Abstract
MicroRNAs (miRNAs) have recently come into focus as key posttranscriptional modulators of gene expression. In this work, we addressed whether in vitro angiogenesis is an miRNA-regulated process. We performed large-scale analysis of miRNA expression in human umbilical vein endothelial cells (HUVECs) and found that 15 highly expressed miRNAs have the receptors of angiogenic factors as putative targets. In particular, we demonstrated that miR-221 and miR-222 affect c-Kit expression and, as a consequence, the angiogenic properties of its ligand stem cell factor. Interaction between miR-222 and c-Kit is likely to be part of a complex circuit that controls the ability of endothelial cells to form new capillaries.
Introduction
Angiogenesis is performed in the adult organism by the mobilization of endothelial cells from preexisting capillaries and requires degradation of extracellular matrix, migration, proliferation, and organization in new capillary networks.1 MicroRNAs (miRNAs) have recently come into focus as a powerful mechanism to control gene expression after transcription. They are short, single-stranded RNA molecules transcribed from noncoding genes, which, after maturation and entry into the RNA interference pathway,2,3 bind by imperfect pairing to the 3′UTR of mRNAs and inhibit translation.4 A growing body of evidence suggests a key regulatory role for miRNAs5-8 during embryogenesis and differentiation of adult tissues, with each differentiation grade characterized by a specific miRNA signature.9
Human umbilical vein endothelial cells (HUVECs) are a valuable model of in vitro angiogenesis because of their ability to form capillarylike structures called tubes in response to appropriate stimuli.10 In this study, we asked whether miRNAs are involved in the posttranscriptional modulation of angiogenesis-related genes. To this end, the miRNA signature of HUVECs was acquired, and several highly expressed miRNAs were found to have angiogenic growth factor receptors as putative targets. In particular, we demonstrated that the miR-221/miR-222 family modulates the angiogenic activity of stem cell factor (SCF) by targeting its receptor, c-Kit.
Study design
Reagents, cells, and culture conditions
Reagents are listed in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Endothelial cells were grown in M199 + 20% FBS, 12.5 U/mL heparin, and 20 ng/mL EGF on 1% gelatin-coated plates (Document S1, isolation protocol and purity assessment). HEK 293T cells were grown in DMEM + 10% FBS. Cells were grown at 37°C in a humidified atmosphere containing 6% CO2.
Transfections
For details on cell transfection, please refer to Document S1.
Molecular assays
miRNome analysis was performed according to Thomson et al.9 For immunodetection of c-Kit by immunohistochemistry, immunofluorescence, or Western blot, RT-PCR, plasmid construction, and EGFP reporter assay, please refer to Document S1.
Cellular assays
Statistical analysis
Reported data are the mean ± SD of at least 3 independent experiments and were compared using the 2-tailed unpaired t test.
Results and discussion
HUVECs, when seeded on Matrigel and stimulated with the appropriate factors, organize themselves into tubes that resemble capillaries. VEGF, bFGF, HGF, SCF, Ang-1, and SDF-1 are angiogenic factors able to induce tube formation and promote survival and migration of HUVECs; only the first 3 increase cell proliferation.10,12,13
To investigate whether the organization into tubes is an miRNA-regulated process, the miRNA signature of HUVECs was obtained and 27 highly expressed miRNAs were identified. Four published prediction algorithms14-17 were used to determine whether these miRNAs are predicted to target the receptors (Flt-1, Flk-1, Nrp-1, Nrp-2, FGF-R, c-Met, c-Kit, Tie-2, CXCR4) of the angiogenic factors listed. Receptors were chosen because they represent the initial step of the signal transduction cascade connecting each growth factor to the final biologic response. Results show (Table 1) that miR-23b, miR-23a, miR-let-7a, miR-let-7b, miR-125b, miR-221, miR-222, miR-125a, miR-24, miR-106a, miR-20, miR-16, miR-let-7c, miR-103, and miR-133a can potentially target 5 of these receptors.
Transmembrane receptors predicted to be targets of miRNAs
miRNA . | Flt-1 . | Nrp-2 . | FGF-R . | c-Met . | c-Kit . |
|---|---|---|---|---|---|
| 21 | — | — | — | — | — |
| 23b | — | — | — | b + d | — |
| 126 | — | — | — | — | — |
| 23a | — | — | — | b + c + d | — |
| 100 | — | — | — | — | — |
| let-7a | — | c | — | — | — |
| let-7f | — | — | — | — | — |
| 99a | — | — | — | — | — |
| let-7b | — | — | — | c | — |
| 125b | c | — | — | — | — |
| 221 | — | — | — | — | a, a, c |
| 223 | — | — | — | — | — |
| 222 | — | c | — | — | a, a |
| 31 | — | — | — | — | — |
| 125a | — | c | — | — | — |
| 24 | b + d | c | c | — | — |
| 29a | — | — | — | — | — |
| 29b | — | — | — | — | — |
| let-7d | — | — | — | — | — |
| 106a | b + c + d | c + d | — | — | — |
| 20 | b + d | c + d | — | — | — |
| let-7e | — | — | — | — | — |
| 16 | — | b + c | — | — | — |
| let-7c | — | — | — | c | — |
| 103 | — | b + c | — | — | — |
| 29c | — | — | — | — | — |
| 133a | — | c | b + d, d | — | — |
miRNA . | Flt-1 . | Nrp-2 . | FGF-R . | c-Met . | c-Kit . |
|---|---|---|---|---|---|
| 21 | — | — | — | — | — |
| 23b | — | — | — | b + d | — |
| 126 | — | — | — | — | — |
| 23a | — | — | — | b + c + d | — |
| 100 | — | — | — | — | — |
| let-7a | — | c | — | — | — |
| let-7f | — | — | — | — | — |
| 99a | — | — | — | — | — |
| let-7b | — | — | — | c | — |
| 125b | c | — | — | — | — |
| 221 | — | — | — | — | a, a, c |
| 223 | — | — | — | — | — |
| 222 | — | c | — | — | a, a |
| 31 | — | — | — | — | — |
| 125a | — | c | — | — | — |
| 24 | b + d | c | c | — | — |
| 29a | — | — | — | — | — |
| 29b | — | — | — | — | — |
| let-7d | — | — | — | — | — |
| 106a | b + c + d | c + d | — | — | — |
| 20 | b + d | c + d | — | — | — |
| let-7e | — | — | — | — | — |
| 16 | — | b + c | — | — | — |
| let-7c | — | — | — | c | — |
| 103 | — | b + c | — | — | — |
| 29c | — | — | — | — | — |
| 133a | — | c | b + d, d | — | — |
MicroRNAs expressed at the highest level and receptors of angiogenic factors are listed in the first column and the first row of the table, respectively. Flt-1/VEGF-R1 receptor and Nrp-2 receptor are bound by vascular endothelial growth factor (VEGF). FGF-R, c-Met, and c-Kit are the receptors of basic fibroblast growth factor (bFGF), hepatocyte growth factor (HGF), and stem cell factor (SCF), respectively. TargetScan14 (a), TargetScanS15 (b), miRanda17 (c), and PicTar16 (d) prediction algorithms were used to identify miRNAs that putatively targeted the 3′UTR of reported receptors. The letter is repeated when the same algorithm predicts more than one target. + indicates that the same target is predicted by more than one algorithm.
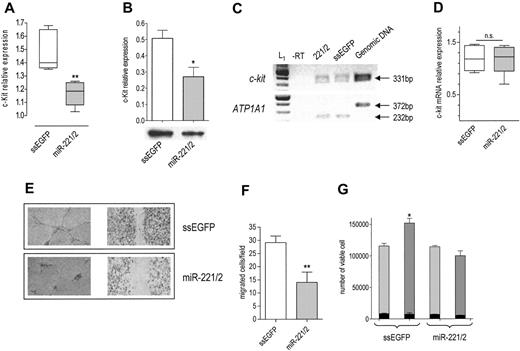
Given that they share the same seed, miR-221 and miR-222 belong to the same family.15 Their genes are located in close proximity on Xp11.3 chromosome.18,19 Interestingly, in c-Kit–positive HUVECs (Figure S1A), they are expressed in a common pri-miRNA (Figure S1B, third panel), suggesting a coordinated transcriptional regulation.20 According to the prediction algorithms considered,14-17 both miR-221 and miR-222 target c-Kit 3′UTR. Given that the activation of c-Kit by SCF promotes the tube formation, migration, and cell survival of HUVECs,10 we sought to determine whether the miR-221/miR-222 family modulates the levels and biologic activity of c-Kit. On transfection of the miR-221/miR-222 mix, c-Kit protein was decreased (Figure 1A-B) without a significant change in mRNA level (Figure 1C-D), a clear indication of posttranscriptional regulation. A decrease in receptor level was expected to dampen the biologic effects of SCF. In keeping with this hypothesis, we found that miR-221/miR-222–transfected cells were no more able to form tubes (Figure 1E, left panel) or to heal wounds (Figure 1E, right panel) in response to SCF. In addition, cell migration potential was reduced (Figure 1F). We also considered the effect of miR-221/miR-222 transfection on SCF-induced cell survival.10 The total number of ssEGFP-transfected cells exposed to SCF for 48 hours was higher than that of miR-221/miR-222–transfected cells cultured in the same conditions but still lower than that expected after one cell doubling (Figure 1G), whereas the fraction of dead cells was similar in both samples before and after SCF addition (Figure 1G, black bars). Taken together these results support the conclusion that miR-221/miR-222 indeed reduced SCF-induced survival without affecting the death rate. Such a deep change in biologic properties caused by a partial protein reduction might be explained by ligand-receptor binding triggering a signal that is highly amplified within the cells, and it confirms the importance of a fine modulation of receptor expression for the final biologic outputs.
To demonstrate that the interaction between miR-221/miR-222 and c-Kit is direct, a heterologous system was set up. On c-kit 3′UTR, miRanda predicts target site 215 for miR-221, and TargetScan predicts target sites 1030 and 1959 for miR-221 and miR-222 (Figure S2A). Target site 1959 is not present in all the published c-kit mRNA sequences. To ensure its presence in HUVECs, we performed reverse transcription–polymerase chain reaction (RT-PCR) with the appropriate pair of primers. Because the sequencing of the PCR product (Figure S2B) demonstrated the presence of the 1959 target site in c-kit mRNA, a c-kit 3′UTR fragment that encompasses all 3 targets was amplified and cloned downstream of EGFP ORF21 within the pEGFP-C1 plasmid to obtain the p3′UTR plasmid (Figure S2C). p3′UTR was transfected in HEK 293T cells, together with the aspecific miR-26a expression plasmid (p26a) or with the specific expression plasmids p221 and p222 (Figure S2D), which showed a strong relative activity on the p3′UTR plasmid (Figure S2E) that was absent when single target sites (p215, p1030, p1959; Figure S2C) were used. These data indicate a direct interaction of miR-221 and miR-222 with c-Kit 3′UTR and suggest that cooperation among the 3 target sites for miR-221 or between the 2 target sites for miR-222 is necessary to obtain a significant effect. However, we cannot rule out the possibility that other unknown target sites for the miR-221/miR-222 family could cooperate with those predicted by the algorithms considered.
Effects of miR-221 and miR-222 transfection in HUVECs. c-Kit protein expression in cells transfected with ssEGFP or with the miR-221/miR-222 mix was detected 48 hours after transfection by (A) FACS (**P < .01) or (B) by Western blot analysis (band intensity normalized to that of tubulin; *P < .05). (C) RT-PCR of c-Kit. (top) c-kit mRNA level was detected by RT-PCR in control and miR-221/miR-222 mix–transfected cells by amplification of a 331-bp fragment. (bottom) A 232-bp fragment of ATP1A1 mRNA was amplified from the same RT samples to ensure equal loading per lane. The absence of the 372-bp fragment in lanes 1 to 3 indicates that the cDNA is not contaminated by genomic DNA. A representative experiment of 3 performed is reported. L1 indicates 100-bp ladder. (D) Quantification of c-Kit mRNA. RT-PCR band intensity of c-Kit and ATP1A1 was quantified using image analysis software, and their ratio was used to compare relative expression. (E) SCF-induced tube formation (left panel): 48 hours after transfection, HUVECs were seeded onto Matrigel and were exposed to 100 ng/mL SCF + 2% FBS for 6 hours. Images of wound healing were taken after staining with crystal violet. An EPSON Expression 1680 Pro scanner (Seiko Epson, Nagano, Japan) equipped with a transparent unit was used to acquire the images. EPSON Twain Network version 2.00E and Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) were used for image acquisition and elaboration. Results of 1 of 3 representative experiments are reported. SCF-induced wound healing (right panel): HUVECs were collected 24 hours after transfection, counted, and seeded at a density of 2 × 105 on a coated 12-well multiplate. Six hours later, the monolayers were wounded and healing was observed after overnight incubation. The tube formation was photographed after staining with crystal violet. Images were acquired with a Wilovert inverted microscope (Hund, Wetzlar, Germany) equipped with 10×/0.25 objective lenses and a CoolPix 4500 digital camera (Nikon, Tokyo, Japan). (F) SCF-induced migration: HUVECs transfected with ssEGFP or miR-221/miR-222 mix were allowed to migrate through type I collagen–coated filters in response to 100 ng/mL SCF for 5 hours. The number of migrated cells per field (magnification, × 200) is reported. **P < .01. (G) Cell viability: cells were collected at the end of transfection (light gray bars) and after 48 hours of exposure to SCF (gray bars), stained with trypan blue, and counted. Total number of viable cells is reported. Dead cells (black bars) in control and miR-221/miR-222–transfected plates are also reported. *P < .05.
Effects of miR-221 and miR-222 transfection in HUVECs. c-Kit protein expression in cells transfected with ssEGFP or with the miR-221/miR-222 mix was detected 48 hours after transfection by (A) FACS (**P < .01) or (B) by Western blot analysis (band intensity normalized to that of tubulin; *P < .05). (C) RT-PCR of c-Kit. (top) c-kit mRNA level was detected by RT-PCR in control and miR-221/miR-222 mix–transfected cells by amplification of a 331-bp fragment. (bottom) A 232-bp fragment of ATP1A1 mRNA was amplified from the same RT samples to ensure equal loading per lane. The absence of the 372-bp fragment in lanes 1 to 3 indicates that the cDNA is not contaminated by genomic DNA. A representative experiment of 3 performed is reported. L1 indicates 100-bp ladder. (D) Quantification of c-Kit mRNA. RT-PCR band intensity of c-Kit and ATP1A1 was quantified using image analysis software, and their ratio was used to compare relative expression. (E) SCF-induced tube formation (left panel): 48 hours after transfection, HUVECs were seeded onto Matrigel and were exposed to 100 ng/mL SCF + 2% FBS for 6 hours. Images of wound healing were taken after staining with crystal violet. An EPSON Expression 1680 Pro scanner (Seiko Epson, Nagano, Japan) equipped with a transparent unit was used to acquire the images. EPSON Twain Network version 2.00E and Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) were used for image acquisition and elaboration. Results of 1 of 3 representative experiments are reported. SCF-induced wound healing (right panel): HUVECs were collected 24 hours after transfection, counted, and seeded at a density of 2 × 105 on a coated 12-well multiplate. Six hours later, the monolayers were wounded and healing was observed after overnight incubation. The tube formation was photographed after staining with crystal violet. Images were acquired with a Wilovert inverted microscope (Hund, Wetzlar, Germany) equipped with 10×/0.25 objective lenses and a CoolPix 4500 digital camera (Nikon, Tokyo, Japan). (F) SCF-induced migration: HUVECs transfected with ssEGFP or miR-221/miR-222 mix were allowed to migrate through type I collagen–coated filters in response to 100 ng/mL SCF for 5 hours. The number of migrated cells per field (magnification, × 200) is reported. **P < .01. (G) Cell viability: cells were collected at the end of transfection (light gray bars) and after 48 hours of exposure to SCF (gray bars), stained with trypan blue, and counted. Total number of viable cells is reported. Dead cells (black bars) in control and miR-221/miR-222–transfected plates are also reported. *P < .05.
Taken together, these results indicate that miR-221 and miR-222 modulate the angiogenic activity of SCF and of the level of its receptor c-Kit. We believe that the interaction between miR-221/miR-222 and c-kit, recently shown to play an important role in erythropoiesis,22 might also have a role in the formation of new vessels that occur in physiological and pathologic conditions (tumor growth). Further validation of the miRNA target pairs reported in Table 1 should help to discover the miRNA network responsible for the fine-tuning of the angiogenic process and, in the long term, to find new therapeutic approaches to modulate angiogenesis.
Prepublished online as Blood First Edition Paper, July 18, 2006; DOI 10.1182/blood-2006-01-012369.
L.P., A.T., and G.R. designed the research. L.P. and A.T. performed the research. L.M., L.C., M.E., A.M., and S.H. contributed analytical tools. L.P., A.T., K.W., S.H., and G.R. analyzed the data. L.P., A.T., and G.R. wrote the paper.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal