Abstract
Brief treatment with transforming growth factor (TGF)–β1 stimulated the migration of macrophages, whereas long-term exposure decreased their migration. Cell migration stimulated by TGF-β1 was markedly inhibited by 10 μg/mL Tat-C3 exoenzyme. TGF-β1 increased mRNA and protein levels of macrophage inflammatory protein (MIP)–1α in the initial period, and these effects also were inhibited by 10 μg/mL Tat-C3 and a dominant-negative (DN)–RhoA (N19RhoA). Cycloheximide, actinomycin D, and antibodies against MIP-1α and monocyte chemoattractant protein-1 (MCP-1) abolished the stimulation of cell migration by TGF-β1. These findings suggest that migration of these cells is regulated directly and indirectly via the expression of chemokines such as MIP-1α and MCP-1 mediated by RhoA in response to TGF-β1. TGF-β1 activated RhoA in the initial period, and thereafter inactivated them, suggesting that the inactivation of RhoA may be the cause of the reduced cell migration in response to TGF-β1 at later times. We therefore attempted to elucidate the molecular mechanism of the inactivation of RhoA by TGF-β1. First, TGF-β1 phosphorylated RhoA via protein kinase A, leading to inactivation of RhoA. Second, wild-type p190 Rho GTPase activating protein (p190RhoGAP) reduced and DN-p190RhoGAP reversed the reduction of cell migration induced by TGF-β, suggesting that it inactivated RhoA via p190 Rho GAP.
Introduction
Transforming growth factor (TGF)–β regulates diverse cellular functions, including tissue differentiation, cell proliferation, and cell migration. Monocytes/macrophages, in particular, secrete TGF-β, which in turn stimulates numerous responses: production of a variety of cytokines, including interleukin-1α (IL-1α) and -β (IL-1β), tumor necrosis factor (TNF)–α, platelet-derived growth factor (PDGF)–BB, and basic fibroblast growth factor (bFGF); recruitment of monocytes to sites of injury or inflammation; phagocytic activity (by up-regulating the expression of cell-surface FcγRIII); and the expression of several integrin receptors on monocytes, including leukocyte function–associated antigen-1 (LFA-1: integrin αLβ2), α3β1, and α5β1, thereby increasing their cell-cell and cell-matrix interactions.1 These observation indicate a proinflammatory function for TGF-β on monocytes.2 In contrast to its activating effects on peripheral blood monocytes, TGF-β reduces the host response to a variety of inflammatory stimuli and is a potent immunosuppressive, anti-inflammatory, and macrophage deactivating agent.3 Resting monocytes express high levels of TGF-β type 1 and 2 receptors, whereas receptor levels decline as cells mature and are then activated by agents such as lipopolysaccharide (LPS) and interferon-γ (IFN-γ).1
The functional complex of TGF-β1 receptors at the cell surface is composed of 2 type 2 (TβRII) and 2 type 1 (TβRI) transmembrane Ser/Thr kinase receptors.4 Receptor-activated Smads (R-smads: Smad1, Smad2, Smad3, Smad5, and Smad 8), which are phosphorylated by type 1 receptors, are released from the receptor complex to form a heterotrimeric complex of 2 R-Smads and a common Smad4 (Co-Smad); the complex then translocates to the nucleus, where it regulates transcription. The structurally distinct Smads, Smad6 and Smad7, act as inhibitory Smads (I-Smads) by competing with R-Smads for receptors.5 The expression of I-Smads is strongly regulated by extracellular signals, and the induction of Smad6 and Smad7 expression by TGF-β1 reveals an inhibitory feedback mechanism for ligand-induced signaling.6 In addition to the R-Smad/Co-Smad activation pathway, TGF-β can activate the extracellular signal-regulated kinase (ERK), c-Jun N–terminal kinase (JNK), and p38MAPK pathways, the last 2 of which are activated via TGF-β–activated kinase 1 (TAK1).4
Rho GTPases regulate the actin cytoskeleton, cell polarity, gene expression, microtubule dynamics, and vesicular trafficking.7 Regulation of the nucleotide-bound state of RhoGTPases, alternative cycling between active GTP- and inactive GDP-bound states, is accomplished by the action of 3 major classes of proteins: guanine nucleotide exchange factors (GEFs), guanine nucleotide dissociation inhibitors (GDIs), and GTPase activating proteins (GAPs).7 Recently, a number of studies have demonstrated that TGF-β1 activates Rho GTPases. It induces lamellipodia and stress fibers in human prostate carcinoma cells, which requires activities of Cdc42 and RhoA.8 In skeletal muscle cells, TGF-β activates Rac1 and Cdc42 but inactivates RhoA.9 In human lens epithelial cells, it activates both RhoA and Rac1.10 Furthermore, TGF-β1 induces epithelial to mesenchymal transdifferentiation via a RhoA-dependent mechanism.11 Thus, TGF-β1 activates different GTPases depending on the cell type. On the other hand, Rho GTPases regulate the phagocytosis of apoptotic cells,12,13 which leads to the resolution of inflammation and is mediated by TGF-β1.14 From these facts, it can be inferred that TGF-β1 signaling is closely linked with Rho GTPase signaling in inflammatory cells.
In particular, TGF-β recruits monocytes to the site of injury or inflammation through several mechanisms. Therefore, it acts as a chemoattractant for monocytes.2 On the other hand, RhoGTPases play key roles in regulating the cellular responses required for cell migration.15 Rho and Rac regulate the polymerization of actin to produce stress fibers and lamellipodia, respectively. In addition to stress fibers, Rho regulates the assembly of focal adhesion complexes. Cdc 42 triggers the formation of the actin-based structure found at the cell periphery, filopodia, which includes localization of lamellipodial activity to the leading edge.16 Leukocyte migration also may be induced by various chemokines. The chemokines are classified mainly into CC and CXC families on the basis of location of N-terminal cysteine residues. In inflammation, the CXC or α-chemokines, including interleukin-8 (IL-8), act mainly on neutrophils, and the CC or β-chemokines, including macrophage inflammatory protein (MIP)–1α and monocyte chemoattractant protein-1 (MCP-1), act mainly on monocytes, lymphocytes, and eosinophils.17,18
In this study, we found that RhoA was activated and inactivated in sequence in response to TGF-β1 and showed that its activity is relevant to cell migration both directly and indirectly via the production of chemokines, including MIP-1α and MCP-1. We focused on the mechanism of RhoA inactivation in response to long-term exposure to TGF-β1 and demonstrated that RhoA is in part inactivated as a result of phosphorylation by protein kinase A (PKA) and in part by activation of p190 Rho GTPase activating protein (p190RhoGAP).
Materials and methods
Materials
PD98059, SB203580, phenylmethylsulfonyl fluoride (PMSF), Triton X-100, glutathione (GSH), formyl Met-Leu-Phe (fMLP), isopropyl-β-d-thiogalactoside (IPTG), LPS, IFN-γ, H89, cycloheximide (CHX), actinomycin D (ActD), cytocalasin D (CytD), ML-7, and Y-27632 were purchased from Sigma Chemical (St Louis, MO). SP600125 was obtained from Tocris (Bristol, United Kingdom). Protein A–agarose bead was from Pierce (Rockford, IL). GSH-Sepharose 4B bead was from Amersham Biosciences (Freiburg, Germany), and Ni-NTA His Bind resin was from Novagen (Madison, WI). Anti-Rac1 antibody was purchased from Upstate Biotechnology (Waltham, MA), and anti-RhoA, anti-Cdc42, anti-Rap1A, anti-RhoGDI, and anti-Smad3 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti–phospho-ERK, anti–phospho-JNK, anti–phospho-p38 MAPK, anti-ERK, anti–p38 MAPK, and anti-JNK antibodies were from Cell Signaling Technology (Beverly, MA).
Cell culture
Mouse macrophage Raw 264 cells were cultured in Dulbecco modified Eagle medium-F12 (DMEM-F12) containing 5% fetal bovine serum (FBS) and antibiotics (100 units/mL streptomycin and 100 units/mL penicillin: Cambrex Bio Science, Walkersville, MD) at 37°C in 5% CO2. HL-60 cells (human promyelocytic leukemia cell) were cultured in RPMI 1640 containing 10% FBS and antibiotics. P815 (mastocytoma cells) and 293 (human kidney epithelial cells) cells were cultured in DMEM containing 10% FBS and antibiotics.
Expression, purification, and transduction of Tat-C3
To introduce C3 exoenzyme efficiently into cells, we fused the HIV-1 Tat transduction domain (amino acid residues 49-57, KKKRRQRRR) to the N-terminus of the C3 exoenzyme containing a His tag (Tat-C3).19,20 Purification of Tat-C3 was performed following a previously described method.19,20 For the transduction of Tat-C3, macrophage cells were grown to confluence in 6-well plates for 4 to 6 hours. The culture medium was replaced with 1 mL serum-free medium, and Raw 264 cells were incubated with 10 μg/mL Tat-C3 for 30 minutes at 37°C.20,21
Transwell motility assays
Assays for migration were performed in transwell cell culture chambers with polycarbonate filters (24-well, 8-μm pore, Corning Costar, Cambridge, MA). The filters were coated with 1 mg/mL fibrinogen for 6 hours and dried, and cells in 200 μL DMEM-12 were added to the upper reservoir. One milliliter of DMEM-12 with or without chemotaxis reagents (1 μg/mL LPS, 50 U/mL IFN-γ, 100 nM fMLP, and 5 ng/mL TGF-β1) was placed in the lower reservoir. After 6 hours of migration, cells remaining on the upper membrane were scraped off, and cells that had migrated to the lower membrane were stained by the Giemsa staining methods (Sigma) and counted under an Axiovert 200 microscope (Zeiss, Göttingen, Germany) with a 40×/0.10 NA Achiroplan objective, and photographed with a Photometrics CoolSNAP camera (Roper Scientific, Tucson, AZ). Images were processed with IPLab 3.65a software (IPLab, Guernsey, United Kingdom) and Adobe Photoshop 7.0 (Adobe Systems, Beaverton, OR). Reagents with effects on cytoskeleton rearrangement (CytD, ML-7 and Y-27632), MAPK inhibitors (PD98059, SB203580, and SP600125), H89 (an inhibitor of PKA), CHX, or ActD were preincubated with cells before TGF-β1 was added to the cells. Cells pretreated with reagents, including TGF-β1, were washed, resuspended in 200 μL DMEM-12, and then loaded in the upper reservoir.
Reverse transcription (RT)-PCR
Total RNA was extracted from macrophages using RNA plus RNA extraction solution (Quantum, Quebec, Canada). Reverse transcription (RT) of mRNA and the polymerase chain reaction (PCR) were carried out with the Superscript II system (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Primer sequences were as follows: MIP-1α, 5′-GCCCTTGCTGTTCTTCTCTGA-3′ (sense), 5′-GGCAATCAGTTCCAGGTCAGT-3′ (antisense)22 ; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-AACGGATTTGGCCGTATT-3′ (sense), 5′-ACTGTGGTCATGAGCCCTT-3′ (antisense). The GAPDH primer pair was used as an internal control. PCR products were separated on 1.5% agarose gel, stained with ethidium bromide, and photographed.
Protein determination
MIP-1α and MCP-1 secreted from Raw 264 cells in response to TGF-β1 were quantitatively determined using enzyme-linked immunosorbent assay (ELISA) technique following manufacturer's instruction (R&D Systems, Minneapolis, MN, and BD Bioscience Pharmingen, San Jose, CA, respectively).
Coimmunoprecipitation (IP)
Macrophages were washed in phosphate-buffered saline (PBS) and lysed in lysis buffer containing 20 mM Tris-HCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA (ethylene glycol tetraacetic acid), 1 mM dithiothreitol, 0.5% deoxycholate, 0.1% SDS, 1.5 mM MgCl2, 150 mM NaCl, pH 8.0. The lysates were centrifuged at 17 000g for 30 minutes at 4°C, and the supernatants were incubated overnight with 2 μg anti-RhoGDI or anti-RhoA antibody. Protein A–conjugated agarose beads (Pierce) were added for 2 hours. The beads were washed 3 times with 1 mL lysis buffer. Bound proteins were eluted by boiling in 2 × Laemmli sample buffer and subjected to Western blot analysis using anti-RhoA, anti-RhoGDI, or anti–phospho-Ser antibodies.
Transient transfection with Smad3/4, RhoA, and p190RhoGAP cDNA constructs
Ninety percent confluent Raw 264 cells in 6-well dishes (Corning) were transiently cotransfected with 4 μg pcDNA3 plasmid constructs encoding Smad3 (wild-type [WT] Smad3 and the dominant-negative [DN] Smad3) and WT Smad4 (5 μg), RhoA (WT, constitutive active [CA]-G14V, DN-T19N), Cdc42 (WT, CA-G12V, and DN-T17N), or p190RhoGAP (WT, and DN: lacking the GTP-binding domain23 ), using lipofectamine 2000 transfection reagent. After 24 hours, the cells were incubated with 5 ng/mL TGF-β for the indicated time periods, resuspended, and lysed for IP and Western blot assays. In some cases, the transfected cells also were used for assay of migration.
GST pull-down assay for activated RhoA, Rac1, Cdc42, and Rap1
We washed 2 × 106 cells in 100-mm plates in ice-cold PBS, harvested them, and lysed them in lysis buffer (50 mM Tris-HCl, pH 7.2, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 500 mM NaCl, 10 mM MgCl2, 5 μg/mL each of leupeptin and aprotinin, and 1 mM PMSF). After centrifugation (16 000g, 15 minutes, 4°C), aliquots of the supernatant were incubated with the GST-Rho binding domain of Rhotekin (GST-RBD), the GST-GTPase binding domain of p21-activated kinase-1 (PAK-1) (GST-PBD), or His-Ral guanine nucleotide dissociation stimulator (GDS). These had been preincubated for 1 hour with 50 μg of GSH-Sepharose beads in the case of GST-fusion proteins or Ni-NTA His-Bind resins for the His-fusion protein. RhoA-GTP also was detected using EZ-Detect Rho activation kit containing GST-RBD (Pierce) following manufacturer's instructions. The beads were incubated with the cell lysates and washed, and the proteins on the beads were run on SDS-PAGE. RhoA, Rac1, Cdc42, and Rap1 were assayed by Western blotting.20,24 Alteration of proteins detected by Western blot was quantified by densitometer.
Statistical analysis
Generally, the experiments were performed in triplicate, each experiment was carried out with duplicate or triplicate samples. Data are presented as mean ± SE. Comparison between 2 groups was tested by t test using GraphPad Prizm program (GraphPad Software, San Diego, CA). Difference between 2 groups was statistically significant if P values were less than .05.
Results
Chemotaxis of Raw 264 cells is first up-regulated and then down-regulated by TGF-β1
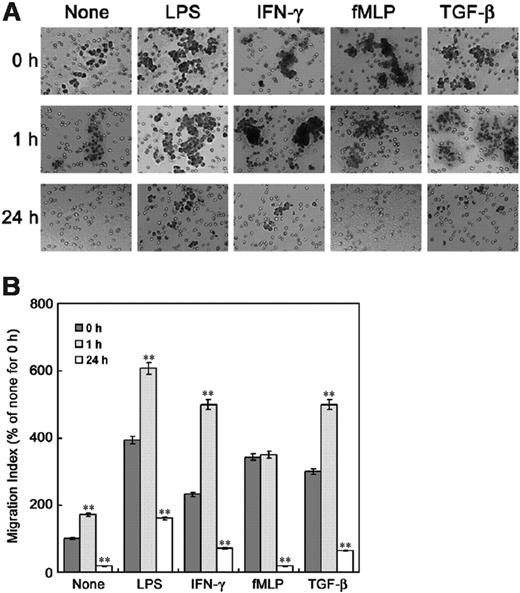
To address the effect of TGF-β1 on chemotaxis of Raw 264 cells, we examined migration of the cells in a transwell chamber assay. In the presence of chemotaxis-inducing agents such as LPS, IFN-γ, and TGF-β1, in the bottom chamber, pretreatment with TGF-β1 for 1 hour stimulated migration, whereas 24 hours' pretreatment with TGF-β1 dramatically reduced migration (Figure 1, Figure 2A). This indicates that TGF-β1 has 2 successive effects on the chemotaxis of macrophages: first stimulation, then inhibition. Although fMLP did not enhance migration of the Raw 264 cells pretreated with TGF-β1 for 1 hour, 24 hours' TGF-β1 treatment dramatically reduced the stimulation of migration by fMLP. Pretreatment with TGF-β1 for 1 hour or 24 hours, however, did not significantly affect chemotaxis of other cell types such as 293 cell (human kidney epithelial cell), HL-60 (human promyelocytic leukemia cell) as neutrophil-like cells, and P815 (mastocytoma cell) as mast cells (data not shown).
Length of incubation period affects the regulation of macrophage chemotaxis by TGF-β1. Assays were performed in transwell cell culture chambers with polycarbonate filters (24-well, 8-μm pore, Corning Costar). Raw 264 cells preincubated with or without 5 ng/mL TGF-β1 for 1 hour or 24 hours were washed, resuspended in 200 μL DMEM-12, and then added to the upper reservoir. One milliliter of serum-depleted DMEM-F12 was added to the lower reservoir with or without chemotaxis reagents (1 μg/mL LPS, 50 U/mL IFN-γ, 100 nM fMLP, and 5 ng/mL TGF-β1). After 6 hours, cells remaining on the upper membrane were scraped off, and cells on the lower membrane surface were stained with (A) Giemsa and (B) counted under a microscope. The results are means ± SE of 3 independent experiments. Each value was compared to the corresponding control at time 0 (**P < .01).
Length of incubation period affects the regulation of macrophage chemotaxis by TGF-β1. Assays were performed in transwell cell culture chambers with polycarbonate filters (24-well, 8-μm pore, Corning Costar). Raw 264 cells preincubated with or without 5 ng/mL TGF-β1 for 1 hour or 24 hours were washed, resuspended in 200 μL DMEM-12, and then added to the upper reservoir. One milliliter of serum-depleted DMEM-F12 was added to the lower reservoir with or without chemotaxis reagents (1 μg/mL LPS, 50 U/mL IFN-γ, 100 nM fMLP, and 5 ng/mL TGF-β1). After 6 hours, cells remaining on the upper membrane were scraped off, and cells on the lower membrane surface were stained with (A) Giemsa and (B) counted under a microscope. The results are means ± SE of 3 independent experiments. Each value was compared to the corresponding control at time 0 (**P < .01).
RhoA is involved in cell migration
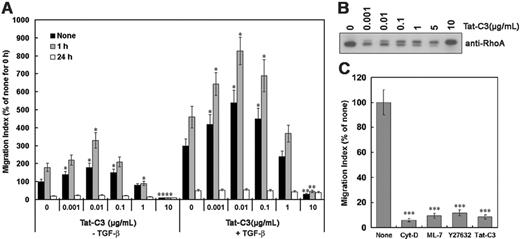
As cell movement requires dynamic reorganization of the actin cytoskeleton, and this is regulated by RhoGTPases,7 we tested whether the migration of cells in response to TGF-β1 requires Rho GTPases. Blockade of endogenous RhoA by 10 μg/mL Tat-C3, which completely modifies the Arg 41 residue of RhoA with ADP-ribosyl group of NAD+,15 markedly inhibited cell migration, whereas 0.001 to 0.1 μg/mL Tat-C3, which modified up to about 50% of RhoA molecules, stimulated migration (Figure 2A,B). The doublet band appears to be unmodified RhoA (lower band) and RhoA modified by Tat-C3 (upper band). In addition, Y-27632 (an inhibitor of ROCK [RhoA-activated kinase]), cytocalasin D (an inhibitor of filamentous actin formation), and ML-7 (an inhibitor of myosin light chain kinase, MLCK) blocked TGF-β1–stimulated cell migration (Figure 2C). These suggest that induction of cell migration by TGF-β1 for 1 hour requires actin-myosin assembly, which is regulated by Rho, ROCK, MLCK, and filamentous actin formation.
Effect of Tat-C3 on cell migration. (A) Tat-C3 was preincubated with Raw 264 cells for 30 minutes, and cell migration was assessed in transwell cell culture chambers in the presence or absence of TGF-β1 in the lower chamber. The results are means ± SE of 3 independent experiments, and each value was compared to the corresponding control (Tat-C3, 0 μg/mL) at the same time scale (0, 1, and 24 hours) (*P < .05 and **P < .01). (B) Raw 264 macrophage cells were incubated with TGF-β1 (0-10 μg/mL) for 1 hour, cell lysates (15 μg/mL of total protein) were run on SDS-PAGE, and Western blotting was performed using anti-RhoA antibody. The doublet band consists of unmodified RhoA (lower band) and RhoA modified by Tat-C3 (upper band). (C) Cells were incubated in the upper reservoir with or without a variety of reagents (1 μM cytochalasin-D, 50 μM ML-7, 50 μM Y27632, 10 μg/mL Tat-C3) for 1 hour. The lower chamber was then filled with 5 ng/mL TGF-β1, and migration measured as described in the legend to Figure 1. The values are means ± SE of 3 independent experiments. Each value was compared to the corresponding control at time 0 (***P < .001).
Effect of Tat-C3 on cell migration. (A) Tat-C3 was preincubated with Raw 264 cells for 30 minutes, and cell migration was assessed in transwell cell culture chambers in the presence or absence of TGF-β1 in the lower chamber. The results are means ± SE of 3 independent experiments, and each value was compared to the corresponding control (Tat-C3, 0 μg/mL) at the same time scale (0, 1, and 24 hours) (*P < .05 and **P < .01). (B) Raw 264 macrophage cells were incubated with TGF-β1 (0-10 μg/mL) for 1 hour, cell lysates (15 μg/mL of total protein) were run on SDS-PAGE, and Western blotting was performed using anti-RhoA antibody. The doublet band consists of unmodified RhoA (lower band) and RhoA modified by Tat-C3 (upper band). (C) Cells were incubated in the upper reservoir with or without a variety of reagents (1 μM cytochalasin-D, 50 μM ML-7, 50 μM Y27632, 10 μg/mL Tat-C3) for 1 hour. The lower chamber was then filled with 5 ng/mL TGF-β1, and migration measured as described in the legend to Figure 1. The values are means ± SE of 3 independent experiments. Each value was compared to the corresponding control at time 0 (***P < .001).
Expression of new genes induced by TGF-β1 plays a role in cell migration
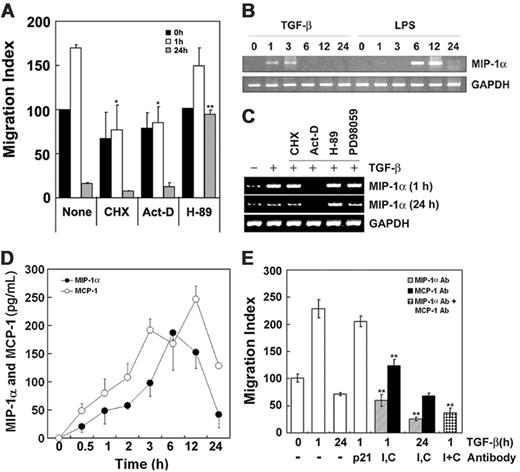
To confirm the involvement in cell migration of chemokines secreted by the cells upon exposure to TGF-β1, protein synthesis induced by TGF-β1 was blocked by the addition of CHX and ActD. As shown in Figure 3A, these agents reduced the stimulation of cell migration by 1-hour treatment with TGF-β1. These observations indicate that both TGF-β1 itself, and, secondarily, chemokines released from the cells by TGF-β1 treatment, may contribute to the stimulation of cell migration. MIP-1α and MCP-1 have been described as strong chemotactic factors for monocytes.17,18 Indeed, we also observed that MIP-1α induced migration of Raw 264 cells (data not shown). We therefore tested whether TGF-β1 induces cell migration by increasing expression of chemokines such as MIP-1α and MCP-1. RAW 264 cells were stimulated with 5 ng/mL TGF-β1 or 1 μg/mL LPS for the indicated times, and MIP-1α mRNA level was determined by RT-PCR. Notably, exposure to TGF-β1 for 1 to 3 hours stimulated MIP-1α gene expression (Figure 3B,C).
TGF-β1 induces cytokines MIP-1α and MCP-1 that participate in cell migration. (A) Cells were incubated with or without 10 μg/mL CHX, 50 ng/mL ActD, and 30 μM H89 for 30 minutes and then with or without TGF-β1 for 1 hour or 24 hours. Cells pretreated with TGF-β1 for 1 hour or 24 hours were washed, resuspended in 200 μL DMEM-12, and then loaded in the upper reservoir. Then the lower reservoir was filled with 1 mL DMEM-12 containing 5 ng/mL TGF-β1, and migration of the cells was determined as described in the legend to Figure 1. The results are means ± SE of 3 independent experiments, and each value was compared to the corresponding control (none, Tat-C3 = 0 μg/mL) at the same time scale in the presence of TGF-β1 as a chemoattractant (*P < .05 and **P < .01). (B) Cells were treated with 5 ng/mL TGF-β1 and 1 μg/mL LPS for indicated times. The levels of MIP-1α and GAPDH mRNAs were measured by RT-PCR. (C) Cells were preincubated with or without 10 μg/mL CHX, 50 ng/mL ActD, 30 μM H89, or 30 μM PD98059 for 30 minutes and then with 5 ng/mL TGF-β1 for 1 hour or 24 hours. Levels of MIP-1α and GAPDH mRNA were determined by RT-PCR. (D) MIP-1α and MCP-1 secreted from Raw 264 cells in response to 5 ng/mL TGF-β1 at various time points were measured by ELISA technique. The results are means ± SE of 3 independent experiments. (E) Antibodies (4 μg/mL) against p21WAF (cyclin-dependent kinase inhibitor), MIP-1α (I), and MCP-1 (C) were added to both upper and lower chambers. Anti-p21WAF antibody was used as a control. Cells pretreated with 5 ng/mL TGF-β1 for 1 hour or 24 hours were loaded in the upper chamber, and then cell migration was estimated in the presence of 5 ng/mL TGF-β1 in lower chamber as in Figure 1. The results are means ± SE of 3 independent experiments (**P < .01).
TGF-β1 induces cytokines MIP-1α and MCP-1 that participate in cell migration. (A) Cells were incubated with or without 10 μg/mL CHX, 50 ng/mL ActD, and 30 μM H89 for 30 minutes and then with or without TGF-β1 for 1 hour or 24 hours. Cells pretreated with TGF-β1 for 1 hour or 24 hours were washed, resuspended in 200 μL DMEM-12, and then loaded in the upper reservoir. Then the lower reservoir was filled with 1 mL DMEM-12 containing 5 ng/mL TGF-β1, and migration of the cells was determined as described in the legend to Figure 1. The results are means ± SE of 3 independent experiments, and each value was compared to the corresponding control (none, Tat-C3 = 0 μg/mL) at the same time scale in the presence of TGF-β1 as a chemoattractant (*P < .05 and **P < .01). (B) Cells were treated with 5 ng/mL TGF-β1 and 1 μg/mL LPS for indicated times. The levels of MIP-1α and GAPDH mRNAs were measured by RT-PCR. (C) Cells were preincubated with or without 10 μg/mL CHX, 50 ng/mL ActD, 30 μM H89, or 30 μM PD98059 for 30 minutes and then with 5 ng/mL TGF-β1 for 1 hour or 24 hours. Levels of MIP-1α and GAPDH mRNA were determined by RT-PCR. (D) MIP-1α and MCP-1 secreted from Raw 264 cells in response to 5 ng/mL TGF-β1 at various time points were measured by ELISA technique. The results are means ± SE of 3 independent experiments. (E) Antibodies (4 μg/mL) against p21WAF (cyclin-dependent kinase inhibitor), MIP-1α (I), and MCP-1 (C) were added to both upper and lower chambers. Anti-p21WAF antibody was used as a control. Cells pretreated with 5 ng/mL TGF-β1 for 1 hour or 24 hours were loaded in the upper chamber, and then cell migration was estimated in the presence of 5 ng/mL TGF-β1 in lower chamber as in Figure 1. The results are means ± SE of 3 independent experiments (**P < .01).
Remarkably, H89 reversed both the inhibition of cell migration (Figure 3A) and the production of MIP-1α mRNA caused by exposure to TGF-β1 for 24 hours (Figure 3C), suggesting PKA is implicated in both processes. Although PD98059 abrogated cell migration induced by TGF-β1 (Figure S1A, available at the Blood website; see the Supplemental Figures link at the top of the online article), it did not block the induction of MIP-1α mRNA by TGF-β1 (Figure 3C), suggesting that ERK is probably essential for cell migration induced by TGF-β1 in accordance with previous literature,25 but the involvement of ERK in cell migration is not relevant to the expression of MIP-1α, although ERK also is implicated in c-fos induction via Elk-1 phosphorylation.26
MIP-1α expression induced by TGF-β1 is mediated through RhoA. (A) Cells were incubated with none, 1 μg/mL LPS, TGF-β1, 10 μg/mL Tat-C3, or 30 μM H89 for 1 hour, and then incubated followed by 5 ng/mL TGF-β1 for 1 hour. (B) Cells were either not transfected (N) or transfected with 4 μg/mL mock vector (M), cDNA constructs encoding WT-RhoA (W), CA-RhoA (C), or DN-RhoA (D). (C) After 36 hours, cells were incubated with or without 5 ng/mL TGF-β1 for 1 hour or 24 hours. The levels of MIP-1α and GAPDH mRNAs were measured by RT-PCR (A-B). Raw 264 cells were transiently transfected with mock vector, DN-N19RhoA, and DN-N17Cdc42, and then MIP-1α secreted from Raw264 cells in response to TGF-β1 for 6 hours was measured by ELISA technique. Data are expressed as mean ± SE of 3 independent experiments (**P < .01).
MIP-1α expression induced by TGF-β1 is mediated through RhoA. (A) Cells were incubated with none, 1 μg/mL LPS, TGF-β1, 10 μg/mL Tat-C3, or 30 μM H89 for 1 hour, and then incubated followed by 5 ng/mL TGF-β1 for 1 hour. (B) Cells were either not transfected (N) or transfected with 4 μg/mL mock vector (M), cDNA constructs encoding WT-RhoA (W), CA-RhoA (C), or DN-RhoA (D). (C) After 36 hours, cells were incubated with or without 5 ng/mL TGF-β1 for 1 hour or 24 hours. The levels of MIP-1α and GAPDH mRNAs were measured by RT-PCR (A-B). Raw 264 cells were transiently transfected with mock vector, DN-N19RhoA, and DN-N17Cdc42, and then MIP-1α secreted from Raw264 cells in response to TGF-β1 for 6 hours was measured by ELISA technique. Data are expressed as mean ± SE of 3 independent experiments (**P < .01).
In addition, protein levels of MIP-1α and MCP-1 secreted from Raw 264 cells in response to TGF-β1 were measured by ELISA: TGF-β1 increased MIP-1α and MCP-1 with a maximum at 6 to 12 hours (Figure 3D). To confirm that secreted chemokines such as MIP-1α and MCP-1 in response to TGF-β1 play a pivotal role in cell migration, the effect of neutralizing the chemokines with specific antibodies was examined: anti–MIP-1α and MCP-1 antibodies significantly reduced cell migration, suggesting that MIP-1α and MCP-1 actually induce migration of Raw 264 cells in response to TGF-β1 (Figure 3E).
Level of MIP1-α transcription is regulated by TGF-β1 and is relevant to RhoA activity
The levels of MIP-1α mRNA induced by TGF-β1 was lowered by Tat-C3 (Figure 4A), suggesting that RhoA also is essential for transcription of these genes in response to TGF-β1. To confirm the requirement for RhoA for TGF-β1–induced MIP-1α transcription, Raw 264 cells were transiently transfected with expression plasmids encoding wild-type (WT)–RhoA, constitutively active (CA)–RhoA (RhoA G14V), and dominant-negative (DN)–RhoA (RhoA T19N) prior to stimulation by TGF-β1. DN-RhoA completely blocked the TGF-β1–induced increase of MIP-1α mRNA, whereas WT-RhoA and CA-RhoA did not affect it (Figure 4B). Furthermore, DN-RhoA reduced a level of MIP-1α protein secreted from Raw 264 cells in response to TGF-β1 (Figure 4C). This result also provides strong evidence that RhoA is required for MIP-1α expression in response to short-term exposure to TGF-β1. Cdc42, a member of the Rho GTPase subfamily, also was examined to determine whether it participates in regulation of MIP-1α expression. In contrast, DN-Cdc42 did not block MIP-1α protein expression in Raw 264 cells in response to TGF-β1 (Figure 4C), suggesting that Cdc42 is not relevant to expression of MIP-1α.
The level of MIP-1α mRNA was increased by exposure to LPS for 6 to 12 hours (Figure 3A) in accord with previous findings.27 Exposure to LPS plus TGF-β1 for 6 hours, however, decreased MIP-1α mRNA levels compared with LPS alone (Figure S2). Thus, TGF-β1 appeared to inhibit induction of MIP-1α expression by LPS. Tat-C3 also reduced expression of MIP-1α in response to LPS (Figure S2), indicating that activated RhoA also is required for transcription of these genes induced by LPS. Remarkably, H89, an inhibitor of PKA, reversed the inhibition of MIP-1α expression by TGF-β1 (Figure S2), suggesting that PKA is required for the inhibition of MIP-1α expression. However, H-89 had no effect on the expression of MIP-1α in response to short-term exposure to TGF-β1 (Figure 4A).
TGF-β1 regulates activities of RhoGTPases
To see whether RhoGTPases are activated by TGF-β1 in macrophage cells, we performed pull-down assays using GST-fusion of the RhoGTPases binding domains of various effectors of RhoGTPases. Cells were treated with TGF-β1 for various times, and GST-PAK-CRIB and GST-Rhotekin-RBD were incubated with the cell lysates to isolate GTP-loaded Rac1, Cdc42, and RhoA of the cells. Rac1 was found to be in the GTP-bound form in the absence of stimulation, and TGF-β1 did not alter its level except marginally at 30 minutes. In contrast, the GTP-bound forms of RhoA and Cdc42 increased from 10 to 60 minutes and were strikingly reduced after 12 hours (Figure 5, Figure 6A).
TGF-β1 inactivates RhoA via p190RhoGAP
GTPase activating proteins (GAPs) function by stimulating GTP hydrolyzing activity of their substrate GTPases, thereby inactivating them.7,22 Among the numerous RhoGAPs, p190RhoGAP accounts for a substantial fraction of the total Rho inhibitory activity in cultured cells.22 Thus, p190RhoGAP appears to be a strong candidate for the RhoA inactivation upon long-term exposure to TGF-β1, and we tried to verify this assumption. Indeed, WT p190RhoGAP abrogated cell migration induced by TGF-β1. DN p190RhoGAP, however, significantly increased migration of the cells pretreated with TGF-β1 for 24 hours compared with that of control cells (Figure 6B). As shown in Figure 6A, TGF-β1 activated RhoA by 30 minutes and thereafter inactivated it. When WT p190RhoGAP was overexpressed, TGF-β1 did not activate RhoA, probably because p190RhoGAP accelerates the hydrolysis of GTP bound to RhoA. However, in the presence of overexpressed DN p190RhoGAP, which lacks the GTP-binding domain22 and therefore lacks GAP activity, the active GTP-bound state of RhoA was sustained, except that it slightly decreased at 24 hours. This suggests that long-term treatment with TGF-β1 may inactivate RhoA via p190RhoGAP.
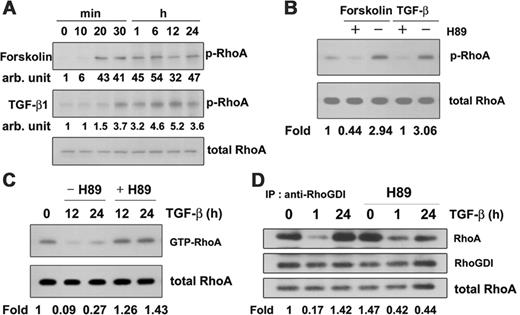
TGF-β1 induces PKA-mediated RhoA phosphorylation, leading to RhoA inactivation
PKA inhibits RhoA activation and facilitates RhoA-RhoGDI complex formation28 during neurite outgrowth29 and inhibition of sustained smooth muscle contraction.30 In addition, TGF-β–induced Smad3/Smad4 complexes directly activate PKA.31 Therefore, we asked whether TGF-β1 induces RhoA phosphorylation and so inactivates RhoA in macrophages. To measure phosphorylation of RhoA by PKA or in response to TGF-β1, cells were treated with 100 μM forskolin to activate adenylate cyclase or with 5 ng/mL TGF-β1, and phosphorylation of RhoA was measured by Western blotting with anti-RhoA antibody after immunoprecipitation of phospho-Ser proteins from cell lysates with anti–phospho-Ser antibody. Forskolin-mediated phosphorylation of RhoA was evident after 20 minutes and sustained for 24 hours. TGF-β1 also caused sustained phosphorylation of RhoA (Figure 7A). To confirm that this phosphorylation of RhoA was mediated by PKA, macrophages were treated with H89 in the absence and presence of TGF-β1. As shown in Figure 7B, both TGF-β1 and forskolin induced phosphorylation of RhoA, and H89 prevented it, suggesting that RhoA is phosphorylated by PKA in response to TGF-β1. To confirm the involvement of PKA in the inactivation of RhoA in response to TGF-β1, H89 was administered to macrophages in the presence of TGF-β1. This reversed the inactivation of RhoA in response to TGF-β1 for 12 to 24 hours, indicating that PKA inactivates RhoA (Figure 7C) and that phosphorylation of RhoA by PKA may be engaged in a regulatory step in TGF-β1 signaling. H89, however, could not reverse Cdc42 inactivation in response to TGF-β1 treatment for 12 to 24 hours (data not shown).
TGF-β1 up-regulates and then down-regulates the GTP-bound form of RhoGTPases. (A) Cells were incubated with or without 5 ng/mL TGF-β1 for the indicated times at 37°C and lysed in a buffer containing 1% Triton X-100. GST-RBD and GST-PBD were mixed with the cell extracts, which were incubated with GSH-Sepharose beads. The beads were washed, and bound proteins were fractionated on SDS-PAGE. Western blotting was performed with anti-RhoA, anti-Rac1, and anti-Cdc42 antibodies. (B) Protein amounts expressed by Western blot were quantitated by densitometry, and relative amounts of GTP-bound form at the indicated time were presented as fold. GTP-bound form/total protein at time 0 was expressed as 1. The values were means ± SE of 3 independent experiments (*P < .05).
TGF-β1 up-regulates and then down-regulates the GTP-bound form of RhoGTPases. (A) Cells were incubated with or without 5 ng/mL TGF-β1 for the indicated times at 37°C and lysed in a buffer containing 1% Triton X-100. GST-RBD and GST-PBD were mixed with the cell extracts, which were incubated with GSH-Sepharose beads. The beads were washed, and bound proteins were fractionated on SDS-PAGE. Western blotting was performed with anti-RhoA, anti-Rac1, and anti-Cdc42 antibodies. (B) Protein amounts expressed by Western blot were quantitated by densitometry, and relative amounts of GTP-bound form at the indicated time were presented as fold. GTP-bound form/total protein at time 0 was expressed as 1. The values were means ± SE of 3 independent experiments (*P < .05).
p190RhoGAP participates in the inhibition of RhoA by TGF-β1 signaling. (A) Cells were transfected with cDNA constructs encoding WT p190RhoGAP or DN p190RhoGAP lacking the GTP-binding domain or not transfected. After 36 hours, they were incubated with or without 5 ng/mL TGF-β1 for the indicated times. GTP-RhoA was determined by GST-RBD pull down, and total RhoA and HA-tagged p190RhoGAP were determined by Western blotting using anti-RhoA and anti-HA antibodies. The values are mean ± SE of 3 (None and DN) and 2 (WT) independent experiments (*P < .05). (B) Ninety percent confluent Raw264.7 cells in 6-well dishes (Corning) were transfected with 4 μg WT and DN p190RhoGAP constructs using the lipofectamine 2000 transfection reagent. After incubated for 24 hours, the cells were treated with 5 ng/mL TGF-β1 for 1 hour and 24 hours and then allowed to migrate in transwell cell culture chambers. Control and 5 ng/mL TGF-β1 pretreated cells were added to the upper reservoir. One milliliter of DMEM-F12 with chemotaxis reagents (5 ng/mL TGF-β1) was added in the lower reservoir. After 6 hours of migration, nonmigrated cells in the upper membrane were scraped off, and transmigrated cells in the lower membrane were stained by Giemsa staining method and then counted under microscopy. The values are means ± SE of 3 independent experiments, and each value was compared with corresponding control at the same time scale (**P < .01; ***P < .001).
p190RhoGAP participates in the inhibition of RhoA by TGF-β1 signaling. (A) Cells were transfected with cDNA constructs encoding WT p190RhoGAP or DN p190RhoGAP lacking the GTP-binding domain or not transfected. After 36 hours, they were incubated with or without 5 ng/mL TGF-β1 for the indicated times. GTP-RhoA was determined by GST-RBD pull down, and total RhoA and HA-tagged p190RhoGAP were determined by Western blotting using anti-RhoA and anti-HA antibodies. The values are mean ± SE of 3 (None and DN) and 2 (WT) independent experiments (*P < .05). (B) Ninety percent confluent Raw264.7 cells in 6-well dishes (Corning) were transfected with 4 μg WT and DN p190RhoGAP constructs using the lipofectamine 2000 transfection reagent. After incubated for 24 hours, the cells were treated with 5 ng/mL TGF-β1 for 1 hour and 24 hours and then allowed to migrate in transwell cell culture chambers. Control and 5 ng/mL TGF-β1 pretreated cells were added to the upper reservoir. One milliliter of DMEM-F12 with chemotaxis reagents (5 ng/mL TGF-β1) was added in the lower reservoir. After 6 hours of migration, nonmigrated cells in the upper membrane were scraped off, and transmigrated cells in the lower membrane were stained by Giemsa staining method and then counted under microscopy. The values are means ± SE of 3 independent experiments, and each value was compared with corresponding control at the same time scale (**P < .01; ***P < .001).
To see whether exposure to TGF-β1 stimulates interaction between RhoA and RhoGDI, we performed coimmunoprecipitation experiments using anti-RhoGDI antibody. The amount of RhoA bound to RhoGDI was higher in cells stimulated with TGF-β1 for 24 hours compared with control cells (Figure 7D), suggesting that RhoA was inactivated by TGF-β1 treatment for 24 hours. However, the stimulation of RhoA-RhoGDI complex formation by TGF-β1 treatment for 24 hours was reduced by H89, indicating that PKA was responsible for RhoA-RhoGDI complex formation.
Discussion
We have shown that short-term treatment with TGF-β1 stimulated the migration of macrophages and raised a level of GTP-RhoA and mRNA and protein levels of MIP-1α, whereas long-term exposure decreased both effects. Furthermore, long-term treatment of TGF-β1 suppressed the mRNA level of MIP-1α induced by LPS, which is known to cause inflammation (Figure S2). These results are in accord with previous evidence that TGF-β1 functions subsequently as a stimulatory and an inhibitory cytokine.1,2
Activation of macrophages by TGF-β1
Although TGF-β1 is a potent chemoattractant for monocytes and macrophages,1,2 the role of activation of RhoA and Cdc42 in response to TGF-β1 is not clear. To demonstrate that TGF-β1 controls macrophage migration via RhoA, we provided evidence that Tat-C3 blocked cell migration (Figure 2) and expression of MIP-1α (Figure 3), that DN-RhoA reduced mRNA and secreted protein levels of MIP-1α, and that TGF-β1 treatment regulated RhoA activity (Figure 5). Thus, it is likely that RhoA is essential for the chemokine production leading to macrophage activation and migration in response to TGF-β1 signaling. Nevertheless, CA-RhoA inhibited cell migration, whereas WT-RhoA and DN-RhoA did not (data not shown). The reason DN-RhoA did not reduce cell migration may be that DN-RhoAcould not completely block endogenous RhoA, probably because of partial transfection: transfection efficiency was about 30% to 40% in Raw 264 cells. Indeed, RhoA inhibited by 0.1 μg/mL Tat-C3 up to 50% did not cause the reduction of cell migration (Figure 2). CA-RhoA may interfere with cell migration because it induces tighter confocal adhesion,32 and it may persist for a long time bound to its effector proteins, preventing cycling between the GTP- and GDP-bound states of RhoA and inhibiting cell migration.
TGF-β1 induces phosphorylation of RhoA facilitating RhoA-RhoGDI complex formation. (A) Cells were treated with 100 μM forskolin or 5 ng/mL TGF-β1 for the indicated times and lysed. Phosphorylated RhoA was immunoprecipitated with anti–phospho-Ser antibody and protein-A–conjugated agarose and detected by Western blotting with anti-RhoA antibody. (B) Cells were treated with 100 μM forskolin or 5 ng/mL TGF-β1 in the presence or absence of 30 μM H89 for 30 minutes and lysed. Immunoprecipitation and immunoblotting were performed as in panels A and B. (C) Cells were preincubated with 30 μM H89 for 30 minutes and then treated with TGF-β1 for 12 hours or 24 hours. GTP-RhoA in cell lysates was bound to GST-RBD and GSH-Sepharose beads and revealed by Western blot using anti-RhoA antibody. (D) Cells were preincubated with 30 μM H89 for 30 minutes, then with TGF-β1 for 1 hour or 24 hours, and lysed. RhoGDI antibody was added to the cell lysates, and immunocomplexes were precipitated with Protein-A–conjugated agarose. Proteins were then fractionated using 2 × Laemmli buffer and analyzed by Western blotting with anti-RhoA and anti-RhoGDI antibodies.
TGF-β1 induces phosphorylation of RhoA facilitating RhoA-RhoGDI complex formation. (A) Cells were treated with 100 μM forskolin or 5 ng/mL TGF-β1 for the indicated times and lysed. Phosphorylated RhoA was immunoprecipitated with anti–phospho-Ser antibody and protein-A–conjugated agarose and detected by Western blotting with anti-RhoA antibody. (B) Cells were treated with 100 μM forskolin or 5 ng/mL TGF-β1 in the presence or absence of 30 μM H89 for 30 minutes and lysed. Immunoprecipitation and immunoblotting were performed as in panels A and B. (C) Cells were preincubated with 30 μM H89 for 30 minutes and then treated with TGF-β1 for 12 hours or 24 hours. GTP-RhoA in cell lysates was bound to GST-RBD and GSH-Sepharose beads and revealed by Western blot using anti-RhoA antibody. (D) Cells were preincubated with 30 μM H89 for 30 minutes, then with TGF-β1 for 1 hour or 24 hours, and lysed. RhoGDI antibody was added to the cell lysates, and immunocomplexes were precipitated with Protein-A–conjugated agarose. Proteins were then fractionated using 2 × Laemmli buffer and analyzed by Western blotting with anti-RhoA and anti-RhoGDI antibodies.
Reversibility of focal complex formation/adhesion is important for cell migration: a high level of integrin-mediated adhesion inhibits cell migration because of the strength of the attachment to the extracellular matrix, and this correlates with high levels of Rho activity.15,33 However, contraction of the bodies of moving cells is dependent on actomyosin contractility and is regulated by RhoA.34 Therefore, reducing RhoA activity has opposing effects: it promotes migration by lowering adhesion, but decreases migration by inhibiting cell body contraction.15
In less-adherent cells lacking focal adhesion, such as macrophages, neutrophils, and various cancer cell lines, RhoA does not affect adhesion but induces cell body contraction, and in these cases RhoA is clearly required for cell migration.15 DN-Cdc42 could not reduce secreted protein levels of MIP-1α (Figure 4C). This indicates that Cdc42 may be relatively less important than RhoA to stimulate cell migration in response to TGF-β1 in macrophages, although short-term treatment of TGF-β1 activates Cdc42 (Figure 5). In macrophages, Rho and Rac are required for the process of cell migration, whereas Cdc42 is essential for cells to respond to a gradient of chemoattractant such as colony stimulatory factor-1 (CSF-1) but not essential for cell locomotion.35
Short-term treatment with TGF-β1 increased MIP-1α and MCP-1 as well as cell migration. The MIP-1α and MCP-1 produced in response to TGF-β1 could act as secondary chemoattractants (Figure 3E). Here, we attempt to explain the signal pathway from TGF-β1 ligand to MIP-1α, MCP-1 expression via RhoA and NF-κB using previously reported documents.
Notably, the activity of the transcription factor NF-κB can be modulated by members of the Rho family small GTPases,36 and RhoA seems to activate NF-κB via a MEKK1-independent mechanism.37 Inactive NF-κB binds to IκB, and NF-κB becomes active and is translocated into the nucleus to play as a transcription factor when IκB is phosphorylated and degraded. We have observed that TGF-β1 induces phosphorylation of IκB and concomitantly reduces its concentration, probably via degradation (data not shown), suggesting that TGF-β1 activates NF-κB. The activated NF-κB may stimulate serum response factor (SRF)–dependent transcription and in turn induce the transcription of c-fos, a component of AP-1 with the aid of ternary complex factors (TCFs).38,39 Indeed, it has been demonstrated that RhoA potentiates AP-1 transcription in T cells.40 Since the promoter of MIP-1α contains 5 AP-1 binding sites41 and the promoter of MCP-1 contains NF-κB, AP-1, and SP-1 binding sites,42 AP-1 seems to be important for the transcription of MIP-1α, and AP-1 and NF-κB are likely to be responsible for transcription of MCP-1. Therefore, TGF-β1 probably induces transcription of MIP-1α and MCP-1 by the following routes: TGF-β1 → RhoA → NF-κB → SRF → c-fos (AP-1) → MIP-1α and MCP-1, and TGF-β1 → RhoA → NF-κB → MCP-1. Hence, it is likely that the activation of RhoA in the first 60 minutes of exposure to TGF-β1 induces the production of chemokines such as MIP-1α and MCP-1 as well, directly stimulating cell migration. Recently, it was documented that TGF-β1 stimulates MCP-1 expression,43 which is regulated by Rho and Rho-kinase.44 Indeed, it also was reported that MIP-1α and MCP-1 are induced by several stimuli via NF-κB activation,45 and TGF-β1 transiently activates NF-κB through TAK1 and IκB kinase (IKK).46
Although ERK stimulates TCFs such as Elk-1, which together with SRF induces transcription of c-fos,39 ERK activated in response to TGF-β1 (Figure S1B) was not involved in the induction of MIP-1α (Figure 3C). Here it should be noted that activated ERK during TGF-β1–induced cell migration is translocated into focal contact,47 although a detail regulatory mechanism of ERK in cell migration is not clear. Furthermore, Rho and ROCK are essential upstream components of ERK1/2, in that pretreatment of Raw 264 cells with Tat-C3 and Y-27632 for 30 minutes inhibited TGF-β1–stimulated ERK1/2 (Figure S1B).
Deactivation of macrophages by TGF-β1
The assumption that TGF-β1 abrogates cell migration by inducing expression of new proteins such as integrin receptors and extracellular matrix proteins so causing excessively tight binding of the cells1,2 can be excluded, in that CHX and ActD blocked the induction of new genes but could not reverse the reduced cell migration in response to long-term exposure to TGF-β1 (Figure 3A). TGF-β1 is known to be an immunosuppressive cytokine with many effects on immune responses.48 Furthermore, administration of TGF-β1 induces Smad 6 and Smad 7 (I-Smads) that in turn negatively regulate TGF-β signaling by interacting with TβRI49,50 and recruiting the E3 ligases Smurf1 and Smurf2, which direct ubiquitin-dependent degradation of TGF-β receptor–Smad7 complexes.51,52
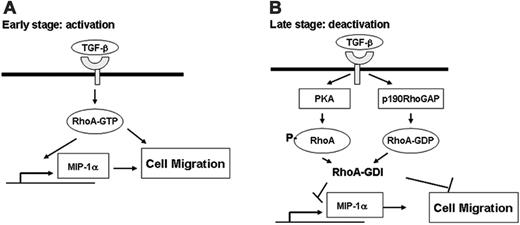
Scheme of the activation and deactivation of macrophage migration in response to TGF-β1. (A) RhoA is activated in response to short-term treatment of TGF-β1 by an unknown mechanism, which induces expression of MIP-1α and MCP-1, probably via NF-κB and AP-1. These cytokines, as well as RhoA, may indirectly and directly, respectively, participate in stimulation of cell migration. (B) Long-term exposure to TGF-β1 inactivates RhoA via phosphorylation by PKA and activation of p190RhoGAP, resulting in inhibition of macrophage cell migration. This scheme shows dual function, in view of chemotaxis, of activation in early phase and deactivation of macrophages in late phase, which is mediated at least in part by RhoA activity.
Scheme of the activation and deactivation of macrophage migration in response to TGF-β1. (A) RhoA is activated in response to short-term treatment of TGF-β1 by an unknown mechanism, which induces expression of MIP-1α and MCP-1, probably via NF-κB and AP-1. These cytokines, as well as RhoA, may indirectly and directly, respectively, participate in stimulation of cell migration. (B) Long-term exposure to TGF-β1 inactivates RhoA via phosphorylation by PKA and activation of p190RhoGAP, resulting in inhibition of macrophage cell migration. This scheme shows dual function, in view of chemotaxis, of activation in early phase and deactivation of macrophages in late phase, which is mediated at least in part by RhoA activity.
The basis of the inhibitory action of TGF-β1 on cell migration (Figure 1), however, has not been elucidated. Here, we showed that inhibition of RhoA is essential for its inhibitory action on cell migration (Figure 2) and that one of the direct causes of the inhibition of cell migration may be inactivation of the RhoA previously activated early upon short-term exposure to TGF-β1. Thus, the mechanism by which RhoA is inactivated during long-term exposure to TGF-β1 needs to be elucidated.
P190RhoAGAP inactivates RhoA. P190RhoGAP may be involved in a mechanism that may be responsible for RhoA inactivation (Figure 6A). DN p190RhoGAP, which lacks a GTP-binding domain, did not inactivate RhoA in response to TGF-β1. If the phosphorylation of RhoA was primarily responsible for RhoA inactivation, the GTP-bound state of RhoA would be reduced even in the presence of DN p190RhoGAP. However, the opposite was the case. Thus, this second mechanism may be the main pathway involved. In particular, DN RhoGAP significantly increased migration of the cells pretreated with TGF-β1 for 24 hours compared with that of control cells (Figure 6B). This also provides strong evidence that the activity of RhoA regulates cell migration induced by TGF-β1. Although p190RhoGAP can be regulated via phosphorylation by Src and protein kinase C,53 it is elusive how p190RhoGAP is activated in response to long-term exposure to TGF-β1.
Phosphorylation of RhoA by PKA inactivates RhoA. The inactivation of RhoA by TGF-β1 was reversed by H89, suggesting that PKA inactivates RhoA (Figure 7C). Indeed, TGF-β1 phosphorylated RhoA (Figure 7A), and as a result the complex of RhoA-RhoGDI might increase during long-term exposure to TGF-β1. H89 also reversed the inhibition of cell migration resulting from long-term exposure to TGF-β1 (Figure 3A). Indeed, the view that PKA is involved in the regulation of cell migration is reinforced by the evidence that PKA inhibits expression of MIP-1α (Figure 3C).27,54 It is not clear whether the activation of PKA in response to TGF-β1 is dependent on cAMP.
It has been independently reported that PKA phosphorylates RhoA and stabilizes the RhoA-RhoGDI complexes55 implicated in protection against endothelial barrier dysfunction56 and in activation of neurite outgrowth of PC12 cells.29 Recently, the molecular mechanism of PKA activation in response to TGF-β1 has been clarified: TGF-β1 treatment leads to the formation of a complex between Smad3/4 proteins and the regulatory subunit of PKA, resulting in release of the PKA catalytic subunit.31 Contrary to our expectation that Smad3/4 would activate PKA and lead to inactivation of RhoA, overexpressed Smad3/4 activated RhoA even in response to long-term administration of TGF-β1 (data not shown). Overexpressed Smad3/4 may compete with and overcome the effect of the I-Smads, resulting in normal transmission of the TGF-β1 signal. Thereafter, TGF-β1 may induce the expression of Net1, a GEF for RhoA, via the overexpressed Smad3, resulting in activation of RhoA.57 PKA also is reported to inhibit AKAP-Lbc, one of the Rho-GEFs, resulting in RhoA inactivation.58
In conclusion, we propose that macrophage migration in response to TGF-β1 is essentially regulated by RhoA activity and that RhoA is eventually inactivated via phosphorylation by PKA and the action of p190RhoGAP (Figure 8). It is to be answered how TGF-β1 allows these factors to inactivate RhoA.
Prepublished online as Blood First Edition Paper, May 16, 2006; DOI 10.1182/blood-2005-10-009191.
J.-G.K., M.-Y.M., C.-Y.J., and H.-Y.W. contributed equally to this work.
Supported by grant R01-2003-000-10568-0 from the Basic Research Program; the Medical Research Center program of Ministry of Science and Technology/the Korea Science and Engineering Foundation (grant no. R13-2005-022-01003-0); and by grant A020007 from the Korea Health 21 R&D Project, Ministry of Health and Welfare, Republic of Korea.
The online version of this article contains a data supplement.
J.-S.K. performed research, analyzed data, and wrote the paper; J.-G.K. performed research and analyzed data; M.-Y.M. performed research and analyzed data; C.-Y.J. performed research and analyzed data; H.-Y.W. performed research and analyzed data; H.-J.K. performed research; Y.-J.J. performed research; J.-Y.S. performed research; J.-I.K. analyzed data; J.K. analyzed data; J.-Y.L. analyzed data; P.-H.K. analyzed data; and J.-B.P. designed research, analyzed data, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We greatly thank Dr J. L. Bos of the University Medical Center Utrecht for providing His-tagged RalGDS-RBD plasmid DNA. We thank Dr S. O. Huh and Dr D. K. Song from the Department of Pharmacology of Hallym University for providing P815 and HL-60 cells, respectively. We also are grateful to Dr J. Settleman of Harvard Medical School for providing the p190RhoGAP cDNAs and to Dr R. Derynck of the University of California at San Francisco for Smad3 and Smad4 cDNAs. In addition, we thank Dr R. Yu of the University of Ulsan, Korea, for an aid to measurement of cytokines.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal