Abstract
We explore the controversial issue of the role of eosinophils in host defense against helminthic parasites using the established Schistosoma mansoni infection model in 2 novel mouse models of eosinophil lineage ablation (ΔdblGATA and TgPHIL). No eosinophils were detected in bone marrow of infected ΔdblGATA or TgPHIL mice, despite the fact that serum IL-5 levels in these infected mice exceeded those in infected wild type by approximately 4-fold. Liver granulomata from infected ΔdblGATA and TgPHIL mice were likewise depleted of eosinophils compared with those from their respective wild types. No eosinophil-dependent differences in granuloma number, size, or fibrosis were detected at weeks 8 or 12 of infection, and differential accumulation of mast cells was observed among the ΔdblGATA mice only at week 12. Likewise, serum levels of liver transaminases, alanine aminotransferase (ALT), and aspartate aminotransferase (AST) increased in all mice in response to S mansoni infection, with no eosinophil-dependent differences in hepatocellular damage observed. Finally, eosinophil ablation had no effect on worm burden or on egg deposition. Overall, our data indicate that eosinophil ablation has no impact on traditional measures of disease in the S mansoni infection model in mice. However, eosinophils may have unexplored immunomodulatory contributions to this disease process.
Introduction
The role of eosinophils in host defense and disease remains controversial, and the debate continues as to whether these cells are active participants or simply bystanders in various pathophysiologic states. This is particularly so with respect to disease caused by helminthic parasites. While it would seem logical to assume that eosinophils should provide a measure of host defense against these important and endemic infections, as they are elicited in large numbers in response to helminth infection, and they degranulate on and cause damage to various forms of the parasitic helminthes in various in vitro settings, the results from numerous experiments performed in vivo have been equivocal.1-3
The cytokine-mediated pathogenesis of the well-characterized mouse model of helminth infection, Schistosoma mansoni, has been described in great detail.4-7 This infection includes a prominent Th2 phase, resulting in an increase in serum interleukin-5 (IL-5) in response to egg deposition in the portal circulation at weeks 6 to 8 after exposure to water-borne cercariae. Increased serum IL-5 results in massive bone marrow and blood eosinophilia. Eosinophils are recruited specifically to the developing liver granulomata, the site of active inflammation and tissue remodeling. Several eosinophil components implicated in debris scavenging and tissue remodeling activity include the eosinophil peroxidase,8 the ribonucleases,9 matrix metalloproteinases,10 and the protease inhibitor, plasminogen activator inhibitor-2 (PAI-2).11 Eosinophils may also play an important role in maintaining the Th2 response to infection via secretion of endogenous IL-4.12,13
Several groups began the exploration of the role of eosinophils in host defense against helminth disease in vivo by using anti–IL-5 and anti–IL-5 signaling blockade strategies.14-18 Since that time there have been many peer-reviewed papers published (see reviews Klion and Nutman1 ; Behm and Ovington2 ; and Meeusen and Balic3 ) documenting the results of in vivo trials with different treatment strategies, pathogens used, and overall perspectives, with no clear consensus emerging. There are many reasons for the lack of clarity. One of the major observations in these studies that may confound the interpretation of the role of eosinophils in disease is that while all of the anti–IL-5 approaches do result in moderate to profound degrees of eosinophil depletion, IL-5 ablation does not eliminate the eosinophil lineage entirely.19 Thus, eosinophil accumulation, albeit reduced, is still a feature of disease. Furthermore, the confounding effect of removing IL-5, as opposed to removing eosinophils directly, remains a significant consideration.19-22 Notably, eosinophils have been shown to contribute to the pathogenesis of asthma and in mouse models of this disorder independently of IL-5.23,24
In this paper, we explore the role of eosinophils in the pathogenesis of helminth infection by using 2 novel models of complete eosinophil lineage ablation.25,26 The ΔdblGATA mice contain an engineered deletion of a palindromic double-enhancer binding site for GATA proteins in the region 5′ to the 1E exon of the gene encoding GATA-1, and are reported as devoid of eosinophils both at baseline and in response to cytokine challenge, without reported effects on other hematopoietic lineages.25 In the TgPHIL model, the lineage–specific eosinophil peroxidase promoter directs the expression of diphtheria toxin A transgene, resulting in the suicide-inactivation of differentiating eosinophils, again leading to mice devoid of eosinophils.26 By using both models, the specificity of the eosinophil-mediated contributions to helminth disease can be determined.
Materials and methods
Mice
Four male ΔdblGATA hemizygous mice (BALB/c background) and genotyping protocols were a generous gift from Dr Alison Humbles and Dr Craig Gerard. Additional male hemizygous and female homozygous mice were derived from breeding with wild-type BALB/c mice (Taconic Laboratories, Rockville, MD). Genotyping was performed on tail-snip DNA by standard methods using primer sequences, sense: 5′-CCC AAT CCT CTG GAC TCC CA-3′; antisense: 5′-CCT ACT GTG TAC CAG GCT AT-3′, with the 459–base pair product indicating the wild-type allele, and the 509–base pair product, the ΔdblGATA allele.25 Wild-type littermates and commercial BALB/c mice were used as controls. Transgenic male TgPHIL mice (C57BL/6 background) are mated with wild-type female mice; transgenic male and female mice are identified by characteristic phenotype and confirmed by polymerase chain reaction (PCR) as described.26 Age-matched transgenic and unaffected male and female littermate mice are used in experiments described.
Infection with Schistosoma mansoni and isolation of serum for cytokine and enzyme determinations
Infected mice were exposed percutaneously to 25 to 40 cercariae of the Puerto Rican strain of Schistosoma mansoni (NMRI) obtained from infected Biomphalaria glabrata snails (Biomedical Research Institute, Rockville, MD) as described.27 Serum for enzyme-linked immunosorbent assay (ELISA) and liver enzyme analysis was obtained by retro-orbital puncture of appropriately anaesthetized animals. The mice were killed at week 8 or week 12 after exposure. Serum IL-4 and IL-5 levels were determined by ELISA (R&D Systems, Minneapolis, MN). Experimental protocols were reviewed by the Animal Care and Use Committee, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), protocol number LPD-16E. Serum chemistries were determined by the clinical laboratories at the Clinical Center at the National Institutes of Health, Bethesda, MD.
Isolation of bone marrow cells for histologic analysis
Mouse bone marrow was collected from femurs and tibiae of S mansoni–infected and uninfected mice by flushing the opened bones with sterile phosphate-buffered saline (PBS). Cells were washed once in PBS + 1% bovine serum albumin (BSA). The bone marrow cells were counted in a hemocytometer, and 104 cells was subjected to cytospin (Thermo Shandon, Pittsburgh, PA). The cytospin preparations were fixed in methanol and stained using Diff Quik (Dade Behring, Dudingen, Switzerland).
Isolation of liver tissue for histologic analysis
Giemsa-stained liver tissue sections were prepared by Histopath of America (Millersville, MD) from liver tissue fixed in 10% phosphate-buffered formalin. Parameters including granuloma size, volume, and eggs per gram were evaluated as previously described.28,29 The eosinophil and mast-cell counts were scored by counting 5 or more granulomata per mouse, 200 to 300 cells per granuloma. Granuloma number was determined by counting 2.25-cm2 areas, 3 slides per mouse, 5 or more granulomata per mouse.
Identification of IL-5+CD4+ T cells from infected liver tissue
Approximately 0.1 to 0.2 g liver tissue was collected from each of 3 to 5 mice after perfusion. Leukocytes were obtained by smashing the tissue between 2 Plexiglas plates, followed by homogenization with syringe plunger through a 100-μm cell strainer (BD discovery Labware; Becton Dickinson, Bedford, MA). The homogenate was washed once and then suspended in 15 mL sterile PBS, mixed with 9 mL isotonic Percoll, and centrifuged at 500g for 15 minutes. The supernatant with hepatocytes was decanted and the leukocyte pellet was washed once with sterile PBS. Following red-cell lysis with ACK solution, the cells were suspended in complete RPMI medium. Viable cells were plated at 3 × 106 per 2 mL in a 24-well plate and stimulated with 10 ng/mL phorbol myristic acid (PMA) + 1 μg/mL ionomycin in the presence of 10 μg/mL brefeldin A for 3 hours at 37°C. The cells were washed, stained for surface CD4, fixed with 2% paraformaldehyde, and permeabilized with 0.1% saponin followed by intracellular staining for IL-5 (BD Pharmingen, San Diego, CA). Viable cells and total lymphocytes were determined by appropriate gating. The samples were evaluated with FACSCalibur (BD, San Jose, CA) and analyzed with Flowjo (Tree Star, Ashland, OR). Experiment no. 1 and experiment no. 2 in the text refer to 2 separate and distinct pools of mouse liver homogenates.
Isolation of bone marrow for RNA preparation
Mouse bone marrow cells isolated as described were suspended in RNazol B (Tel-Test, Friendswood, TX) at a concentration of 1 mL per 106 cells (15-25 × 106 cells total), and extraction proceeded as per the manufacturer's instructions. The precipitated RNA was harvested by centrifugation, washed in 70% ethanol, dried, and resuspended in diethyl-pyrocarbonate (DEPC)–treated sterile water. RNA concentration was measured spectrophotometrically at optical density (OD) 260, with typical yields of 60 μg total RNA at OD 260/OD 280 ratios of 2.0. Equal amounts of bone marrow RNA were pooled from 5 to 6 mice per condition prior to complementary RNA (cRNA) and cDNA synthesis.
Isolation of liver tissue for RNA preparation
Livers from mice were immersed in RNAlater (Ambion, Austin, TX) followed by blade homogenization in 7 mL RNAzol B reagent (Tel-Test). After chloroform was added (1:10 vol/vol), the specimen was mixed thoroughly and incubated on ice for 15 minutes. After a centrifugation at 13 600g for 20 minutes at 4°C, the aqueous layer was transferred to fresh tubes. Equal volumes of ice-cold isopropanol were added, and RNA was precipitated at –20°C. Total RNA was pelleted by centrifugation, washed twice in 80% ethanol, dried, and resuspended in diethyl pyrocarbonate–treated water. RNA was quantitated spectrophotometrically.
Gene microarray analysis
RNA samples from S mansoni–infected BALB/c and ΔdblGATA mice (n = 5-6 each) were pooled and subjected to gene microarray at the Microarray Core Facility in Rochester, NY as described previously.30 Data analysis was performed using the M-430 mouse genome chip and data were analyzed with GeneSpring 7.0 software (Silicon Genetics, Redwood City, CA) in ratio mode using the Cross Gene Error Model.
Quantitative RT-PCR
Pooled RNA (2 μg) prepared as described was subjected to DNAse I treatment (Invitrogen, Carlsbad, CA) and reverse transcribed using a First Strand cDNA Synthesis Kit for RT-PCR (AMV; Roche Diagnostics, Indianapolis, IN). cDNA (1 μL) was subjected to Taqman (Q) PCR using custom Fam-labeled probe and primers to mouse plasminogen activator inhibitor 2 (ABI catalog no. Mm00 440 905_m1), eosinophil peroxidase (ABI catalog no. Mm00514768_m1), major basic protein (ABI catalog no. Mm00435905_m1), interleukin 5 receptor alpha (ABI catalog no. Mm00434284_m1), flavin monooxygenase 2 (ABI catalog no. Mm00490159_m1), and rodent GAPDH (Vic-labeled probe, ABI catalog no. 4308313) (Applied Biosystems, Foster City, CA), using an Applied Biosystems 7700 PRISM instrument (50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute). Other transcripts were detected using the SYBR green detection method (SYBR green master mix, ABI catalog no. 4309155) and primers as follows: collagen I: forward, 5′-ACTGGACTGTCCCAACCCC-3′ and reverse, 5′-TCCCTCGACTCCTACATCTTCTG-3′; collagen III: forward, 5′-AACCTGGTTTCTTCTCACCCTTC-3′ and reverse, 5′-ACTCATAGGACTGACCAAGGTGG-3′; collagen VI: forward, 5′-CGCCCTTCCCACTGACAA-3′ and reverse, 5′-GCGTTCCCTTTAAGACAGTTGAG-3′; and beta-actin31 : forward 5′-AAGTCCCTCACCCTCCCAAAAG-3′ and reverse, 5′-AAGCAATGCTGTCACCTTCCC-3′.
Detection of eosinophils in bone marrow of Schistosoma mansoni–infected wild-type and eosinophil lineage–ablated ΔdblGATA and TgPHIL mice. (A) Cells from bone marrow from S mansoni–infected mice, including wild type (BALB/c, C57BL/6), eosinophil lineage–ablated ΔdblGATA (BALB/c background), and TgPHIL (C57BL/6 background). Arrows indicate examples of eosinophils. (B) Percent eosinophils (± SEM) in bone marrow at t = 8 weeks after exposure to cercariae. Mice are infected (+) or uninfected controls (–), including wild types, and eosinophil lineage–ablated ΔdblGATA and TgPHIL as in panel A; n = 4 to 5 mice per group.
Detection of eosinophils in bone marrow of Schistosoma mansoni–infected wild-type and eosinophil lineage–ablated ΔdblGATA and TgPHIL mice. (A) Cells from bone marrow from S mansoni–infected mice, including wild type (BALB/c, C57BL/6), eosinophil lineage–ablated ΔdblGATA (BALB/c background), and TgPHIL (C57BL/6 background). Arrows indicate examples of eosinophils. (B) Percent eosinophils (± SEM) in bone marrow at t = 8 weeks after exposure to cercariae. Mice are infected (+) or uninfected controls (–), including wild types, and eosinophil lineage–ablated ΔdblGATA and TgPHIL as in panel A; n = 4 to 5 mice per group.
All experiments include no reverse transcriptase and no template controls.
Photography and image analysis
All microscopic images were visualized on a Zeiss Axiophot II microscope (Carl Zeiss, Thornwood, NY) and photographed with a Coolsnap HQ camera (Photometrics, Tucson, AZ); digital processing was done using IP Lab 3.6 Scanalytics software (BD Biosciences Bioimaging, Rockville, MD). Composites were assembled in Microsoft Office Powerpoint 2003 (Microsoft, Seattle, WA).
Statistical analysis
Datasets were analyzed by Student t test or Mann-Whitney U test as appropriate. Statistical analysis of hepatic fibrosis in the different mouse strains was done by covariance analysis using the log of the total liver eggs as the covariate and the log of hydroxyproline per egg.
Results
Bone marrow eosinophilia in response to S mansoni infection
Cells were isolated from bone marrow of S mansoni–infected wild-type and eosinophil lineage–ablated mice at 8 weeks after exposure to cercariae. Eosinophilic myelocytes and promyelocytes with characteristic red-staining cytoplasmic granules are prominent among the cells isolated from the wild-type mice (Figure 1A). No cells with these staining properties were detected among those isolated from either of the eosinophillineage–ablated ΔdblGATA or TgPHIL mice. Percent eosinophils and eosinophilic precursors reached 32% ± 1.7% in the bone marrow of infected BALB/c mice, and 25% ± 2.1% eosinophils were detected among the bone marrow cells of the infected wild-type C57BL/6 mice (Figure 1B). No eosinophils or precursors were detected in any of the eosinophil lineage–ablated bone marrow cytospin preparations evaluated.
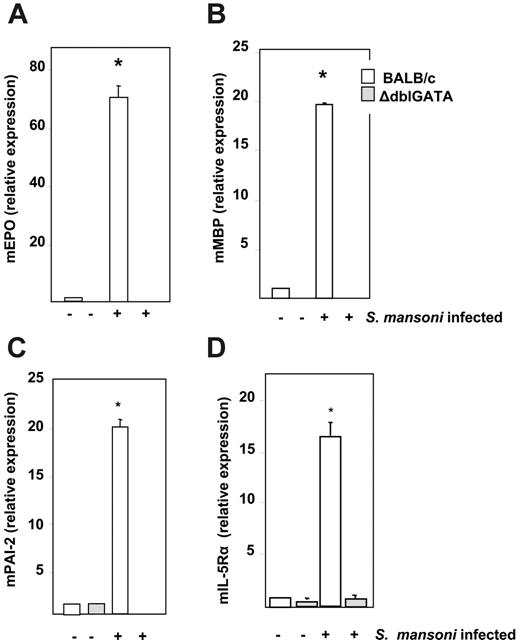
Relative expression levels of transcripts encoding mouse eosinophil peroxidase (mEPO), major basic protein (mMBP), interleukin-5 receptor alpha subunit (IL-5Rα), and plasminogen activator inhibitor-2 (mPAI-2) were determined by quantitative RT-PCR from bone marrow RNA from uninfected and infected (t = 8 weeks) wild-type BALB/c and ΔdblGATA mice (Figure 2). The expression level of each transcript in uninfected wild-type BALB/c mice was normalized to 1.0. Expression of transcripts encoding mEPO, mMBP, IL-5Rα, and mPAI-2 all increased in wild-type mice in response to S mansoni infection to +70-, +19-, +16-, and +20-fold, respectively, over the levels detected in uninfected wild-type mice. No transcripts encoding mEPO or mMBP were detected in either infected or uninfected ΔdblGATA mice. Expression levels of transcripts encoding mIL-5Rα and mPAI-2 remained at or below the baseline expression determined for uninfected wild-type mice in both infected and uninfected ΔdblGATAmice. Comparable results for these 4 transcripts were obtained by gene microarray analysis of bone marrow from uninfected and infected wild-type C57BL/6 and eosinophillineage–ablated TgPHIL mice (data not shown).
Relative expression of transcripts encoding eosinophil proteins. Relative expression of (A) mouse eosinophil peroxidase (mEPO), (B) mouse eosinophil major basic protein (mMBP), (C) mouse plasminogen activator inhibitor-2 (mPAI-2), and (D) mouse interleukin-5 receptor alpha (mIL-5Rα) from bone marrow RNA of uninfected and S mansoni–infected wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice at t = 8 weeks after exposure to cercariae. Values are mean fold change ± SEM with the expression in BALB/c uninfected mice normalized to 1.0; n = 4 mice per group; *P < .01 vs all other data points shown.
Relative expression of transcripts encoding eosinophil proteins. Relative expression of (A) mouse eosinophil peroxidase (mEPO), (B) mouse eosinophil major basic protein (mMBP), (C) mouse plasminogen activator inhibitor-2 (mPAI-2), and (D) mouse interleukin-5 receptor alpha (mIL-5Rα) from bone marrow RNA of uninfected and S mansoni–infected wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice at t = 8 weeks after exposure to cercariae. Values are mean fold change ± SEM with the expression in BALB/c uninfected mice normalized to 1.0; n = 4 mice per group; *P < .01 vs all other data points shown.
Serum Th2 cytokine determinations. Serum levels of (A) interleukin-5 in wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice, (B) interleukin-5 in wild-type C57BL/6 and eosinophil lineage–ablated TgPHIL mice, (C) interleukin-4 in wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice, and (D) interleukin-4 in wild-type C57BL/6 and eosinophil lineage–ablated TgPHIL mice at time points indicated. Values are average ± SEM; n = 5 mice per group (uninfected); n = 12 to 13 mice per group (infected); *P < .01 versus uninfected mice.
Serum Th2 cytokine determinations. Serum levels of (A) interleukin-5 in wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice, (B) interleukin-5 in wild-type C57BL/6 and eosinophil lineage–ablated TgPHIL mice, (C) interleukin-4 in wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice, and (D) interleukin-4 in wild-type C57BL/6 and eosinophil lineage–ablated TgPHIL mice at time points indicated. Values are average ± SEM; n = 5 mice per group (uninfected); n = 12 to 13 mice per group (infected); *P < .01 versus uninfected mice.
Detection of Th2 cytokines in serum response to S mansoni infection
Interleukin-5 was detected at 151 ± 27 pg/mL in the infected BALB/c mice and at 561 ± 47 pg/mL in the infected ΔdblGATA mice at week 7 of infection (3.7-fold difference, *P < .001; Figure 3A). Similarly, interleukin-5 was detected at 128 ± 2 pg/mL in infected C57BL/6 mice and 551 ± 20 pg/mL in infected TgPHIL mice at week 8 of infection (4.3-fold difference, *P < .001; Figure 3B). These elevated serum IL-5 levels in the eosinophil lineage–ablated mice persisted through week 11; no IL-5 was detected in sera from any uninfected mice. Interleukin-4 was detected in sera from infected mice only; no differential expression in wild-type versus eosinophil lineage–ablated mice was observed (Figures 3C and 3D). The fraction of IL-5–producing CD4+ T lymphocytes present in liver tissue of infected wild-type and eosinophil lineage–ablated mice varied (Table 1), but not in a consistent pattern that would account for the elevated serum IL-5 levels; the intensity of IL-5 staining per cell (MFI) was equivalent in all cases (data not shown). The absence of eosinophils may result in the elimination of an IL-5 receptor–dependent feedback inhibition loop, or a relative deficiency in soluble IL-5 receptor,32 or a rebound effect similar to that observed in the clinical setting immediately after the discontinuation of anti–IL-5 therapy.33
Identification of IL-5–producing CD4+ T cells in liver tissue of S mansoni–infected wild-type and eosinophil lineage–ablated mice
. | %lymphocytes . | %CD4+ . | %IL-5+ CD4+ . | IL-5+ CD4+/total CD4+ cells . |
|---|---|---|---|---|
| BALB/c | 0.13 | |||
| Experiment no. 1 | 17.9 | 5.5 | 0.73 | |
| Experiment no. 2 | 15.7 | 4.8 | 0.63 | |
| Average | 16.8 | 5.2 | 0.68 | |
| ΔdbIGATA | 0.11 | |||
| Experiment no. 1 | 38.3 | 13.9 | 1.6 | |
| Experiment no. 2 | 38.6 | 12.6 | 1.4 | |
| Average | 38.5 | 13.3 | 1.5 | |
| C57BL/6 | 0.32 | |||
| Experiment no. 1 | 38.7 | 11.8 | 3.9 | |
| Experiment no. 2 | 36.3 | 8.6 | 2.6 | |
| Average | 37.5 | 10.2 | 3.3 | |
| TgPHIL | 0.32 | |||
| Experiment no. 1 | 23.9 | 7.5 | 2.5 | |
| Experiment no. 2 | 26.8 | 6.9 | 2.1 | |
| Average | 25.4 | 7.2 | 2.3 |
. | %lymphocytes . | %CD4+ . | %IL-5+ CD4+ . | IL-5+ CD4+/total CD4+ cells . |
|---|---|---|---|---|
| BALB/c | 0.13 | |||
| Experiment no. 1 | 17.9 | 5.5 | 0.73 | |
| Experiment no. 2 | 15.7 | 4.8 | 0.63 | |
| Average | 16.8 | 5.2 | 0.68 | |
| ΔdbIGATA | 0.11 | |||
| Experiment no. 1 | 38.3 | 13.9 | 1.6 | |
| Experiment no. 2 | 38.6 | 12.6 | 1.4 | |
| Average | 38.5 | 13.3 | 1.5 | |
| C57BL/6 | 0.32 | |||
| Experiment no. 1 | 38.7 | 11.8 | 3.9 | |
| Experiment no. 2 | 36.3 | 8.6 | 2.6 | |
| Average | 37.5 | 10.2 | 3.3 | |
| TgPHIL | 0.32 | |||
| Experiment no. 1 | 23.9 | 7.5 | 2.5 | |
| Experiment no. 2 | 26.8 | 6.9 | 2.1 | |
| Average | 25.4 | 7.2 | 2.3 |
CD4+ T lymphocytes producing IL-5 were identified from single-cell suspensions from liver tissue by flow cytometric analysis as described in “Materials and methods.” Values shown represent percentages of total viable leukocytes isolated analyzed in 2 separate experiments of 3 to 5 mice each.
Liver pathology in wild-type versus eosinophil lineage–ablated mice
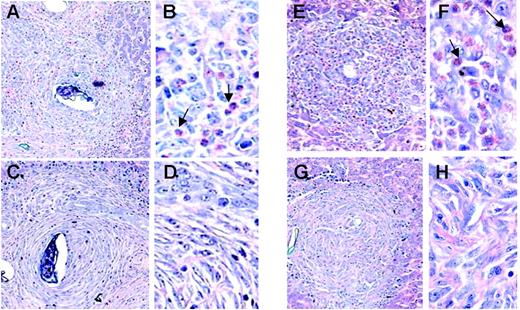
The granulomatous inflammatory response that develops around the egg is a well-characterized lesion associated with schistosome infection in the mouse model17,18 (Figure 4). The granulomata of the ΔdblGATA mice are devoid of eosinophils, while approximately 30% of the cells in the wild-type BALB/c granulomata were identified as eosinophils at week 12 of infection. Similarly, eosinophils were not detected in the granulomata of the TgPHIL mice, in contrast to those of the wild-type C57BL/6 (50% eosinophils). Gene microarray data of liver RNA from ΔdblGATAmice are consistent with these findings; we observe preferential expression of the eosinophil-specific and related transcripts, major basic protein (mMBP, +11.0-fold) and eosinophil peroxidase (mEPO, +13.5-fold), eosinophil associated-ribonuclease-1 (mEAR-1, +16.7-fold), arachidonate-15-lipoxygenase (+19.6-fold), and sialic-acid binding Ig-like lectin (Siglec-F, +4.3-fold).34 A slight but statistically significant difference in mast-cell numbers at week 12 was observed among the ΔdblGATA mice only (Table 2). However, it is difficult to interpret this result in light of the role played by the GATA-1 transcription factor as a regulator of mast-cell differentiation.35 Very few neutrophils were detected overall. Microscopic evaluation revealed an equivalent number of granulomata per unit area, equivalent volume, and degree of fibrosis at weeks 8 and 12 after exposure as determined by hydroxyproline assay. Hepatic fibrosis is related to number of eggs in a nonlinear fashion, as hepatic fibrosis per egg decreases with increasing intensity of infection. As such, analysis of covariance was performed (“Materials and methods”), which showed no significant differences between the infected eosinophil lineage–ablated mice and their respective controls at the 12-week time point evaluated.
Properties of liver granulomata of S mansoni–infected mice
Wk of infection/mouse strain . | Granulomata* . | Volume, mm3 × 10-3 . | Hydroxyproline, μM/g . | Hydroxyproline, μM/104 eggs . | Eosinophils, % total cells . | Mast cells, no. per granuloma . |
|---|---|---|---|---|---|---|
| 8 | ||||||
| BALB/c | nd | 28.6 ± 2.5 | nd | nd | 10.1 ± 2.3 | 7.2 ± 1.2 |
| ΔdblGATA | nd | 26.2 ± 1.5 | nd | nd | 0† | 8.5 ± 2.0 |
| 12 | ||||||
| BALB/c | 150 ± 12 | 27.4 ± 2.2 | 16.6 ± 0.9 | 2.5 ± 1.4 | 33.2 ± 4 | 3.1 ± 0.5 |
| ΔdblGATA | 130 ± 8 | 27.6 ± 3.2 | 16.3 ± 1.1 | 4.7 ± 0.7 | 0† | 5.5 ± 0.4† |
| C57BL/6 | 228 ± 16 | 20.1 ± 1.4 | 10.6 ± 0.6 | 2.4 ± 1.2 | 51.1 ± 2.9 | 1.1 ± 0.3 |
| TgPHIL | 219 ± 20 | 16.7 ± 1.5 | 11.4 ± 0.5 | 3.5 ± 1.0 | 0†‡ | 2 ± 0.4 |
Wk of infection/mouse strain . | Granulomata* . | Volume, mm3 × 10-3 . | Hydroxyproline, μM/g . | Hydroxyproline, μM/104 eggs . | Eosinophils, % total cells . | Mast cells, no. per granuloma . |
|---|---|---|---|---|---|---|
| 8 | ||||||
| BALB/c | nd | 28.6 ± 2.5 | nd | nd | 10.1 ± 2.3 | 7.2 ± 1.2 |
| ΔdblGATA | nd | 26.2 ± 1.5 | nd | nd | 0† | 8.5 ± 2.0 |
| 12 | ||||||
| BALB/c | 150 ± 12 | 27.4 ± 2.2 | 16.6 ± 0.9 | 2.5 ± 1.4 | 33.2 ± 4 | 3.1 ± 0.5 |
| ΔdblGATA | 130 ± 8 | 27.6 ± 3.2 | 16.3 ± 1.1 | 4.7 ± 0.7 | 0† | 5.5 ± 0.4† |
| C57BL/6 | 228 ± 16 | 20.1 ± 1.4 | 10.6 ± 0.6 | 2.4 ± 1.2 | 51.1 ± 2.9 | 1.1 ± 0.3 |
| TgPHIL | 219 ± 20 | 16.7 ± 1.5 | 11.4 ± 0.5 | 3.5 ± 1.0 | 0†‡ | 2 ± 0.4 |
Analysis based on measurements from more than 100 granulomata (n = 8-13 mice per group) from Giemsa-stained liver tissue; methodology was as described.28,29 Data are presented as plus or minus standard error of the mean.
nd indicates not determined.
Number of single-egg granulomata was scored per 2.25 cm2 of infected liver tissue.
Significant differences between BALB/c and ΔdblGATA at given time point.
Five eosinophils were identified in one granuloma of one TgPHIL mouse.
Microscopic pathology of hepatic granulomata of S mansoni–infected wild-type and eosinophil lineage–ablated ΔdblGATA and TgPHIL mice. Giemsa-stained liver tissue sections featuring granulomata from S mansoni–infected BALB/c (A-B), ΔdblGATA (C-D), C57BL/6 (E-F), and TgPHIL (G-H) mice all at 12 weeks of infection. Arrows indicate examples of eosinophils. Original magnifications × 10 (A-B,E-F) and × 40 (C-D,G-H).
Microscopic pathology of hepatic granulomata of S mansoni–infected wild-type and eosinophil lineage–ablated ΔdblGATA and TgPHIL mice. Giemsa-stained liver tissue sections featuring granulomata from S mansoni–infected BALB/c (A-B), ΔdblGATA (C-D), C57BL/6 (E-F), and TgPHIL (G-H) mice all at 12 weeks of infection. Arrows indicate examples of eosinophils. Original magnifications × 10 (A-B,E-F) and × 40 (C-D,G-H).
Differential expression of fibrosis-related genes
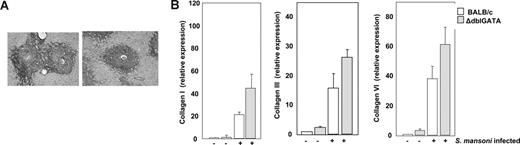
Picrosirius red–stained fibrous tissues within the liver granulomata of infected wild-type and ΔdblGATA mice (week 8 of infection) are shown in Figure 5A. Transcripts encoding collagens I, III, and VI were present on the gene microarray comparison of liver mRNAs from infected wild-type versus ΔdblGATA mice (t = 8 weeks of infection), but no significantly different levels of expression were determined (fold changes of +1.2, +1.3, and +1.4, respectively, wild type vs ΔdblGATA). A more comprehensive and sensitive analysis of gene transcription was performed by quantitative RT-PCR on liver mRNAs from both uninfected and S mansoni–infected BALB/c and ΔdblGATA mice (Figure 5B). Expression of collagens I, III, and VI increased with S mansoni infection, although no eosinophil-dependent differential expression was appreciated at this time point.
Analysis of liver enzymes in serum
We observed approximately 5-fold and approximately 3-fold elevations in serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST), respectively, in response to S mansoni infection, but no statistically significant differences between the infected wild-type and infected eosinophil lineage–ablated mice were detected (Table 3). The transaminases remained elevated through week 11, although at diminished levels throughout, and without any consistent, eosinophil-dependent pattern. Minor elevations over baseline levels (1.4-fold) were observed for lactate dehydrogenase in response to infection among the BALB/c and ΔdblGATA mice only.
Serum chemistries of wild-type and eosinophil lineage–ablated mice infected withS mansoni
Mouse strain/wk of infection . | ALT, U/L . | Fold ↑ . | AST, U/L . | Fold ↑ . | Alk phos, U/L . | CK, U/L . | LD, U/L . |
|---|---|---|---|---|---|---|---|
| BALB/c | |||||||
| 0 | 46.8 ± 6.5 | 1.0 | 78.3 ± 5.3 | 1.0 | 94.8 ± 3.7 | 427 ± 114 | 581 ± 59 |
| 8 | 217 ± 29* | 4.6† | 202 ± 20* | 2.6† | 98.5 ± 4.8 | 602 ± 92 | 817 ± 62‡ |
| 11 | 124 ± 13‡ | 2.6† | 132 ± 9‡ | 1.7† | 115 ± 6 | 237 ± 33 | 579 ± 48 |
| ΔdblGATA | |||||||
| 0 | 33.4 ± 1.8 | 1.0 | 80.6 ± 10.5 | 1.0 | 88 ± 5.1 | 497 ± 137 | 640 ± 59 |
| 8 | 152 ± 44* | 4.6† | 163 ± 22‡ | 2.0† | 54.5 ± 4.0 | 505 ± 87 | 896 ± 95‡ |
| 11 | 101 ± 11‡ | 3.0† | 124 ± 12 | 1.5† | 81 ± 6 | 218 ± 31 | 591 ± 62 |
| C57BL/6 | |||||||
| 0 | 39 ± 3.5 | 1.0 | 63.5 ± 7.5 | 1.0 | 82 ± 5.3 | 184 ± 59 | 290 ± 32 |
| 8 | 194 ± 13* | 5.0† | 177 ± 12* | 2.8† | 56 ± 2 | 240 ± 22 | 457 ± 30 |
| 11 | 148 ± 16‡ | 3.8† | 154 ± 17‡ | 2.4 | 78 ± 3 | 137 ± 23 | 410 ± 33 |
| TgPHIL | |||||||
| 0 | 58 ± 9 | 1.0 | 74 ± 9 | 1.0 | 115 ± 11 | 159 ± 37 | 252 ± 10 |
| 8 | 113 ± 9‡ | 1.9† | 132 ± 8‡ | 1.8† | 58 ± 4 | 219 ± 26 | 371 ± 20 |
| 11 | 93 ± 12 | 1.6† | 111 ± 13 | 1.5† | 73 ± 6 | 120 ± 24 | 253 ± 19 |
Mouse strain/wk of infection . | ALT, U/L . | Fold ↑ . | AST, U/L . | Fold ↑ . | Alk phos, U/L . | CK, U/L . | LD, U/L . |
|---|---|---|---|---|---|---|---|
| BALB/c | |||||||
| 0 | 46.8 ± 6.5 | 1.0 | 78.3 ± 5.3 | 1.0 | 94.8 ± 3.7 | 427 ± 114 | 581 ± 59 |
| 8 | 217 ± 29* | 4.6† | 202 ± 20* | 2.6† | 98.5 ± 4.8 | 602 ± 92 | 817 ± 62‡ |
| 11 | 124 ± 13‡ | 2.6† | 132 ± 9‡ | 1.7† | 115 ± 6 | 237 ± 33 | 579 ± 48 |
| ΔdblGATA | |||||||
| 0 | 33.4 ± 1.8 | 1.0 | 80.6 ± 10.5 | 1.0 | 88 ± 5.1 | 497 ± 137 | 640 ± 59 |
| 8 | 152 ± 44* | 4.6† | 163 ± 22‡ | 2.0† | 54.5 ± 4.0 | 505 ± 87 | 896 ± 95‡ |
| 11 | 101 ± 11‡ | 3.0† | 124 ± 12 | 1.5† | 81 ± 6 | 218 ± 31 | 591 ± 62 |
| C57BL/6 | |||||||
| 0 | 39 ± 3.5 | 1.0 | 63.5 ± 7.5 | 1.0 | 82 ± 5.3 | 184 ± 59 | 290 ± 32 |
| 8 | 194 ± 13* | 5.0† | 177 ± 12* | 2.8† | 56 ± 2 | 240 ± 22 | 457 ± 30 |
| 11 | 148 ± 16‡ | 3.8† | 154 ± 17‡ | 2.4 | 78 ± 3 | 137 ± 23 | 410 ± 33 |
| TgPHIL | |||||||
| 0 | 58 ± 9 | 1.0 | 74 ± 9 | 1.0 | 115 ± 11 | 159 ± 37 | 252 ± 10 |
| 8 | 113 ± 9‡ | 1.9† | 132 ± 8‡ | 1.8† | 58 ± 4 | 219 ± 26 | 371 ± 20 |
| 11 | 93 ± 12 | 1.6† | 111 ± 13 | 1.5† | 73 ± 6 | 120 ± 24 | 253 ± 19 |
Serum samples were taken from mice prior to infection (week 0, n = 8-13 mice) and at weeks 8 and 11 (n = 12-13 mice) after exposure to cercariae. Although elevations in serum ALT and AST were observed in all mice response to infection, no consistent differences were observed between wild type and eosinophil lineage—ablated mice.
Data are presented as plus or minus standard error of the mean.
ALT indicates alanine aminotransferase; AST, aspartate aminotransferase; Alk phos, alkaline phosphatase; CK, creatine kinase; and LD, lactate dehydrogenase.
P < .01 versus uninfected mice of the same genotype.
Ratios of infected to uninfected values at points indicated.
P < .05 versus uninfected mice of the same genotype.
Quantitation of parasites and eggs in wild-type and eosinophil-deficient mice
Although there were clearly more total worms present overall in the C57BL/6 and TgPHIL mice, no eosinophil-dependent effects could be discerned (Table 4). Likewise, the presence or absence of eosinophils had no impact on the number of eggs per gram of liver tissue, or on the percentage of eggs deposited within liver tissue.
Parasites and eggs detected inS mansoni–infected wild-type and eosinophil lineage–ablated mice
. | Worms . | . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Mouse strain . | Males* . | Females* . | Pairs . | Total . | Eggs/g liver tissue, × 10-3 . | Eggs in liver tissue, % . | |||
| BALB/c | 2.1 ± 0.9 | 0 | 4.8 ± 0.4 | 11.7 ± 0.8 | 22.0 ± 1.4 | 83 ± 3 | |||
| ΔdblGATA | 1.4 ± 0.5 | 0 | 4.3 ± 0.5 | 9.9 ± 1.1 | 23.5 ± 2.6 | 81 ± 3 | |||
| C57BL/6 | 5.6 ± 1.0† | 0 | 5.6 ± 0.53 | 17.2 ± 1.0† | 16.7 ± 2.0 | 90 ± 2 | |||
| TgPHIL | 5.3 ± 1.5‡ | 0.4 ± 0.2 | 7.1 ± 1.3 | 19.6 ± 2.5‡ | 21.2 ± 3.0 | 92 ± 0.7 | |||
. | Worms . | . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Mouse strain . | Males* . | Females* . | Pairs . | Total . | Eggs/g liver tissue, × 10-3 . | Eggs in liver tissue, % . | |||
| BALB/c | 2.1 ± 0.9 | 0 | 4.8 ± 0.4 | 11.7 ± 0.8 | 22.0 ± 1.4 | 83 ± 3 | |||
| ΔdblGATA | 1.4 ± 0.5 | 0 | 4.3 ± 0.5 | 9.9 ± 1.1 | 23.5 ± 2.6 | 81 ± 3 | |||
| C57BL/6 | 5.6 ± 1.0† | 0 | 5.6 ± 0.53 | 17.2 ± 1.0† | 16.7 ± 2.0 | 90 ± 2 | |||
| TgPHIL | 5.3 ± 1.5‡ | 0.4 ± 0.2 | 7.1 ± 1.3 | 19.6 ± 2.5‡ | 21.2 ± 3.0 | 92 ± 0.7 | |||
Analysis based on measurements taken at week 12 after exposure to cercariae of S mansoni as described in “Materials and methods.” Data are presented as plus or minus standard error of the mean.
Males and females refer to worms detected as singles.
P < .001 versus BALB/c.
P < .005 versus ΔdblGATA.
Discussion
Eosinophils have long been associated with helminth infection, although the nature and specifics of their role in this disease remain unclear. Earlier studies used anti–IL-5 methodology in order to reduce blood and tissue eosinophilia characteristic of the Th2 response to helminth infection.14-18 Among the earliest of these studies, Sher et al17 administered the anti–IL-5 monoclonal antibody TRFK-536 to C3H/HEN mice and demonstrated that this cytokine (and by extension, eosinophils) had no impact on worm burden, hepatic fibrosis, or granuloma formation characteristic of S mansoni infection. Brunet et al18 studied S mansoni infection in interleukin-5 gene–deleted C57BL/6 mice, and likewise concluded that hepatic pathology and susceptibility to infection were indistinguishable between wild-type and gene-deleted strains. One major difference between these 2 IL-5 depletion studies and the work presented here using ΔdblGATA and TgPHIL eosinophil lineage–ablated mice is that in the former studies, the eosinophil counts both in the periphery and in the granulomata could not be reduced completely. Sher et al17 reduced the percentage of eosinophils in the granulomata from approximately 40% to 1% to 2%, and Brunet et al18 found that the granulomata of the IL-5 gene–deleted mice contained 7.5% eosinophils compared with 55% in the wild type; many argued that it was possible that even a few eosinophils could provide substantial protection. Indeed, residual tissue eosinophilia (that remaining after attenuation of the effects of IL-5) has been shown to contribute to remodeling of the airways in asthmatic patients and to contribute to functional changes in airway responsiveness in mouse models of disease.37-41 Furthermore, neither of the aforementioned studies could eliminate the confounding factor of removing the cytokine IL-5 from the overall pathophysiologic picture.
Hepatic fibrosis in response to S mansoni infection. (A) Picrosirius red staining of fibrous tissue in granulomata of wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice at week 8 of infection. (B) Differential expression of transcripts encoding collagen I, collagen III, and collagen VI in liver RNA from uninfected and S mansoni–infected BALB/c and ΔdblGATA mice. Values are mean fold change ± SEM with expression in uninfected BALB/c mice normalized to 1.0.
Hepatic fibrosis in response to S mansoni infection. (A) Picrosirius red staining of fibrous tissue in granulomata of wild-type BALB/c and eosinophil lineage–ablated ΔdblGATA mice at week 8 of infection. (B) Differential expression of transcripts encoding collagen I, collagen III, and collagen VI in liver RNA from uninfected and S mansoni–infected BALB/c and ΔdblGATA mice. Values are mean fold change ± SEM with expression in uninfected BALB/c mice normalized to 1.0.
In this work, we explore the role of eosinophils using the traditional parameters for determining their role in antihelminth host defense, which involves enumeration of worms, eggs, and granulomatous responses, deriving largely from the early appreciation of eosinophils in their “kamikaze” roles in vitro,42 as they degranulate and ostensibly reduce the number of organisms and their byproducts via the actions of secretory toxins.43-45 Using these functional parameters in the eosinophil lineage–ablated ΔdblGATA and TgPHIL mice, we find that eosinophils have no impact on worm burden, egg deposition, or granuloma formation other than the eosinophil depletion itself.
Among the issues and caveats to be considered in the interpretation of these results, it is important to note that there are clear and discernible differences between human and mouse eosinophils. Among the major distinctions between human and mouse eosinophils, one must consider the evolutionary divergence of the secretory ribonucleases,46 the presence (in humans) or absence (in mice) of Charcot-Leyden crystal protein (galectin-10),47 and, perhaps most important, the differences in propensity to degranulate.42,48 As such, human and mouse eosinophils may not have interchangeable roles in health and disease. Similarly, while it can use the mouse effectively to complete the mammalian phase of its life cycle, S mansoni is not strictly a natural rodent pathogen. However, it is clear that human and mouse eosinophils do play important roles in immune responses independent of their ability to degranulate, as they also regulate T-cell responses directly and secrete of a range of proinflammatory mediators and cytokines49-52 ; the recent report by Voehringer et al53 indicates that eosinophils play a role in the prevention of secondary infection in the Nippostrongylus brasiliensis infection model. An evaluation of these eosinophil-mediated activities in the setting of acute and/or chronic schistosome infection is certainly worthy of further consideration.
However, if eosinophils are in fact playing some as-yet-to-be-identified role in the pathophysiology of helminth infection, what exactly might that be? Eosinophils are recruited specifically to the granulomata in response to Th2 stimuli, and they form a significant component of this structure; as shown in this paper, 30% to 50% of the cells at week 11 are eosinophils. Granulomata are generally understood as protective; mice with structurally insufficient granulomata can develop acute hepatotoxicity in response to infection.54 Yet eosinophil depletion alone has apparently minimal impact in the acute setting. We observe weight loss at 6 to 9 weeks among infected ΔdblGATA mice (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article), but no evidence for increased hepatocellular damage in either infected ΔdblGATA or TgPHIL eosinophil lineage–ablated mice (Table 3).
If they are not contributing to or reducing histopathology directly, why would eosinophil recruitment be an essential feature of liver granulomata? Among the novel hypotheses to consider is that presented by Lee and Lee42 who have suggested that the primordial and perhaps essential function of the eosinophil is as a metabolic scavenger. Large numbers of eosinophils might be recruited to the liver granulomata, sites of rapid remodeling of metabolically, enzymatically rich tissue, in order to assist in clearance and detoxification of cellular debris. There already exists a considerable literature on the role of eosinophils in remodeling in asthmatic lung tissue37-41,55 (note: that occurs without active degranulation in mouse models). The role of IL-5 and potentially eosinophils in hepatic fibrosis in schistosome disease has already been noted at later time points than those addressed in this study.56 Alternatively, the role of eosinophils as antigen-presenting cells has been considered in the literature,57-59 and has only recently been explored in the context of helminth infection.60
In summary, using 2 distinct models of eosinophil lineage–ablation, we find that eosinophils have no direct impact on traditional measures of helminth disease in the well-characterized mouse model of S mansoni infection. However, the recruitment of large numbers of eosinophils to the granulomata in both humans and mice suggests that this is very unlikely to be a redundant process. A detailed temporal and metabolic analysis may be required in the wild-type and eosinophil-ablated models to disclose discrete and subtle immunomodulatory contributions of this granulocyte to the disease process.
Prepublished online as Blood First Edition Paper, June 13, 2006; DOI 10.1182/blood-2006-04-015933.
Supported by Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), funds to T.A.W. and H.F.R.
J.M.S. and K.D.D. contributed equally to this work.
T.A.W. and H.F.R. contributed equally to this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful to Dr Alison Humbles and Dr Craig Gerard, Children's Hospital, Harvard Medical School, for sharing the ΔdblGATA mice with us; Ms Sandy White, LPD, NIAID, for assistance with the Schistosoma mansoni infections; and Ms Shauna Everett and Mr Rick Dreyfuss of Medical Arts, NIH, for assistance with image preparation. We also thank Dr Jonas Byström and Dr Takeaki Nitto, LAD, NIAID, for careful reading and helpful comments on this paper.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal