Abstract
Most lymphomas arise by transformation of germinal center (GC) B cells. TCL1, a proto-oncogene first recognized for its role in T-cell transformation, also induces GC B-cell malignancies when dysregulated in pEμ-B29-TCL1 transgenic (TCL1-tg) mice. Clonal B-cell lymphomas develop from polyclonal populations with latencies of 4 months or more, suggesting that secondary genetic events are required for full transformation. The goals of this study were to determine the GC-related effects of TCL1 dysregulation that contribute to tumor initiation and to identify companion genetic alterations in tumors that function in disease progression. We report that compared with wild-type (WT) cells, B cells from TCL1-tg mice activated in a manner resembling a T-dependent GC reaction show enhanced resistance to FAS-mediated apoptosis with CD40 stimulation, independent of a B-cell antigen receptor (BCR) rescue signal. Mitogenic stimulation of TCL1-tg B cells also resulted in increased expression of Aicda. These GC-related enhancements in survival and Aicda expression could underlie B-cell transformation. Supporting this notion, no B-cell lymphomas developed for 20 months when TCL1-tg mice were crossed onto an Oct coactivator from B cell (OCA-B)–deficient background to yield mice incapable of forming GCs. Spectral karyotype analyses showed that GC lymphomas from TCL1-tg mice exhibit recurrent chromosome translocations and trisomy 15, with corresponding MYC overexpression. We conclude that pEμ-B29-TCL1 transgenic B cells primed for transformation must experience the GC environment and, for at least some, develop genome instability to become fully malignant.
Introduction
Most common B-cell lineage lymphomas in humans originate by transformation of germinal center (GC) B cells. These include classes of non-Hodgkin lymphoma (NHL) designated follicular lymphoma (FL), diffuse large B-cell lymphoma (DLBCL), and Burkitt lymphoma (BL) as well as classical Hodgkin lymphoma.1 FL and BL are characterized by recurring reciprocal chromosome (Chr) translocations that fuse Ig genes either with the BCL2 cell survival gene or with the MYC proto-oncogene, respectively.2,3 Also, reciprocal Chr translocations involving the BCL6 transcriptional repressor gene and more than 20 partner loci, including the Ig genes, typify DLBCL.4 These and additional tumor-promoting genes are subsequently expressed under the control of Ig regulatory sequences rather than their native regulatory elements, resulting in dysregulated expression. Interestingly, MYC, BCL6, and other genes, such as TCL1, are also expressed at high levels in many NHLs in the absence of activating translocations. In these cases, inappropriately high expression is a consequence of other genetic or possibly epigenetic alterations and is likely contributory to the transformation process.5-7
The TCL1 proto-oncogene encodes a 14-kDa intracellular protein that associates in a multimeric complex with the serine/threonine kinase AKT.7-11 Interactions with TCL1 enhance AKT activation and, by an unknown mechanism, TCL1 stimulates the protein kinase C–mitogen-activated protein kinase–extracellular signal–related kinase (PKC-MAPK-ERK) signal transduction pathway, promoting cell survival and proliferation.8,12,13 In human T cells, TCL1 is normally expressed only by immature cortical thymocytes and by activated peripheral T cells.13,14 However, dysregulated expression from Chr rearrangements between TCL1 and T-cell receptor (TCR) loci results in persistent high-level expression in mature T-cell leukemia/lymphomas.15 A direct, initiating role for heightened TCL1 expression in T-cell transformation is suggested by studies of transgenic mice with T-cell lineage–restricted TCL1 expression that develop mature T-cell leukemias following an extended premalignant phase of polyclonal expansion.16
Despite its original identification in T-cell leukemias, TCL1 seems to have an even more prominent role in B cells and is differentially expressed during normal B-cell maturation. Expression is robust from pre-B through follicular B-cell development, but is down-regulated during the GC reaction and is silenced in post-GC memory B and plasma cells.17 High levels of TCL1 expression have been documented for human B-cell lymphomas originating from pre-GC and GC B cells and in AIDS-related lymphomas of post-GC cell origin.17-19 The suggestion that inappropriately high expression in the GC could contribute to GC B-cell lymphomagenesis is strongly supported by studies of mice bearing a pEμ-B29-TCL1 transgene expressed in both T and B cells, which develop mature BCL6-positive B-cell lymphomas with mutated Ig genes, indicating a likely origin from GC B cells.12
B-cell lymphomas in pEμ-B29-TCL1 transgenic (TCL1-tg) mice develop after long latencies. This indicates that high expression of TCL1 is insufficient to drive transformation and that other genetic or epigenetic changes, presumably occurring in the GC, are required. Aberrant DNA break repair associated with the normal GC processes of Ig somatic hypermutation (SHM) and class switch recombination (CSR) serve as possible sources for complementing mutations to facilitate complete transformation. While cooperation of MYC with various companion genes, such as BCL2, promotes B-cell malignancy, it is unclear which genes partner with TCL1 to facilitate B lymphomagenesis. In mature human T-cell lymphomas that have frequent TCL1 rearrangements, abnormalities of Chr 8, which contains MYC at 8q24, are observed in 70% to 80% of cases, but the nature of the specific complementing genes required for transformation has not been defined.20,21 For some human B-cell lymphomas with elevated TCL1 levels, such as BL, heightened expression of MYC caused by specific activating translocations is frequently observed in altered gene expression profiles.22
In the present study, we have used this TCL1-tg lymphoma model to examine how TCL1 alters GC-related functions that may contribute to B-cell transformation and to determine whether genome instability and/or altered MYC expression may complement TCL1 dysregulation in mature B-cell lymphomas.
Materials and methods
Mice
Strain-matched wild-type (WT), TCL1-tg, Oct coactivator from B cell (OCA-B)–deficient (OCA-B–/–), and TxO (TCL1+/–, OCA-B–/–) mice were housed and mated in a specific pathogen–free facility as previously described.12,23 Cells used were from 6- to 9-week-old age- and sex-matched mice with no evidence of lymphoproliferative disorder or lymphoma, in compliance with approved animal use guidelines.
Lymphocyte isolation
B and T cells were isolated by magnetic sorting using an AUTOMACS cell sorter (Miltenyi Biotech, Auburn, CA) and negative selection with anti-CD4–PE, anti-CD8–PE, anti-B220–PE, and anti-Gr1–PE antibodies (Abs) (BD Pharmingen, San Diego, CA) and anti-PE microbeads (Miltenyi Biotech), as appropriate and according to the manufacturer's instructions.
Proliferation analyses
Isolated spleen cells were cultured at 1 × 105 cells per well in 96-well plates in RPMI medium 1640 with 10% fetal calf serum (FCS) with the indicated amounts of anti–immunoglobulin M (IgM), concanavalin A, or plate-bound anti-CD3ϵ plus 1 μg/mL anti-CD28 monoclonal Ab (mAb). At 48 hours, cells were pulsed with 0.037 MBq (1 μCi) of [3H]-thymidine for 12 hours, harvested onto glass fiber filters, and the counts per minute (cpm) were determined.
FASL-induced apoptosis
Splenic B cells (1 × 106, > 95% pure) were cultured with anti-CD40 Ab (1.5 μg/mL; BD Pharmingen) for 48 hours with or without addition of anti-IgM Ab (10 μg/mL; Jackson ImmunoResearch Laboratories, Bar Harbor, ME) during the last 24 hours. After treatment with FAS ligand (FASL, 1:25; Upstate Biotechnology, Lake Placid, NY) for 4 hours, cells were harvested and stained with Annexin V–FITC and propidium iodide (BD Pharmingen) followed by flow cytometry. FASL-induced death was determined as: % death = 100 × (% viable untreated cells – % viable cells with FASL) ÷ % viable untreated cells.
RT-PCR
Total RNA was isolated using Trizol reagent (GIBCO-BRL, Gaithersburg, MD). First-strand cDNA was synthesized from 1 μg of RNA with SuperScriptII reverse transcriptase (GIBCO-BRL). Reverse transcriptase–polymerase chain reaction (RT-PCR) primers for activation-induced cytidine deaminase (Aicda) and Gapdh are previously described.24 A PCR protocol for Aicda amplification of 30, 35, and 40 cycles consisting of 94°C for 2 minutes and 59°C for 30 seconds was followed by an elongation step at 72°C for 1 minute.
Western analysis
Protein was extracted from cell pellets using NP-40 lysis buffer containing protease inhibitors, and 20 μg per sample was fractionated by SDS-PAGE followed by immunoblotting, as previously described.17 Rabbit anti–mouse MYC antiserum (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), goat anti–mouse β-actin antiserum (1:5000; Sigma-Aldrich, St Louis, MO), or rabbit anti-TCL1 antiserum (1:15 000) were used to detect immunoreactive bands with electrochemiluminescence (ECL) reagent (Amersham Biosciences, Piscataway, NJ). Densitometry was performed using SynGene Analysis software (SynGene, Upland, CA).
Histology and tumor classification
Fixed tissues were thin sliced and stained with hematoxylin and eosin. Tumors were classified using morphologic, flow cytometric, immunohistochemical, and clonality data in accordance with Mouse Models of Human Cancer Consortium (MMHCC) criteria.25
Flow cytometry
Cell suspensions were depleted of red blood cells (RBCs), blocked with anti-FcγII/III receptor mAb, and surface-stained with PE- or FITC-conjugated Abs. Data were collected on a FacsCalibur (BD Pharmingen). Data were analyzed using FCS EXPRESS (De Novo Software, Thornhill, ON, Canada). Primary Abs used include anti-B220–PE, anti-IgMa–FITC, anti-IgMb–FITC, anti-CD4–PE, anti-CD5–PE, anti-CD8–FITC, and anti-Fas–PE (all from BD Pharmingen). Peanut agglutinin (PNA)–biotin (Vector Laboratories, Burlingame, CA) and antibiotin-streptavidin secondary Ab (Southern Biotech, Atlanta, GA) were also used.
Results
Enhanced proliferation by TCL1 in GC and non-GC B cells
The proliferative advantage conferred by TCL1 to peripheral lymphocytes responding to various B- and T-cell stimuli8,12,13 may increase the opportunity for mature lymphocyte transformation events. In contrast, this growth advantage is not observed for TCL1-expressing thymocytes and immature T-cell transformation is not linked with aberrant TCL1 expression.13 Since TCL1 mediates the transformation of GC B cells, it is possible that TCL1 has distinct proliferative effects on GC and non-GC B-cell populations. To investigate whether increased proliferation of TCL1-tg B cells occurs in both B-cell subsets in response to stimulation through the B-cell antigen receptor (BCR), purified splenic B cells were labeled with CFSE, stimulated with anti-IgM plus interleukin-4 (IL-4) for 72 hours and then stained with Ab to B220 and PNA. IgM stimulation was chosen since almost all GC tumors that form in TCL1-tg mice express surface IgM.12 IgM+ GC and non-GC B cells respond to this stimulation, with B220+PNA+ staining of activated B cells in vitro capturing both IgM+ GC B cells and those that acquire a GC B-cell immunophenotype.26 Higher percentages of both GC (B220+PNA+) and non-GC (B220+PNA–) B cells from TCL1-tg mice had undergone multiple rounds of division compared with WT controls (Figure S1, available on the Blood website; see the Supplemental Material link at the top of the online article). More than 35% of TCL1-tg non-GC B cells showed more than 2 cell divisions compared with 25% for WT non-GC B cells (ratio, approximately 1.4:1). Similarly, more than 40% of TCL1-tg GC B cells showed more than 2 cell divisions compared with less than 25% for WT GC B cells (ratio, approximately 1.6:1). These findings suggested that TCL1 does not provide a GC B-cell–specific proliferative advantage and that other functions related to the GC also contribute to the transformation of GC B cells.
B cells with heightened TCL1 expression are more resistant to CD40-mediated, FASL-induced apoptosis
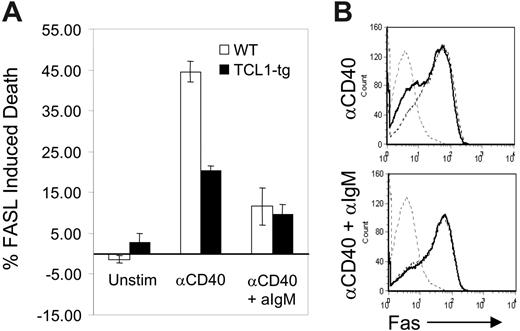
Transformed B cells from TCL1-tg mice exhibit features of GC rather than pre-GC or post-GC B cells. This occurrence cannot be explained by the enhanced proliferation of nontransformed TCL1-tg B cells, regardless of their tendency to adopt a GC or non-GC fate. This prompted us to assess the effects of heightened TCL1 expression on a prominent feature of the GC reaction, cell death induced by the FAS/FASL signaling pathway. Naive, mature primary mouse B cells express little or no FAS.27 CD40-expressing B cells recruited into the GC engage CD40L on CD4+ T cells, which induces B-cell activation and the surface expression of FAS.28 CD40L-engaged GC B cells then become susceptible to FASL-induced cell death. Similarly, stimulation of naive B cells with CD40L (CD154) in vitro induces FAS expression on B cells and FASL sensitivity to cell death.28 Subsequent engagement of the BCR by antigen (Ag) protects CD40-activated B cells from FAS-induced death by increasing the expression of BCL2L1 (Bcl-XL) and CFLAR (Flip).29-31 This BCR-mediated rescue signal probably acts to reinforce appropriate B-cell–T-cell interactions and eliminate illegitimate, non-Ag–driven B-cell expansion in the GC.32,33 For this study, purified B cells from TCL1-tg and WT mice were activated with anti-CD40 and then treated with FASL in the presence or absence of anti-IgM Ab. Cells from TCL1-tg mice treated with anti-CD40 and FASL in the absence of BCR ligation had less than half the sensitivity of WT B cells to induction of apoptosis (Figure 1A). This disparity could not be attributed to differences in expression of surface FAS induced by anti-CD40, as comparable levels were detected on CD40-activated cells of either genotype (Figure 1B). Interestingly, B cells from mice of both genotypes were rescued to comparable extents by BCR ligation. These results demonstrated that enhanced expression of TCL1 induced increased resistance to FAS-mediated death in the absence of BCR engagement and rescue signaling. This survival advantage conferred by TCL1 could result in the persistence of GC B cells otherwise destined to die, and these cells might be the targets of mutagenic events that result in transformation.
TCL1 protects CD40-stimulated B cells from FASL-induced apoptosis. (A) Isolated B220+ spleen B cells from WT (□) or TCL1-tg (▪) mice were treated with 1.5 μg/mL anti-CD40 followed by incubation with FASL (1:50) with or without 10 μg/mL anti-IgM. The percentage of death induced with each condition was determined as described in “Materials and methods.” (B) Surface FAS expression was evaluated in purified B cells isolated from WT and TCL1-tg mice. Cells were either unstimulated (gray hatched line, both WT and TCL1-tg) or treated with anti-CD40 (1.5 μg/mL) with or without 10 μg/mL anti-IgM (WT, hatched line; TCL1-tg, solid black line). The data are representative of 3 separate experiments, and error bars represent standard deviation between experiments.
TCL1 protects CD40-stimulated B cells from FASL-induced apoptosis. (A) Isolated B220+ spleen B cells from WT (□) or TCL1-tg (▪) mice were treated with 1.5 μg/mL anti-CD40 followed by incubation with FASL (1:50) with or without 10 μg/mL anti-IgM. The percentage of death induced with each condition was determined as described in “Materials and methods.” (B) Surface FAS expression was evaluated in purified B cells isolated from WT and TCL1-tg mice. Cells were either unstimulated (gray hatched line, both WT and TCL1-tg) or treated with anti-CD40 (1.5 μg/mL) with or without 10 μg/mL anti-IgM (WT, hatched line; TCL1-tg, solid black line). The data are representative of 3 separate experiments, and error bars represent standard deviation between experiments.
Aicda expression is augmented in stimulated TCL1-tg B cells
Expression of AICDA has been reported for GC B cells,24,34,35 and among human NHL expression is mainly confined to those of GC phenotype.35-38 Human DLBCL exhibit a series of genetic alterations that can result from inappropriate repair of DNA double-strand breaks caused by AICDA during CSR and SHM. These changes include Chr translocations and activating mutations in the 5′ regulatory regions of proto-oncogenes such as BCL6, MYC, and PIM1.39 In addition, mice overexpressing Aicda from a transgene develop T-cell lymphomas with high levels of mutations in TCR and MYC genes, with no recurring gross Chr anomalies, and increased CSR in stimulated GC B cells.40,41
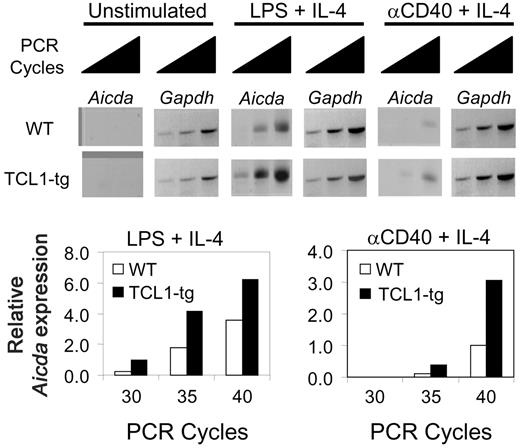
TCL1 enhances B220+PNA+ GC B-cell proliferation (Figure S1) and increased Aicda expression has recently been linked to an increased rate of cell division.41 To determine whether Aicda expression is elevated in TCL1-tg B cells as a possible contributor to lymphoma development, we examined Aicda expression in purified B cells of TCL1-tg and WT mice using semiquantitative RT-PCR. Transcripts were not detected in rested and unstimulated cells from mice of either genotype but were induced in cells coactivated with lipopolysaccharide (LPS) plus IL-4 or anti-CD40 plus IL-4 (Figure 2).42 When normalized to expression of Gapdh, Aicda transcripts were approximately 2- to 3-fold higher in TCL1-tg than WT B cells activated by either approach. These results demonstrated that heightened Aicda expression in TCL1-tg B cells from exogenous stimulation could have a role in GC B-cell transformation by a predicted increase in genome instability.43,44
A GC microenvironment is required for TCL1-tg mice to develop lymphomas
The findings thus far indicate that several effects of TCL1 on B cells could set the stage for development of B-cell lymphomas of GC phenotype: increased responsiveness to proliferative stimuli for both B220+PNA+ GC B cells (Figure S1) and supporting CD4+ T cells;13 decreased sensitivity to CD40-mediated FASL-induced apoptosis (Figure 1); and increased expression of Aicda with enhanced cell proliferation (Figure 2). While we hypothesize that the GC is required for TCL1-mediated mature B-cell transformation, the fact that TCL1-tg lymphomas express BCL6 and exhibit SHM of Ig genes suggests, but does not prove, that they require the GC environment to develop. To test this important hypothesis, TCL1-tg mice were crossed with mice deficient in the transcriptional coactivator OCA-B (OCA-B–/–), to generate mice (TxO) incapable of forming GC. Previous studies have shown varying effects of OCA-B deficiency on the development of bone marrow, B1, B2, and marginal zone B cells, with at least some differences being strain dependent. However, a lack of GC development with an intact B-cell response to T-independent antigen stimulation and terminal differentiation to antibody-secreting cells were constant findings.45,46 Interestingly, the generation of T-dependent antibody-secreting cells was blocked and caused the accumulation of GC-phenotype founder B cells that express BCL6 and Aicda and which can undergo SHM and CSR.46 These T-dependent OCA-B–/– GC-like B cells resemble the B cells that form GC B-cell lymphomas in TCL1-tg mice.12
TCL1 enhances Aicda expression in B cells. Isolated B220+ spleen B cells from WT or TCL1-tg mice were treated with LPS (10 μg/mL) plus IL-4 (50 U/mL) or anti-CD40 (1.5 μg/mL) plus IL-4 (50 U/mL), followed by 30, 35, and 40 cycles of RT-PCR to detect relative Aidca and Gapdh mRNA levels. The data are representative of 2 repeat experiments.
TCL1 enhances Aicda expression in B cells. Isolated B220+ spleen B cells from WT or TCL1-tg mice were treated with LPS (10 μg/mL) plus IL-4 (50 U/mL) or anti-CD40 (1.5 μg/mL) plus IL-4 (50 U/mL), followed by 30, 35, and 40 cycles of RT-PCR to detect relative Aidca and Gapdh mRNA levels. The data are representative of 2 repeat experiments.
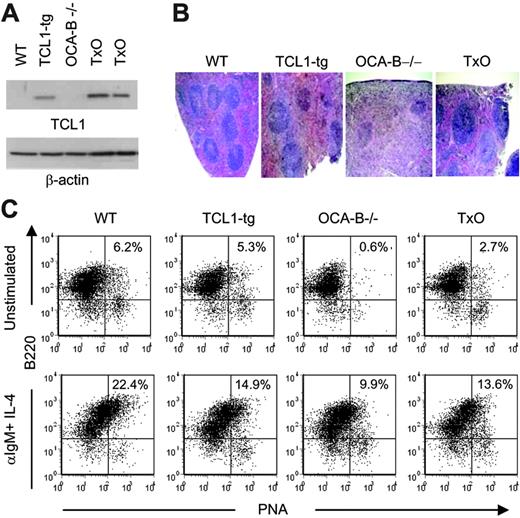
To determine whether a deficiency in OCA-B would affect TCL1-mediated lymphocyte responses in the TCL1-tg mice, we first studied TCL1 protein levels in spleens of TCL1-tg and TxO mice. OCA-B could affect expression of the TCL1 transgene that is regulated by OCA-B–sensitive B29 promoter and Eμ enhancer elements.47 However, Western blot analyses showed that the levels of TCL1 protein expression were similar in the spleens of TxO and TCL1-tg mice (Figure 3A) and less than seen in T-cell leukemias with TCL1 gene rearrangements.13 TxO bone marrow did not show the developmental block that characterizes early B-cell differentiation in OCA-B–/– mice, and TxO marrow had similar to slightly higher frequencies of immature, transitional, and recirculating B cells than did OCA-B–/– bone marrow (Table S1).48
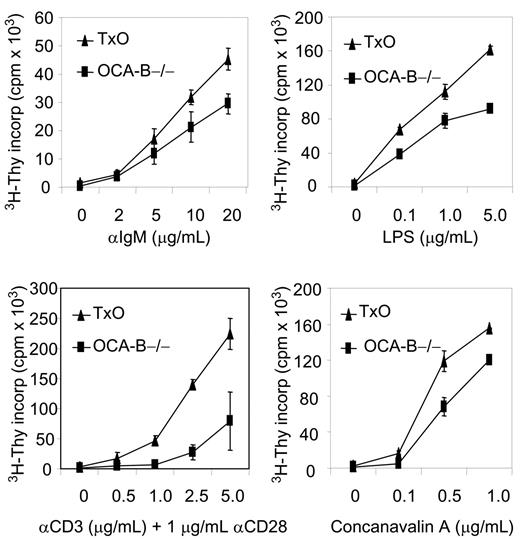
Histologically, the spleens of TxO mice, like those of OCA-B–/– mice, were devoid of GCs (Figure 3B). TxO spleens differed from those of OCA-B–deficient mice in having much larger primary follicles. This effect could be ascribed to expression of the TCL1 transgene, since the primary follicles of TCL1-tg mice were considerably larger than those of WT animals (Figure 3B). Consistent with a lack of GCs, the frequencies of B cells with a B220+PNA+ GC phenotype in spleens from OCA-B–/– mice were markedly reduced when compared with WT spleens (Figure 3C). In TxO spleens, the frequencies of B220+PNA+ cells were increased more than 4-fold from the levels in OCA-B–/– spleens. This suggests that heightened expression of TCL1 might promote the expansion of cells with the B220+PNA+ GC phenotype49 that are primed to enter nascent GCs, even in mice incapable of GC formation. Further comparisons of TxO and OCA-B–/– mice showed that TCL1 increased the proliferation responses of splenocytes to BCR ligation, LPS, concanavalin A, or anti-CD3 plus anti-CD28 (Figure 4). These results indicated that the TCL1 transgene was well expressed in the OCA-B–deficient background, and that the follicular B cells of OCA-B–deficient mice, while responsive to the proliferative effects of TCL1, were still unable to form GCs.
Characterization of 6- to 8-week-old TxO mice. (A) TCL1 expression is shown by Western blot for WT, TCL1-tg, OCA-B–/–, and TxO spleen cells. β-actin expression is shown in the bottom panel. (B) H&E staining of WT, TCL1-tg, OCA-B–/–, and TxO spleens. Original magnification, ×40. Visualization was performed using an Olympus BX40 microscope with a UPlan 4×/0.13 NA air objective (Olympus America, Melville, NY). Images were collected with an Olympus DP10 camera and processed with Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA). (C) Detection of B220+PNAhi B cells by flow cytometry. Cells were untreated (top panels) or were stimulated with anti-IgM plus IL-4 (bottom panels).
Characterization of 6- to 8-week-old TxO mice. (A) TCL1 expression is shown by Western blot for WT, TCL1-tg, OCA-B–/–, and TxO spleen cells. β-actin expression is shown in the bottom panel. (B) H&E staining of WT, TCL1-tg, OCA-B–/–, and TxO spleens. Original magnification, ×40. Visualization was performed using an Olympus BX40 microscope with a UPlan 4×/0.13 NA air objective (Olympus America, Melville, NY). Images were collected with an Olympus DP10 camera and processed with Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA). (C) Detection of B220+PNAhi B cells by flow cytometry. Cells were untreated (top panels) or were stimulated with anti-IgM plus IL-4 (bottom panels).
TCL1 enhanced growth of stimulated OCA-B–/– non-GC B and T cells. Isolated OCA-B–/– and TxO splenocytes were incubated with the listed factors and assayed at 48 hours for tritiated thymidine incorporation. Error bars represent standard deviation in triplicate measurements at each assay point.
TCL1 enhanced growth of stimulated OCA-B–/– non-GC B and T cells. Isolated OCA-B–/– and TxO splenocytes were incubated with the listed factors and assayed at 48 hours for tritiated thymidine incorporation. Error bars represent standard deviation in triplicate measurements at each assay point.
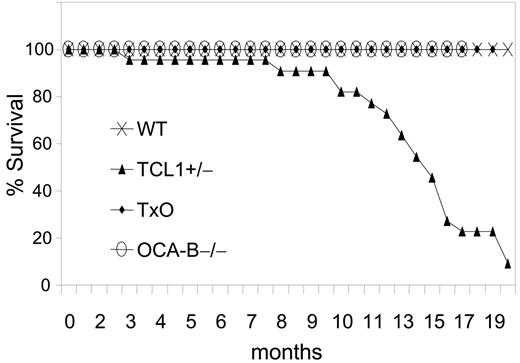
The GC is required for B-cell lymphomagenesis in pEμ-B29-TCL1 transgenic mice. Kaplan-Meier survival plot is shown. WT (n = 20), TCL1-tg (n = 22), OCA-B–/– (n = 23), and TxO (n = 25) mice were monitored up to 20 months or until they became moribund, followed by killing and necropsy.
The GC is required for B-cell lymphomagenesis in pEμ-B29-TCL1 transgenic mice. Kaplan-Meier survival plot is shown. WT (n = 20), TCL1-tg (n = 22), OCA-B–/– (n = 23), and TxO (n = 25) mice were monitored up to 20 months or until they became moribund, followed by killing and necropsy.
We then compared TCL1-tg, TxO, OCA-B–/–, and WT mice for tumor development (Figure 5). TCL1-tg mice first became ill at around 3 to 4 months of age, and by 20 months, 86% of the mice (19 of 22) had been killed when moribund with advanced disease. In contrast, none of the WT, OCA-B–/–, or TxO mice became ill during this time period. At necropsy, TCL1-tg mice had greatly enlarged spleens that most often contained monomorphic populations of IgM+B220+CD5dull B cells (Figure S2; Table S2) that were clonal for IgH gene rearrangements by Southern blot analysis (Figure S3). Lymphadenopathy was common due to diffuse subcortical accumulations of the same cells found in the spleen, often with accompanying histiocytosis. Hepatomegaly was also frequent with perivascular infiltrates of malignant lymphoid cells and histiocytes. Some cases exhibited lymphomatous infiltrates of the lung, kidney, or salivary gland. By histopathologic criteria, the lymphomas that developed were all of GC or post-GC origin, being comprised of FL, DLBCL, BLL, and plasmablastic lymphomas. Two cases, 1 plasmablastic and 1 BLL, had associated leukemic phases, and many were associated with prominent accumulations of TCL1-negative histiocytes (Figure S4). Most lymphomas exhibited intense nuclear staining for PAX5 (Figure S2) and BCL6, as shown previously.12 Overall, these results indicate that B cells of TCL1-tg mice were critically dependent on exposure to the GC environment for neoplastic transformation.
TCL1-tg BLLs are marked by chromosomal alterations and enhanced MYC expression
Recurring genetic alterations in human B-cell lymphomas include chromosomal translocations as well as point mutations in regulatory sequences of MYC, PIM1, and BCL6 genes that can result in cell activation. The long latency for lymphoma development in TCL1-tg mice indicated that TCL1 dysregulation was insufficient for transformation, and that other genetic alterations were required for the appearance of overt lymphomas. To determine whether gross chromosome changes might be involved, we used spectral karyotype (SKY) analysis to examine metaphase spreads from 5 primary lymphomas comprising 4 BLLs and 1 histiocyte-associated DLBCL (Table 1; Figure S5). All BLLs had reciprocal Chr translocations, including 3 involving Chr 5: 2 T(2;5) and 1 T(5;12). In addition, all BLLs were trisomic for Chr 15 (trisomy 15 [Ts(15)]). The single DLBCL tested by SKY had neither translocations nor Ts(15).
SKY and MYC expression of TCL1-tg B-cell lymphomas
Tumor . | MMHCC . | Translocation . | No. metaphases (no. abnormal metaphases) . | Trisomy 15 . | MYC . |
|---|---|---|---|---|---|
| K1 | DLBCL | ND | ND | ND | 0.7 |
| K2 | BLL | ND | ND | ND | 1.2 |
| K7 | BLL | T(11;17) | 16 (16) | + | 3.2 |
| K9 | DLBCL | None | 11 (0) | − | 2.2 |
| K10 | BLL | T(2;5),T(X;7),T(7;9),T(9;X) | 9 (8) | + | 1.8 |
| K14 | DLBCL | ND | ND | ND | 1.0 |
| K15 | BLL | ND | ND | ND | 1.8 |
| K18 | BLL | T(5;12) | 14 (12) | + | 2.0 |
| P3 | BLL | der(5),T(2;5) | 8 (8) | + | 3.1 |
| WT | NA | ND | ND | ND | ∼0 |
Tumor . | MMHCC . | Translocation . | No. metaphases (no. abnormal metaphases) . | Trisomy 15 . | MYC . |
|---|---|---|---|---|---|
| K1 | DLBCL | ND | ND | ND | 0.7 |
| K2 | BLL | ND | ND | ND | 1.2 |
| K7 | BLL | T(11;17) | 16 (16) | + | 3.2 |
| K9 | DLBCL | None | 11 (0) | − | 2.2 |
| K10 | BLL | T(2;5),T(X;7),T(7;9),T(9;X) | 9 (8) | + | 1.8 |
| K14 | DLBCL | ND | ND | ND | 1.0 |
| K15 | BLL | ND | ND | ND | 1.8 |
| K18 | BLL | T(5;12) | 14 (12) | + | 2.0 |
| P3 | BLL | der(5),T(2;5) | 8 (8) | + | 3.1 |
| WT | NA | ND | ND | ND | ∼0 |
The fact that Myc is located on Chr 15, and that Ts15 found in mouse thymic T-cell lymphomas are associated with overexpression of Myc,50,51 suggested that TCL1-associated BLL might also express MYC at increased levels. We examined this possibility by performing western analyses of MYC levels in extracts from 6 BLLs and 3 DLBCLs. The results (Figure 6; Table 1) revealed heightened expression of MYC in cases with and without Ts(15). MYC expression is aberrant and not passively increased from TCL1-enhanced proliferation since IgM+ centroblasts, from which TCL1-tg IgM+ BLLs probably arise, are the most rapidly proliferating cells in humans, and yet they do not express MYC.52 Further supporting this notion, nonstimulated TCL1-tg B cells do not have a proliferative advantage over WT B cells in culture (Figure S1), and the level of MYC expression in in vitro–activated TCL1-tg B cells is unchanged compared with WT B cells (data not shown).
Mutations found in the 5′ regulatory sequences of the MYC proto-oncogene in human GC lymphomas have features strongly suggesting that they are caused by AICDA.39 In support of this notion, Aicda transgenic mice develop T-cell lymphomas with heavily mutated sequences 5′ of the Myc gene.40 Since enhanced Aicda expression is associated with TCL1 expression (Figure 2), it seemed possible that heightened expression of MYC in the TCL1-tg lymphomas could be due to mutations 5′ to the gene. To test this possibility, we sequenced the 5′ regions in 2 lymphomas that exhibited SHM of IgH, 1 with and 1 without Ts(15) (Table S3). In neither tumor was the frequency of mutations increased over that found in WT spleen cells, despite the fact that most TCL1-tg tumors exhibit moderate to high-levels of Aicda expression (Figure S6).
Discussion
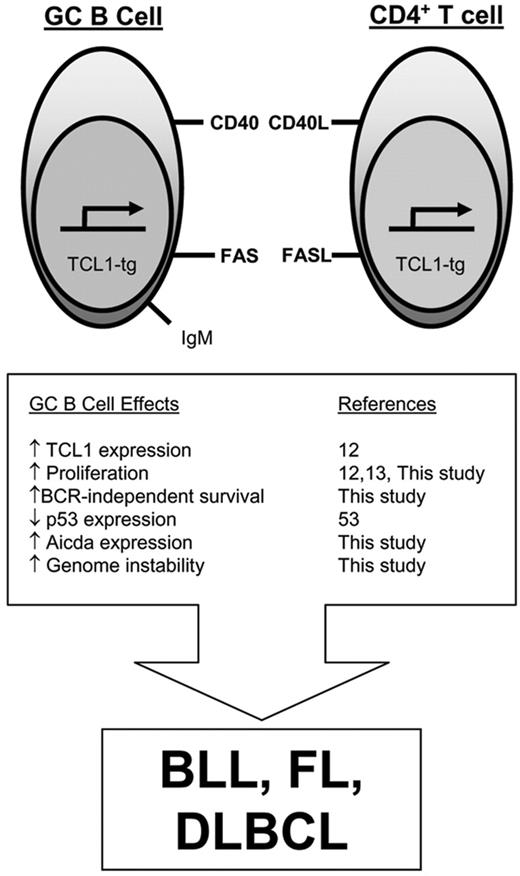
The findings presented here provide new insights into the mechanisms by which TCL1 dysregulation sets the stage for B-cell transformation, the cellular interactions required for evolution of the transformed phenotype, and the role of genomic instability in the emergence of malignant lymphomas (Figure 7).
Elevated expression of MYC protein in TCL1-tg DLBCL and BLL. A Western blot for MYC and β-actin expression is shown for BLL (K7, P3, and K18), DLBCL (K9, K14) and control (WT) spleen cells.12 The ratio of MYC to β-actin expression was determined by densitometry.
Elevated expression of MYC protein in TCL1-tg DLBCL and BLL. A Western blot for MYC and β-actin expression is shown for BLL (K7, P3, and K18), DLBCL (K9, K14) and control (WT) spleen cells.12 The ratio of MYC to β-actin expression was determined by densitometry.
TCL1 is normally expressed in bone marrow B-cell progenitors, migrants to secondary lymphoid compartments, and mature cells in the mantle zone, but is down-regulated during the GC reaction and is silenced in post-GC memory B and plasma cells.17 A role for TCL1 in differentiation is not known, but its physiologic function is likely tied to its ability to enhance the signaling activity of AKT and PKC-MAPK-ERK pathways,8,9,13 thereby affecting both cell survival and proliferation. Direct evidence for this came from studies here of isolated TCL1-tg B cells that demonstrated enhanced sensitivity to GC-related proliferative signals and increased resistance to CD40L-dependent, FAS-mediated apoptosis. Inhibition of CD40-dependent cell-death pathways may contribute importantly to GC B-cell transformation as widespread FAS-dependent apoptosis is a striking feature of GC dark zone centroblasts.52
Enhanced activation of AKT is unlikely to be sufficient for transformation, however, as mice with B cells deficient in PTEN, a negative regulator of AKT, did not develop B-cell lymphomas, and a constitutively active AKT caused cell senescence.54,55 Constitutive activation of AKT in mice with PTEN-deficient B cells also results in decreased and not increased expression of Aicda and reduced CSR.54 Aicda repression from senescence-inducing PTEN-deficiency contrasts sharply with enhanced Aicda expression from proliferation-inducing TCL1 expression shown here. This key distinction links TCL1 but not PTEN deficiency to cell-cycle–dependent Aicda expression41 and, more importantly, provides a mechanistic explanation for why TCL1 expression but not PTEN deficiency promotes mature B-cell transformation by increasing rather than decreasing genome instability.43,44 Genome alterations are usually opposed by a p53-dependent checkpoint, which protects the genome by forcing damaged cells into cell-cycle arrest or apoptosis. However, GC B cells present a special situation in which p53 is normally repressed by BCL6, possibly to permit Aicda-dependent SHM and CSR without deleting antigen-selected B cells.53 TCL1 expression might further reinforce p53 silencing in this transgenic model by activating AKT and MAPK signaling pathways, which phosphorylate Mdm2 and reduce p53 expression.56 By a similar AKT-dependent mechanism, PTEN deficiency could also decrease p53 expression, but a simultaneous reduction in Aicda would be predicted to protect GC B cells from enhanced DNA damage and impair tumor production.54,57 These findings suggest that additional AKT-dependent or alternative AKT-independent mechanisms downstream of TCL1 contribute to malignant transformation, perhaps through activation of the PKC-MAPK-ERK signaling pathway.13 Supporting this notion, a recent study has shown a role for MAPK activation in controlling Aicda expression in mouse B cells.58
In addition to providing a critical distinction between TCL1 expression and PTEN deficiency that could control GC B-cell transformation, a striking conclusion from our studies was that development of lymphomas in TCL1-tg mice was absolutely dependent on expression of the transcriptional coactivator OCA-B. Although a deficiency in OCA-B has been shown to affect several aspects of B-cell development in a somewhat strain-dependent manner, an OCA-B knockout on any strain background is incapable of generating GCs.45 We showed that the effect on lymphoma development could not be ascribed to major changes in early B-cell development, altered frequencies of follicular B cells, or inhibition of TCL1 transgene expression in OCA-B–/– B cells. From this, we infer that signals inherent to the GC microenvironment are critical to the transformation of B cells with aberrant regulation of TCL1, and that Aicda expression is necessary43,44 but not sufficient, since activated OCA-B–/– B cells46 express Aicda and do not form cancers. One source of these signals in our model could be T cells that also express the TCL1 transgene. This possibility is suggested by the observation that FL appearing in mice bearing a BCL2 transgene expressed in both B and T cells fail to develop if the mice are depleted of CD4+ T cells, which could be required to sustain GC expansion until additional B-cell–transforming events emerge.59 It should be noted that marginal zone B cells are variably reduced in number and function in OCA-B–deficient mice, and that marginal zone B-cell lymphomas (MZLs) with features of centroblasts have been described.45,60 MZLs with features of BLL, a common class of TCL1-tg lymphoma, have not been described, making it less likely that the OCA-B–/– effect on lymphoma development can be ascribed to changes in the marginal zone. It is noteworthy that histiocytes feature prominently in many TCL1-tg lymphomas, and recent studies have identified the tumor microenvironment and host inflammatory responses mediated by products of macrophages and dendritic cells as defining features in a subset of human DLBCL.61 Since histiocytes do not express the TCL1 transgene (Figure S4), their association with TCL1-tg lymphomas suggests they are recruited to the tumors in response to signals originating from TCL1-expressing B or T cells.
Model for TCL1-tg effects on GC B cells that promote malignant transformation. Aberrantly elevated pEμ-B29-TCL1 expression in the GC enhances lymphocyte proliferation, increases resistance to FAS-induced cell death, and augments Aicda expression. Since GC B cells have reduced p53 expression,53 these tumorigenic features occur in a setting that is relatively permissive for usual (SHM, CSR) and abnormal genomic alterations, increasing the likelihood for GC B-cell lymphoma development.
Model for TCL1-tg effects on GC B cells that promote malignant transformation. Aberrantly elevated pEμ-B29-TCL1 expression in the GC enhances lymphocyte proliferation, increases resistance to FAS-induced cell death, and augments Aicda expression. Since GC B cells have reduced p53 expression,53 these tumorigenic features occur in a setting that is relatively permissive for usual (SHM, CSR) and abnormal genomic alterations, increasing the likelihood for GC B-cell lymphoma development.
Analyses of TCL1-tg lymphomas by SKY provided additional insights into the mechanisms that complement TCL1 dysregulation for transformation. Recurrent Ts(15) associated with heightened expression of MYC was perhaps most revealing and provided the first reported alterations in tumors induced by dysregulated TCL1 family gene expression in transgenic mice with strong parallels to human B-cell malignancies. Dysregulated expression of MYC secondary to Ig-MYC translocations is the hallmark feature of human BL and BLL. Additionally, dysregulated MYC has been shown to impair double-stranded DNA break repair.62,63 In some cell types, these changes are insufficient to induce transformation, indicating a need for complementing mutations. Studies of MYC transgenic mice using proviral insertional mutagenesis to tag interacting oncogenes identified several complementation groups for transformation. These include Pal/Bmi1/Gfi1 in 1 set, members of the Pim family of serine threonine kinases in a second, and Runx2 in a third.64 Although Tcl1 was not identified in these screens, it is possible that MYC and TCL1 form yet another set of complementing genes. This suggestion gains weight from the observation that MYC is overexpressed in human T-cell lymphomas bearing TCL1 translocations20,65 and could be tested by crossing transgenics in which both MYC and TCL1 are targeted to the B-cell lineage.
SKY analyses of the TCL1-tg lymphomas also revealed recurring translocations involving Chr 5 and others involving Chr 2, 7, 9, 11, 17, and X. Although the significance of these events is currently not known, the T(5;12) may provide an opportunity to clone the gene at the Chr 5 breakpoint if the IgH locus on Chr 12 is involved in the translocation. The fact that stimulated TCL1-tg B cells are protected from death and express increased levels of Aicda may be relevant to the frequency with which translocations were observed in these lymphomas. In the plasmacytomas of IL-6 transgenic mice or in retrovirally transduced primary B cells, for example, it is clear that Aicda is absolutely required but may not be sufficient for the development of Igh/Myc translocations.43,66 Furthermore, in a pristane-induced model of lymphomagenesis, Aicda was found to be required for the outgrowth of transformed cells.44 Therefore, increased levels of Aicda in our model suggest a role for TCL1 in supporting the development of genetic lesions and maintaining the cells that harbor them.
Taken together, these results establish that pEμ-B29-TCL1 lymphomas require the GC to develop and strongly suggest that transformation arises as a result of tumorigenic changes to key GC-related functions and pathways. Our data further point to TCL1 effects on BCR signaling and CD40L-dependent FAS-induced death pathways that set the stage for GC-associated increases in Aicda expression in p53-suppressed B cells, leading to tolerated genetic errors including aneuploidy and aberrant MYC activation.
Prepublished online as Blood First Edition Paper, May 25, 2006; DOI 10.1182/blood-2006-02-001354.
Supported by NIH Grants T32A107126 (R.R.S), CA113872 (R.G.R.), CA109901 and CA92625 (F.W.A.), CA90571 and CA107300 (M.A.T.); and the Institute for Cell Mimetic Space Exploration (CMISE), a National Aeronautics and Space Administration University Research and Engineering Training Institute (NASA URETI) Institute Award NCC 2-1364.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of the issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Tom Rothstein (Boston University School of Medicine) for Fas-related reagents and insightful discussions, David Dawson, Samuel French, and Ali Kuraishy (UCLA) for helpful discussions, and Jerrold M. Ward and Torgny N. Fredrickson (NIH) for histological analyses.
F.W.A. is an Investigator of the Howard Hughes Medical Institute. M.A.T. is a member of the Institute for Cell Mimetic Studies, Institute for Stem Cell Biology and Medicine, and California NanoSystems Institute at UCLA and a Scholar of the Leukemia and Lymphoma Society.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal