1.2+ DN thymocytes for CD44 and CD25 expression. (C) Reduction of DN and DP thymocytes in IgHP5ki mutant mice at the age of 3 weeks. Absolute cell numbers (× 106) are shown for the indicated mice (at least 4/genotype). (D) GFP expression of the IgHP5ki allele (gray area) in the different thymocyte subsets. Black lines indicate the absence of GFP expression in wild-type thymocytes. (E) Variegated phenotype in a 6-week-old IgHP5ki/+ mouse. The CD44/CD25 expression profile is shown for gated GFP− and GFP+ cells of the Thy1.2+ DN thymocyte population.
Abnormal T-cell development in IgHP5ki mutant mice. (A) Decreased cellularity of the thymus in IgHP5ki/+ and IgHP5ki/P5ki mice. The average number of thymocytes (× 106) was determined at 3, 6, and 12 weeks with at least 3 mice analyzed per genotype and time point. Error bars represent standard deviations of the mean. (B) Aberrant T lymphopoiesis in IgHP5ki mutant mice. Total Thy1.2+ thymocytes of 3-week-old mice were examined for CD4 and CD8 expression and the Lin−Thy1.2+ DN thymocytes for CD44 and CD25 expression. (C) Reduction of DN and DP thymocytes in IgHP5ki mutant mice at the age of 3 weeks. Absolute cell numbers (× 106) are shown for the indicated mice (at least 4/genotype). (D) GFP expression of the IgHP5ki allele (gray area) in the different thymocyte subsets. Black lines indicate the absence of GFP expression in wild-type thymocytes. (E) Variegated phenotype in a 6-week-old IgHP5ki/+ mouse. The CD44/CD25 expression profile is shown for gated GFP− and GFP+ cells of the Thy1.2+ DN thymocyte population.
Development of immature T-lymphoblastic lymphoma in IgHP5ki mutant mice
Kaplan-Meier survival analysis demonstrated that half of the IgHP5ki/P5ki mice died within 4.4 months (Figure 4A) As shown by macroscopic or histologic analysis, the homozygous IgHP5ki mutant mice developed disseminated lymphoma with complete penetrance by the age of 10 months. Lymphomas also occurred in heterozygous IgHP5ki/+ mice, although with a longer latency and at a lower incidence, because only 16% of all heterozygous mice died within 1 year (Figure 4A). All diseased mice showed splenomegaly and enlarged lymph nodes, whereas the majority of moribund mice also had a huge thymus (Figure 4B-D). Histologic analysis of the thymus, spleen, and lymph nodes revealed complete disruption of the normal architecture by a monomorphic population of medium-sized lymphoid cells, as shown for the thymus of a diseased mouse (Figure 4F) compared to a wild-type mouse (Figure 4E). Tumor cells were also found outside the capsule of the thymus and lymph nodes, where they infiltrated into adjacent tissues (Figure S4A). Extensive tumor infiltrates were furthermore detected in the lung (Figure 4H), liver (Figure 4I), kidney (Figure 4K), salivary gland, testis, and heart of moribund mice, which demonstrates efficient spreading of the tumor cells into nonlymphoid organs. Cytologically, the tumor cells displayed a high nuclear-to-cytoplasmic ratio; their nuclei were round to ovoid in shape and contained a predominantly fine chromatin (Figure 4G). The tumor cells furthermore showed high mitotic and apoptotic activity (Figure 4G).
Lymphoma development in IgHP5ki mutant mice. (A) Kaplan-Meier survival curves. The survival of IgHP5ki/+ mice (n = 153) and IgHP5ki/P5ki mice (n = 46) was followed over time. Lymphomas were detected in all dead IgHP5ki/P5ki mice, which were subjected to macroscopic or histologic analysis. Because we were unable to analyze 36% of the IgHP5ki/P5ki mice at death, we conclude that at least 64% of all homozygous mutant mice died due to lymphoma development. (B-D) Gross aspect of the thymus (B), spleen (C), and lymph nodes (D) of the moribund IgHP5ki/P5ki (ki/ki) tumor mouse 11 and a wild-type (+/+) littermate. (E-G) Disrupted architecture of the thymus in the IgHP5ki/P5ki tumor mouse 11. Histologic sections of the thymus from a wild-type mouse (E) and the IgHP5ki/P5ki tumor mouse (F-G) were stained with eosin and hematoxylin. The cytologic details of the tumor cells are shown at higher magnification in panel G. CO indicates cortex; ME, medulla. (H-K) Infiltration of the lung (H), liver (I), and kidney (J) by lymphoma cells in the diseased IgHP5ki/P5ki tumor mouse 11.
Lymphoma development in IgHP5ki mutant mice. (A) Kaplan-Meier survival curves. The survival of IgHP5ki/+ mice (n = 153) and IgHP5ki/P5ki mice (n = 46) was followed over time. Lymphomas were detected in all dead IgHP5ki/P5ki mice, which were subjected to macroscopic or histologic analysis. Because we were unable to analyze 36% of the IgHP5ki/P5ki mice at death, we conclude that at least 64% of all homozygous mutant mice died due to lymphoma development. (B-D) Gross aspect of the thymus (B), spleen (C), and lymph nodes (D) of the moribund IgHP5ki/P5ki (ki/ki) tumor mouse 11 and a wild-type (+/+) littermate. (E-G) Disrupted architecture of the thymus in the IgHP5ki/P5ki tumor mouse 11. Histologic sections of the thymus from a wild-type mouse (E) and the IgHP5ki/P5ki tumor mouse (F-G) were stained with eosin and hematoxylin. The cytologic details of the tumor cells are shown at higher magnification in panel G. CO indicates cortex; ME, medulla. (H-K) Infiltration of the lung (H), liver (I), and kidney (J) by lymphoma cells in the diseased IgHP5ki/P5ki tumor mouse 11.
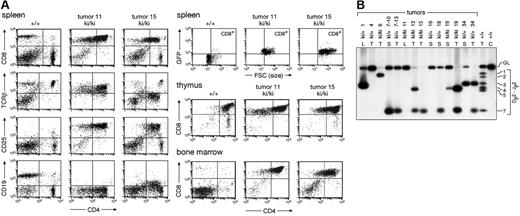
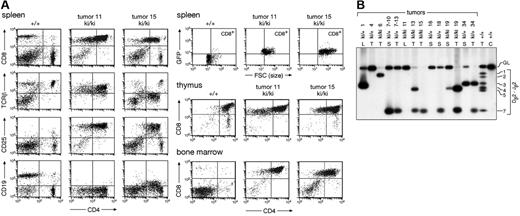
Immunophenotyping of the lymphoma cells isolated from the thymus, bone marrow, spleen, and lymph nodes of homozygous and heterozygous IgHP5ki mutant mice revealed that most tumor cells were CD4+CD8+ in all lymphoid organs with a varying contribution of CD8 SP cells (Figure 5A; data not shown). In about 20% of moribund mice, the lymphoma cells in the bone marrow and peripheral lymphoid organs consisted mainly of CD8 SP T cells, although the corresponding lymphoma in the thymus predominantly contained DP T cells. Detailed flow cytometric analysis demonstrated that the DP and CD8 SP lymphoma cells expressed TCRβ at a low (tumor 15) to intermediate (tumor 11) level, were large in size, and had an activated CD25+ phenotype (Figure 5A). Importantly, the lymphomas expressed GFP and thus Pax5 (Figure 5A). Moreover, some of the tumors also expressed the Pax5 target gene CD19 at a low level, as exemplified by tumor 15 (Figure 5A). We conclude, therefore, that forced Pax5 expression in thymocytes induces the formation of immature T-cell lymphomas.
Ectopic Pax5 expression results in immature T-cell lymphomas. (A) Immunophenotype of lymphoma cells. The spleen, thymus, and bone marrow of a wild-type (+/+) and 2 IgHP5ki/P5ki (ki/ki) tumor mice were analyzed for CD4 and CD8 expression; TCRβ, CD19, CD25, and GFP expression is also shown for the splenocytes. (B) Clonal TCRβ rearrangements in Pax5-induced T-cell lymphomas. Dβ2-Jβ2 rearrangements were analyzed by PCR amplification of tumor DNA isolated from the thymus (T), lymph node (L), or spleen (S) of IgHP5ki/+ and IgHP5ki/P5ki mice. The different tumor mice are indicated by their numbers. The designations 7-10 and 7-13 refer to secondary lymphomas in the spleen of the recipient mice 10 and 13, which were intravenously injected with lymphoma cells from the spleen or thymus of tumor mouse 7, respectively. C indicates control tail DNA. Numbers to the right refer to the different Jβ2 segments and GL to the germline PCR product.
Ectopic Pax5 expression results in immature T-cell lymphomas. (A) Immunophenotype of lymphoma cells. The spleen, thymus, and bone marrow of a wild-type (+/+) and 2 IgHP5ki/P5ki (ki/ki) tumor mice were analyzed for CD4 and CD8 expression; TCRβ, CD19, CD25, and GFP expression is also shown for the splenocytes. (B) Clonal TCRβ rearrangements in Pax5-induced T-cell lymphomas. Dβ2-Jβ2 rearrangements were analyzed by PCR amplification of tumor DNA isolated from the thymus (T), lymph node (L), or spleen (S) of IgHP5ki/+ and IgHP5ki/P5ki mice. The different tumor mice are indicated by their numbers. The designations 7-10 and 7-13 refer to secondary lymphomas in the spleen of the recipient mice 10 and 13, which were intravenously injected with lymphoma cells from the spleen or thymus of tumor mouse 7, respectively. C indicates control tail DNA. Numbers to the right refer to the different Jβ2 segments and GL to the germline PCR product.
Cell lines of different lymphomas could be readily established in vitro within 2 to 3 weeks in coculture with stromal ST2 cells. These cell lines largely maintained the phenotype of the original tumor (Figure S5). Moreover, the tumor cells of heterozygous and homozygous IgHP5ki mutant mice rapidly gave rise to secondary lymphomas in different organs within 7 weeks after intravenous injection into wild-type syngeneic mice (Figures S4D-F and S5). We next studied the clonal origin of the different tumors. PCR analysis of the TCRβ recombination status of IgHP5ki/+ and IgHP5ki/P5ki lymphomas revealed that each tumor was clonal because it contained maximally 2 distinct Dβ2-Jβ2 rearrangements (Figure 5B). Every tumor was therefore derived from a single T cell that, in addition to ectopic Pax5 expression, had to acquire further genetic alterations to undergo malignant transformation. We identified the same clonal origin for the lymphomas in the spleen and thymus of tumor mouse 7 (Figure 5B). Surprisingly however, the rearrangement pattern differed for the lymphomas present in the same 2 organs of tumor mouse 19 or 34 (Figure 5B; Figure S6A-B). This difference could result from 2 independent oncogenic events giving rise to distinct monoclonal tumors in different organs of these mice. Alternatively, the different rearrangement patterns may arise from ongoing V(D)J recombination in CD4+CD8+ lymphoma cells due to continuous RAG protein expression. Together these data demonstrate that ectopic Pax5 expression in the T-lymphoid lineage leads to the development of aggressive immature T-cell lymphomas.
IkPax5/+ mutant mice also die of aggressive T-lymphoblastic lymphomas
Previously, we have shown that Pax5 expression under the control of the Ikaros (Ik) locus results in grossly abnormal T-cell development in IkPax5/+ mice.4 Already at 2 weeks, the thymic cellularity and the number of DP thymocytes were more strongly reduced in these mice4 compared to IgHP5ki/P5ki mice (Figure 3A). Consistent with more severe developmental abnormalities, all IkPax5/+ mice died between 2 and 4 months (data not shown). Moribund IkPax5/+ mice had enlarged thymus, spleen, and lymph nodes, and histologic analysis of these lymphoid organs revealed diffuse infiltration by lymphoblastic lymphoma (Figure 6A). Extensive tumor infiltrates were found in many nonlymphoid organs such as the heart (Figure 6B), lung (Figure 6C), liver, kidney, intestine, and testis. The tumor cells in the thymus and spleen were monoclonal and mostly CD25+CD19+TCRβintCD4+CD8− (Figure 6D-E), although some tumors predominantly consisted of either DN or DP T cells (data not shown). Immature T-cell lymphomas even developed in Ikneo/+lck-cre mice (Figure 6D-E), where ectopic Pax5 expression was exclusively activated within the T-lymphoid lineage by Cre-mediated deletion of a floxed neomycin stop cassette.4 Together these data indicate that ectopic Pax5 expression in IkPax5/+ thymocytes also results in the formation of immature T-cell lymphomas, although with shorter latency compared to IgHP5ki mutant mice.
Development of immature T-cell lymphomas in IkPax5 mutant mice. (A-C) Histologic analysis of the lymphomas in the IkPax5/+ tumor mouse 80. A hematoxylin and eosin-stained section of the thymus (A) reveals the characteristic appearance of lymphoblastic lymphoma. The heart (B) and lung (C) are massively infiltrated by lymphoma cells. (D-E) Immunophenotype of immature T-cell lymphomas in the thymus (D) and spleen (E) of IkPax5/+ and Ikneo/+lck-cre mice.
Development of immature T-cell lymphomas in IkPax5 mutant mice. (A-C) Histologic analysis of the lymphomas in the IkPax5/+ tumor mouse 80. A hematoxylin and eosin-stained section of the thymus (A) reveals the characteristic appearance of lymphoblastic lymphoma. The heart (B) and lung (C) are massively infiltrated by lymphoma cells. (D-E) Immunophenotype of immature T-cell lymphomas in the thymus (D) and spleen (E) of IkPax5/+ and Ikneo/+lck-cre mice.
Gene expression changes in Pax5-expressing thymocytes
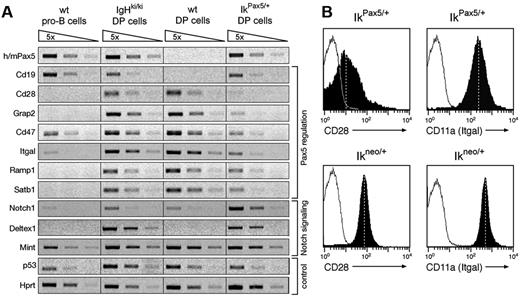
To gain molecular insight into the oncogenic role of Pax5 in T cells, we investigated the transcriptional changes in Pax5-expressing thymocytes. To this end, we compared the expression of candidate genes by semiquantitative reverse transcription–PCR (RT-PCR) in sorted GFP+ DP thymocytes from IgHP5ki/P5ki and IkPax5/+ mice with DP thymocytes and pro-B cells from wild-type mice. PCR conditions, which detect the human and mouse Pax5 mRNAs with equal efficiency, revealed that the human PAX5 minigene of the IgHP5ki and IkPax5 alleles was expressed in DP thymocytes at a similar level compared to the endogenous Pax5 gene in wild-type pro-B cells (Figure 7A). Because we previously measured a 5-fold higher expression level of the IkPax5 alleles in DN thymocytes,22 we confirmed this result by flow cytometric analysis of GFP expression, indicating that the transcription of the IkPax5 allele is down-regulated during the transition from DN to DP thymocytes (Figure S7). Previous expression analyses revealed that ectopic Pax5 expression activates the transcription of several B-cell–specific genes in IkPax5/+ DN thymocytes.22 Consistent with this finding, the Pax5 target gene CD19 was also expressed in DP thymocytes of IkPax5/+ and IgHP5ki/P5ki mice (Figure 7A). We next focused our attention on T-cell–specific genes that are repressed by Pax5 in lymphoid progenitors during B-cell commitment.7 The Pax5-repressed genes Cd28, Grap2 (Gads), Cd47, Itgal (Cd11a), Ramp1, and Satb1 were similarly expressed or only minimally affected in DP thymocytes of IgHP5ki/P5ki mice compared to wild-type mice (Figure 7A). However, the same genes were efficiently down-regulated in DP thymocytes of IkPax5/+ mice (Figure 7A), suggesting that the 5-fold higher Pax5 expression in IkPax5/+ DN thymocytes may be essential for initiating gene repression in early T-cell development. Flow cytometric analysis furthermore confirmed that the expression of the cell surface proteins CD28 and CD11a (Itgal) was down-regulated on DP thymocytes of IkPax5/+ mice relative to control Ikneo/+ mice (Figure 7B). This experiment also demonstrated that ectopic Pax5 expression rather than Ikaros heterozygosity is responsible for the down-regulation of CD28 and CD11a (Figure 7B). Surprisingly, Notch1 transcription, which is normally repressed by Pax5 in the B-cell lineage,4 was even increased by ectopic Pax5 expression in DP thymocytes of IgHP5ki/P5ki and IkPax5/+ mice (Figure 7A). The Mint gene coding for a repressor of Notch signaling was, however, similarly expressed in DP cells irrespective of the genotype (Figure 7A). In contrast, Deltex1, a direct target of Notch signaling,23 was expressed in DP thymocytes of all mutant mice compared to wild-type controls, suggesting that ectopic Pax5 expression promotes Notch signaling within the T-cell lineage. Hence, Pax5 appears to contribute to the rapid development of T-cell lymphomas in IkPax5/+ mice by activating B-cell–specific genes, repressing T-lymphoid genes and possibly enhancing Notch signaling.
Repression of T-lymphoid genes in Pax5-expressing thymocytes. (A) RT-PCR analysis of the indicated genes in sorted pro-B cells from wild-type (wt) bone marrow and in sorted DP thymocytes from wild-type, IgHP5ki/P5ki, and IkPax5/+ mice. The cDNA was normalized for equal expression of the hypoxanthine phosphoribosyltransferase (Hprt) gene prior to semiquantitative PCR of 5-fold serial cDNA dilutions. (B) CD28 and CD11a (Itgal) expression on gated DP thymocytes of IkPax5/+ and control Ikneo/+ mice (filled areas) as compared with wild-type DP thymocytes (black lines). The mean fluorescence is indicated by vertical dashed lines.
Repression of T-lymphoid genes in Pax5-expressing thymocytes. (A) RT-PCR analysis of the indicated genes in sorted pro-B cells from wild-type (wt) bone marrow and in sorted DP thymocytes from wild-type, IgHP5ki/P5ki, and IkPax5/+ mice. The cDNA was normalized for equal expression of the hypoxanthine phosphoribosyltransferase (Hprt) gene prior to semiquantitative PCR of 5-fold serial cDNA dilutions. (B) CD28 and CD11a (Itgal) expression on gated DP thymocytes of IkPax5/+ and control Ikneo/+ mice (filled areas) as compared with wild-type DP thymocytes (black lines). The mean fluorescence is indicated by vertical dashed lines.
Discussion
Four of the 9 developmental control genes of the human PAX family have been associated with specific chromosomal translocations in human tumors. These translocations generate novel transcription factors by fusing the N-terminal DNA-binding domain of the Pax protein with the C-terminal regulatory sequences of a second transcription factor.28,29 PAX3-FKHR and PAX7-FKHR translocations are found in alveolar rhabdomyosarcoma, a pediatric muscle tumor,28 the PAX8-PPARG1 translocation in thyroid follicular carcinoma,30 and the PAX5-ETV6 translocation in B-progenitor acute lymphoblastic leukemia.31 Where analyzed, the fusion protein proved to be a more potent transcriptional activator than the corresponding wild-type Pax protein, suggesting that the fusion proteins contribute to oncogenesis by deregulating the expression of Pax target genes.28 PAX5 also participates in a second, regulatory translocation, t(9;14)(p13;q32), which leads to deregulated expression of a structurally intact PAX5 gene under the control of the IgH locus in a subset of B-NHLs. Ectopic expression of PAX5 was also observed in medulloblastoma,32 glioblastoma,33 and neuroblastoma,34 although the molecular mechanisms causing PAX5 deregulation in these common tumors of the nervous system are still unknown. Despite the compelling association of PAX genes with human tumors, the oncogenic potential of these genes has not yet been demonstrated in transgenic mouse models in vivo.29
Here we have described the characterization of an IgHP5ki knock-in mouse that reproduces the specific structural and regulatory features of the t(9;14)(p13;q32) translocation identified in the human KIS-1 lymphoma.9,10,24 One important difference between this knock-in mouse and the corresponding human lymphoma is the fact that the PAX5 insertion in the IgHP5ki allele corresponds to a germline mutation, whereas the IgH-PAX5 translocation in human lymphomas arises in germinal center B cells during late B-cell development.9-11 Consequently, the inserted PAX5 gene is transcribed in all IgH-expressing cells of the knock-in mouse, which contrasts with the exclusive expression of the endogenous Pax5 gene in the B-lymphoid lineage of wild-type mice.14 The expression of the IgHP5ki allele is initiated in multipotent hematopoietic progenitors and is subsequently expressed in DCs and in NK, T, and B cells. Interestingly, the T-lymphoid lineage proved to be particularly sensitive to the oncogenic action of Pax5 because the IgHP5ki/+ and IgHP5ki/P5ki mice die of immature T-lymphoblastic lymphoma. Although the IgHP5ki knock-in mouse unequivocally identifies Pax5 as an oncogene, the development of T-lymphoblastic lymphomas was surprising because the PAX5 gene has not been implicated in the genesis of human T-cell tumors so far.35 One possible reason for the absence of PAX5 in chromosomal translocations involving the TCRB or TCRA loci is provided by the observation that all 3 genes are present in the same centromere-to-telomere orientation in the human genome. Hence, any head-to-head rearrangement between PAX5 and a TCR locus would result in a translocation chromosome lacking any centromere and thus in its rapid loss in contrast to the IgH-PAX5 translocation. However, the transcription factor genes TAL1(SCL), LYL1, LMO1, LMO2, HOX11, and HOX11L2 are activated as T-cell oncogenes by translocations involving the TCRA or TCRB locus.35 Importantly, the forced T-cell–specific expression of these transcriptional regulators, which like Pax5 are normally not expressed in the T-lymphoid lineage, is the most prevalent cause for human T-cell leukemia.35
The substitution of the JH gene segments by the PAX5 minigene interferes with V(D)J recombination of the IgH gene and thus arrests B-cell development at the pro-B cell stage in homozygous IgHP5ki/P5ki mice similar to previously published mice lacking the JH domain.36 The absence of B cells can lead to opportunistic infections, which may explain the significantly higher mortality of IgHP5ki/P5ki mice compared to heterozygous IgHP5ki/+ mice. However, we could detect lymphomas in all dead IgHP5ki/P5ki mice analyzed, indicating that death by opportunistic infections is an unlikely reason for the lower survival of these mice. An alternative explanation is provided by the observed variegated expression of the IgHP5ki allele within the T-lymphoid lineage. T cells, which fail to express the IgHP5ki allele and thus evade the oncogenic action of Pax5, are more abundant in heterozygous IgHP5ki/+ mice compared to homozygous IgHP5ki/P5ki mice. The variegated expression of the IgHP5ki allele could reflect the preferential location of the IgH locus at the nuclear periphery of thymocytes, which constitutes a repressive chromatin compartment.22 In addition, the IgH locus may be subject to classic position-effect variegation in T cells in agreement with the finding that the Eμ enhancer is able to induce variegated expression of a CD2 transgene in T lymphocytes.37 The probability that 1 of the 2 IgH alleles escapes the negative influence of repressive chromatin is thus increased in homozygous IgHP5ki/P5ki mice, which could explain their higher tumor incidence.
Pax5 expression from the IgHP5ki allele interferes with T-cell development prior to lymphoma formation, because DN and DP thymocytes are reduced in IgHP5ki/+ and IgHP5ki/P5ki mice. In particular, Pax5 expression leads to an incomplete block at the earliest DN1 progenitor cell stage of T-cell development. This early block is a likely reason that the IgHP5ki/+ HSCs in competition with wild-type HSCs fail to give rise to T-cell development in bone marrow chimeras. T lymphopoiesis is even more severely affected in IkPax5/+ mice,4 which express Pax5 from the Ikaros locus at approximately a 5-fold higher level in early T-cell development compared to the IgHP5ki allele22 (Figure S7). In agreement with a higher Pax5 expression level, the IkPax5/+ mice develop aggressive T-cell lymphomas with a shorter latency than IgHP5ki/P5ki mice. T cells of heterozygous Ik+/− mice are known to be hyperproliferative,38 and homozygous Ik−/− mice develop T-cell lymphomas, which identified Ikaros as a tumor suppressor gene of the T-lymphoid lineage.39 Hence, the IkPax5 allele provides 2 oncogenic hits, that is, ectopic expression of Pax5 and inactivation of one Ikaros gene, which together facilitate the more rapid development of T-cell lymphomas in IkPax5/+ mice.
Molecular analyses previously revealed that the IkPax5/+ pro-T (DN) cells activate the B-lymphoid transcription factor gene EBF in addition to Pax5, which leads to the expression of multiple B-cell–specific genes in early thymocytes.22 Consistent with this finding, B-lymphoid genes, as exemplified by CD19, are also expressed to varying degrees in IgHP5ki mutant thymocytes. An important aspect of B-cell commitment is the Pax5-dependent repression of B-lineage inappropriate genes, some of which are normally expressed in the T-lymphoid lineage.2,7 Here, we have shown that Pax5 represses several of these T-lymphoid genes with particularly high efficiency in IkPax5/+ thymocytes. Some of the repressed genes are essential for normal T-cell development or function because they code for the costimulatory receptor CD28,40 the integrin receptor LFA-1 (Itgal),41 the integrin-associated protein CD47,42 the signaling adaptor protein Grap2 (Gads),43 and the nuclear architectural protein Satb1.44 Paradoxically, Pax5 is known to interfere with the entry of lymphoid progenitors into the T-cell lineage by down-regulating Notch1 transcription4 and, yet within the T-lymphoid lineage, Pax5 may enhance Notch signaling as evidenced by increased Deltex1 expression. Activating mutations of NOTCH1 are known to be a frequent cause for human T-lymphoblastic leukemia.45 Hence, Pax5 appears to contribute to T-cell lymphoma development by activating B-cell–specific genes, repressing T-lymphoid genes, and possibly enhancing Notch signaling.
The BCL6 gene, coding for a transcriptional repressor involved in germinal center B-cell development, is activated as an oncogene by translocation to the IgH locus in human diffuse large B-cell lymphoma (DLBCL) similar to PAX5 in B-NHL.46 A mouse model recapitulating the IgH-BCL6 translocation has recently been generated by insertion of a BCL6 minigene downstream of the Iμ promoter of the endogenous IgH locus.47 Contrary to the IgHP5ki allele, the IμBCL6 allele appears to be expressed only in B cells, where it results in a lymphoproliferative disease that progresses to DLBCL with a latency period of more than 1 year.47 We have monitored the IgHP5ki/+ mice colony maximally for 1 year and identified only a single GFP+CD19+ plasmacytoma in addition to many T-cell lymphomas (data not shown). B-cell tumors may either develop after 1 year in IgHP5ki/+ mice or may require normal functional T-helper cells to promote germinal center formation in contrast to the aberrant T-cell development observed in IgHP5ki/+ mice. To bypass this problem and to prevent T-cell lymphoma formation, we have recently generated a conditional IgHP5ki allele, which additionally contains a floxed transcription stop cassette in PAX5 intron 1. Deletion of this stop cassette by an Aid-cre transgene activates PAX5 expression in germinal center B cells, where the IgH-PAX5 translocation normally arises in human patients. Hence, the conditional IgHP5ki/+ mouse has the potential to more faithfully recapitulate the IgH-PAX5 translocation characteristic of human B-NHLs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: A.S. generated and characterized the IgHP5Ki knock-in mouse, W.J. performed the histologic analyses of the tumors, and M.B. contributed to the planning of the experiments and wrote the manuscript.
This work was supported by Boehringer Ingelheim and by the Austrian Industrial Research Promotion Fund. We thank K. Rajewsky for providing the IgH targeting vector pIVhL2neor, M. Forsberg for the PAX5 minigene construct, C. Cobaleda for help in tumor analysis, and C. Theussl for blastocyst injection.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal