Chronic myeloid leukemia (CML) is characterized by the presence of the constitutively active BCR-ABL protein tyrosine kinase. Using a multipotent hemopoietic cell line, FDCP-Mix, expressing BCR-ABL tyrosine kinase, we investigated the initial effects of this kinase in primitive hematopoietic stem cells. We identified down-regulation of a novel gene, CCN3, as a direct consequence of BCR-ABL kinase activity. CCN3 has been reported to function as a tumor suppressor gene in solid tumors. Northern and Western blotting plus immunocytochemical analysis confirmed CCN3 expression is decreased and is tyrosine-phosphorylated in BCR-ABL kinase active FDCP-Mix cells. Decreased cellular CCN3 correlated with increased CCN3 secretion in BCR-ABL kinase active cells. In vitro treatment of human CML cell lines with imatinib or siRNA directed against BCR-ABL significantly reduced BCR-ABL while increasing CCN3 expression. Cells from patients responding to imatinib showed a similar decrease in BCR-ABL and increase in CCN3. CML CD34+ cells treated with imatinib in vitro demonstrated increased CCN3 protein. Transfecting CCN3 into BCR-ABL+ cells inhibited proliferation and decreased clonogenic potential. CCN3 plays an important role in internal and external cell-signaling pathways. Thus, BCR-ABL can regulate protein levels by governing secretion, a novel mechanism for this tyrosine kinase.
Introduction
Chronic myeloid leukemia (CML) is a clonal disorder of pluripotent hematopoietic cells that is characterized by the presence of the Philadelphia chromosome (Ph+), the result of a reciprocal translocation between chromosomes 9 and 22. The translocation encodes a chimeric protein, BCR-ABL, which is a constitutively activated protein tyrosine kinase (PTK)1 and which has an essential role in the molecular pathology of CML. CML is a progressive disease with an initial chronic phase in which there is a marked expansion in the late myeloid cell population (reviewed by Clarkson and Strife2 ). CML progenitor cells possess only a subtle defect in maturation, while retaining their requirements for bone marrow stromal cells or cytokines to survive and proliferate.3 This chronic phase, which is of variable duration, is usually followed by an accelerated phase leading to blast crisis. The blast crisis is the most severe manifestation of the disease in which differentiation is apparently blocked. Imatinib (Gleevec or STI571), an Abl tyrosine kinase inhibitor, is the first line and gold standard for treating CML. Nonetheless, there is clear evidence of resistance to this drug. The development of other approaches to treat the disease with small-molecule inhibitors to new targets has been, and will be, a focus for future research into treatment of imatinib-resistant patients and those in accelerated phase or blast crisis.4-7 Thus, understanding the process of leukemogenesis driven by this oncogene is still an important research objective.
Most cell-line models expressing BCR-ABL PTK are already differentiation blocked, and assume a growth factor-independent phenotype.8-11 In this respect they mirror the properties of cells from patients with CML in blast crisis. However, we have established a cell-line model for CML that has properties of chronic-phase progenitor cells that progressively change to those resembling accelerated phase by using an inducible temperature-sensitive BCR-ABL PTK (ts-BCR-ABL FDCP-Mix).12-14 DNA microarray technology has been used to identify differential expression as a result of the initial activation of BCR-ABL. Activation of BCR-ABL in FDCP-Mix cells has identified transcriptional and secretory events regulated by BCR-ABL. These effects are observed to be due to BCR-ABL in primary CML cells.
Materials and methods
Cell lines
FDCP-Mix cells transfected with the retroviral vector pM5-neo carrying a p210 ts-BCR-ABL cDNA were used as a model system for CML.12 The p210 ts-BCR-ABL cDNA encodes a temperature-sensitive mutant of the p210 BCR-ABL that is kinase active when the cells are grown at the permissive temperature of 32°C. Cells transfected with vector only were used as a control. Cells were routinely cultured in Fisher medium supplemented with preselected batches of horse serum (20% vol/vol), interleukin-3 (IL-3; 5% vol/vol) and G418 (50 μg/μL). The cells were subcultured twice weekly to maintain a log phase culture of 2 × 105 cells/mL and maintained at 39°C in 5% CO2 in air. Cells were transferred to the permissive temperature of 32°C for a minimum of 4 hours prior to experimentation unless otherwise indicated.
HL-60 cells were obtained from the European Collection of Cell Cultures (Salisbury, United Kingdom). KU812 and K562 cells were obtained from Deutsche Sammlung von Mikrorganismen und Zellkulturen (DSMZ GmbH, Braunschweig, Germany). All 3 of these cell lines were maintained in RPMI-1640 medium supplemented with 10% fetal calf serum (Gibco BRL, Paisley, United Kingdom).
LAMA84 imatinib-resistant and sensitive clones (LAMA84-r and LAMA84-s) were a gift from Prof Junia Melo (Imperial College, London, United Kingdom). LAMA84-r cells overexpress BCR-ABL and the multidrug resistance P-glycoprotein (Pgp).15 Imatinib-resistant cells were grown in RPMI-1640-10% fetal calf serum supplemented with 1 μM imatinib. Parental sensitive cell lines were maintained in parallel cultures without imatinib.
Primary CML samples and normal controls
Bone marrow aspirate samples were obtained from healthy donors and patients with CML at diagnosis or following treatment (as indicated). All human samples were obtained with ethical approval from the Research Ethics Committee Northern Ireland, and those involved gave their informed consent for participation in accordance with the Declaration of Helsinki. A complete hematological response (HR) is defined as white cell count lower than 10 × 109/L with a normal differential count and less than 5% circulating immature cells, platelet count of lower than 450 × 109/L, and the disappearance of all signs and symptoms related to CML activity. A major cytogenetic response (MCR) is defined as less than or equal to 35% Philadelphia-positive (Ph+) marrow metaphases and a complete cytogenetic response (CCR) as no detectable Ph+ marrow metaphases.16,17 Aspirates were collected in RPMI-1640 supplemented with 10% fetal calf serum and containing 100 IU preservative-free heparin (Leo Laboratories, Princes Risborough, United Kingdom). Mononuclear cells were separated over Ficoll-Hypaque (Pharmacia Biotech, Uppsala, Sweden) using standard procedures.18 CD34+ cells were prepared using the MACS Direct CD34 Progenitor Cell Isolation kit (Miltenyi Biotech, Bisley, United Kingdom).
DNA microarray analysis
All experiments were performed using Affymetrix Mu 6500 oligonucleotide arrays (Affymetrix, Santa Clara, CA), as described at http://www.affymetrix.com/products/arrays. ts-BCR-ABL FDCP-Mix and control cells were grown at the permissive temperature for 3 hours, 6 hours, 12 hours, and 24 hours to capture all events elicited by the oncogene. RNA was isolated by the guanidinium thiocyanate/acid phenol method using TRIzol reagent (Gibco BRL). Total RNA from each sample was used to prepare biotinylated target RNA, with minor modifications from the manufacturer's recommendations (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). RNA was purified using RNeasy (Qiagen, Crawley, United Kingdom) and mRNA extracted using Oligotex (Qiagen). Briefly, 10 μg of mRNA from each of the timepoints was pooled and used to generate first-strand cDNA by using a T7-linked oligo(dT) primer. After second-strand synthesis, in vitro transcription was performed with biotinylated UTP and CTP (Enzo Diagnostics, Farmingdale, NY), resulting in approximately 100-fold amplification of RNA. The target cDNA generated from each sample was processed per manufacturer's recommendation using an Affymetrix GeneChip Instrument System. The biotinylated transcripts were then hybridized to an Affymetrix Mu6500 array.
Controls were added to 10 μg fragmented cDNA before overnight hybridization. Arrays were then washed and stained with streptavidin-phycoerythrin, before being scanned on an Affymetrix GeneChip scanner. Ratios (3′/5′) for GAPDH and beta-actin were confirmed to be within acceptable limits (0.78-0.94), and BioB spike controls were found to be present on all chips, with BioC, BioD, and CreX also present in increasing intensity. When scaled to a target intensity of 100 (using Genespring array analysis software; Agilent Technologies, Palo Alto, CA), scaling factors for all arrays were within acceptable limits (0.53-1.23), as were background, Q values, and mean intensities.
Northern blot analysis
RNA was extracted from ts-BCR-ABL FDCP-Mix and control cells after 24 hours in culture using TRIzol reagent (Gibco BRL). Samples were prepared from 3 separate passages of cells to allow the confirmation of gene expression in triplicate. Northern blotting was performed using a 32P radiolabeled probe by standard techniques.19 The probe for CCN3 was synthesised by reverse transcription-polymerase chain reaction (RT-PCR) using RNA prepared from the control FDCP-Mix cell line. RNA was reverse transcribed (Moloney murine leukemia virus [M-MuLV] reverse transcriptase; Gibco BRL) and cDNA was amplified using the primers 5′AAGTCAAGTCTCTGCATCTCTGC3′ and 5′CTGAGCACCTGTTAAATTTCTCC3′ designed from the sequence X96585 (GenBank; www.ncbi.nlm.nih.gov). cDNA was denatured for 10 minutes at 94°C and then amplified more than 35 cycles using the following parameters: 1 minute at 94°C, 1 minute at 59°C, and 1 minute at 72°C. A final step of 72°C for 10 minutes was undertaken to ensure all transcripts were full length. PCR products were visualized after electrophoresis on agarose gels stained with ethidium bromide. Full-length product was excised and extracted (QIA Quick Gel Extraction kit; Qiagen) and used for Northern blotting following labeling with radioisotope (Amersham Pharmacia, Chalfont St Giles, United Kingdom; and NEB, Hitchin, United Kingdom).
Western blot analysis
Protein was extracted from cells by suspending in RIPA buffer (1 × phosphate-buffered saline [PBS], 1% Nonidet NP-40, 0.1% SDS) containing a cocktail of protease inhibitors (Complete Mini Cocktail; Roche Diagnostics, Lewes, United Kingdom) at a concentration of 107 cells/mL and lysing for 10 minutes on ice. Samples were sonicated for 10 seconds to ensure complete lysis, centrifuged at 10 000g at 4°C for 10 minutes, and then the supernatant was removed for analysis. Total protein content was determined by the Bradford protein method using the BCA protein assay kit (Pierce, Cramlington, United Kingdom). Protein (15 μg) was loaded onto a precast Bis-Tris polyacrylamide gel (10%) using the Novex mini gel system (Invitrogen, Paisley, United Kingdom) and subsequently transferred to a PVDF membrane. CCN3 expression was detected using an antibody raised against the C terminus of CCN3.20 Equivalent protein loading was controlled by monitoring actin expression using a panactin antibody (Cell Signaling Technology, Beverly, MA). Immunoblots were visualized by enhanced chemiluminescence (Supersignal; Pierce, Rockford, IL). Optical densitometry was performed using the Autochemi Sytem (Ultra-Violet Products, Cambridge, United Kingdom) and corrected for protein loading.
CCN3 protein is glycosylated and binds to heparin. Medium in which ts-BCR-ABL FDCP-Mix or control cells had been grown for 24 hours was harvested and enriched for CCN3 protein using heparin sepharose.21 Medium in which CD34+ cells had been grown for 72 hours in the presence or absence of imatinib (1 μM) was harvested and treated in a similar manner. Protein was also extracted from whole cells (2 × 106) from the same cultures and treated in an identical manner. These preparations are representative of secreted and cellular CCN3, respectively.
The phosphorylation status of CCN3 was examined by immunoprecipitation of CCN3 from whole-cell lysates and staining with an antibody to phosphotyrosine. Whole-cell lysates (200 μg) were allowed to couple with L59 CCN3 antibody22 at room temperature for 1 hour. Protein A/G (20 μL; Santa Cruz Biotechnology, Santa Cruz, CA) was added and incubated for 1 hour at room temperature; then, the protein-bound protein A/G complexes were pelleted. The complexes were washed 3 times in lysis buffer, once in PBS, and then resuspended in 50 μL gel-loading buffer. Phosphorylated protein was detected using the PY99 antibody (Santa Cruz Biotechnology).
Protein stability was examined by growing ts-BCR-ABL FDCP-Mix and control cells in the presence or absence of 1 μg/mL cycloheximide (Sigma, Poole, United Kingdom). Cells were harvested after 24, 48, or 72 hours, and protein was extracted.
Confocal microscopy
Cells were fixed in 50% (vol/vol) solution of 4% paraformaldehyde in PBS overnight at 4°C. Cells were then pelleted and resuspended in PBS, and 2 × 105 cells were allowed to attach to a silanized slide by evaporation. Slides were transferred to 0.5% (vol/vol) Triton X-100 (Sigma) in PBS (PBS-T) for 1 hour and then 5% (vol/vol) goat serum (Sigma) in PBS-T for 30 minutes to block antibody binding to nonspecific sites. Primary antibody (K19M, 1:250)20 in PBS-T was allowed to bind for 1 hour; unbound antibody was removed by 4 × 15-minute washes with PBS-T. Slides were immersed in 5% (vol/vol) goat serum for 30 minutes prior to treatment with secondary antibody, Alexa 488 (1:500; Molecular Probes, Eugene, OR) in PBS-T for 1 hour. Unbound antibody was removed by 4 × 15-minute washes in PBS-T and treated with 5% (vol/vol) goat serum in PBS-T for 30 minutes prior to staining with propidium iodide (1 ng/mL; Calbiochem, Nottingham, United Kingdom) for 30 minutes. Excess stain was removed by 2 × 15-minute washes in PBS before being mounted in Vectashield (Vector Laboratories, Burlingame, CA) and visualized by a Bio-Rad Microradiance confocal laser-scanning microscope equipped with a 40×/0.75 numeric aperture (NA) or 63×/1.4 NA oil immersion objective (Bio-Rad Laboratories, Hercules, CA).
Inhibition of BCR-ABL gene expression by siRNA
K562 cells were nucleofected following the manufacturer's instructions using the Cell Line Nucleofector Kit V, program T-16 (Amaxa GmbH, Cologne, Germany) and transfected with either the appropriate small interfering RNA (siRNA), a scrambled siRNA sequence, or a nonsilencing fluorescently labeled siRNA (Amaxa GmbH) to monitor transfection efficiency. A 21-base siRNA directed against the fusion sequence of BCR-ABL was used to specifically reduce gene expression (sense 5′GCAGAGUUCAAAAGCCCUUdTdT 3′, antisense 5′AAGGGCUUUUGAACUCUGCdTdT 3′),23 and a scrambled siRNA sequence was used as control (sense 5′UUGUACGGCAUCAGCGUUAdTdT 3′, antisense 5′UUACGCUGAUGCCGUACAAdTdT 3′). Real-time PCR was performed as previously described to determine the reduction in BCR-ABL mRNA.24,25
RQ-PCR
Real-time quantitative PCR (RQ-PCR) was performed with an ABI PRISM 7700 Sequence Detector, which exploits TaqMan probe-based chemistry (Applied Biosystems, Foster City, CA). The 5′ reporter used was FAM, and the 3′ quencher was TAMRA. Primers and probes for CCN3 were designed against GenBank published sequences in association with Primer Express (Applied Biosystems). Primer and probe sets for BCR-ABL were used as recommended by the Europe Against Cancer protocol.24,25 The amplification reactions (12.5 μL) contained 59 ng cDNA equivalents (or control), 1 × Taqman universal PCR master mix, final concentrations of 5 mM MgCl2, 0.2 mM dATP/dCTP/dGTP, 0.4 dUTP, 0.125 U AmpliTaq Gold, 2 μM primers (forward and reverse) and 200 nM TaqMan probe. Amplifications were performed following an initial 2-minute incubation at 50°C to allow UNG to destroy any contaminating RNA, followed by treatment at 95°C for 10 minutes to inactivate the UNG enzyme and activate the AmpliTaq Gold DNA polymerase. This was followed by 40 to 45 cycles of denaturing at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute. Data were collected and analyzed with Sequence Detector v1.6.3 software (Applied Biosystems). Relative quantitative (Q-PCR) data were calculated based on the δδCT method.26,27
Expression of CCN3 in K562 cells
K562 cells were nucleofected following the manufacturer's instructions using the Cell Line Nucleofector Kit V, program T-16 (Amaxa GmbH) with 5 μg of either vector (pCb6+; Invitrogen) or vector containing full-length CCN3 sequence. Transfection was performed in triplicate and the functional consequences of CCN3 expression were evaluated 24 hours after transfection (mean transfected cells, 97.9% ± 0.4%). Protein was extracted and Western blotting performed to confirm CCN3 expression. Clonogenicity was determined by plating cells (1 × 105) in triplicate in methyl cellulose (Stem Cell Technologies, Vancouver, BC, Canada) and counted after 7 days on an inverted microscope (Olympus model 214034; Olympus, Tokyo, Japan; magnification, ×40 [NA 0.65]). Cells (5 × 105) were also fixed in 70% ethanol for flow cytometric analysis of cell-cycle status. Fixed cells were resuspended in PBS and incubated in RNase A (100 μg/mL; Qiagen) and propidium iodide (40 μg/mL; Calbiochem) for 30 minutes at 37°C. Samples were then analyzed on a Coulter Epics Elite Cell Sorter (Beckman Coulter, High Wycombe, United Kingdom). Data were collected on the basis of peak signal versus integral signal to exclude doublets. Analysis was performed on 20 000 events using winMDI software (http://facs.scripps.edu/software.html), and data were generated and then ranked from a common gate in the sub-G0 area.
Effect of exogenous CCN3 on K562 cells
K562 cells (1 × 106 cells/well) were treated with recombinant CCN3 (100 ng/mL; Peprotech EC, London, United Kingdom) in vitro for 48 hours. Cells were then fixed in 70% ethanol for flow cytometric analysis of cell-cycle status as before.
Statistical analysis
Statistical analysis was performed on data from at least 3 experiments using SPSS analytical software10.0 (http://www.spss.com). Experiments that involved the FDCP-Mix cell line model (kinase active and control cells) were run in parallel and data were analyzed using the paired samples t test. Data collected from patient samples were analyzed using the nonparametric independent samples method or Mann-Whitney test.
Results
CCN3 is down-regulated as a consequence of BCR-ABL kinase activity
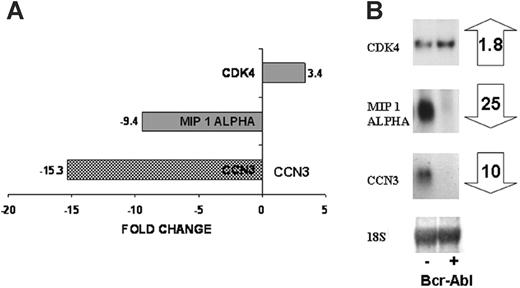
Our aim was to identify a candidate gene whose expression may be targeted by the BCR-ABL protein tyrosine kinase at an early stage in transformation. We therefore performed microarray analysis to identify genes whose expression is altered as a consequence of 3 to 24 hours of expression of BCR-ABL PTK activity in multipotent FDCP-Mix cells. These cells show a suppression of apoptosis over this period, but no autonomous proliferation, and no IL-3 production.12 The DNA microarray analysis identified differential expression of 300 genes as result of BCR-ABL activity and included MIP1A (CCL3; down-regulated 9.4-fold) and CDK4 (up-regulated 3.4-fold), which have previously been associated with leukemogenesis.28,29 Expression of CCN3 was down-regulated by 15-fold as a consequence of BCR-ABL kinase activity by DNA microarray analysis (Figure 1A). Northern blot analysis confirmed these observations, demonstrating 25-fold down-regulation of MIP1A, 1.8-fold up-regulation of CDK4, and 10-fold down-regulation of CCN3 (Figure 1B).
The expression of CCN3 protein is decreased as a consequence of BCR-ABL activity
We next considered whether CCN3 protein expression was also decreased as a consequence of BCR-ABL kinase activity. Western blotting of whole-cell lysates demonstrated the presence of a doublet corresponding to p50 CCN3, consistent with expression of the full-length CCN3 forms (Figure 2A). CCN3 levels were significantly reduced in the BCR-ABL kinase active cells compared with control cells, with a 55% decrease in expression at 24 hours (mean percentage of integrated optical densitometry units [IOD] 47.6 ± 18 and 102.8 ± 7.3, respectively; n = 3, P = .026).
Differential gene expression as a result of BCR-ABL kinase activity. DNA microarray analysis was performed on FDCP-Mix control cells and cells expressing BCR-ABL kinase activity. The microarray identified genes differentially expressed as a result of the BCR-ABL kinase activity, a small subset including CDK4, MIP 1α, CCN3 is shown (A). Northern blotting was used to confirm expression of these genes in control cells (-) and BCR-ABL kinase-active cells (+) after 24 hours in culture. Fold changes in gene expression are shown (B).
Differential gene expression as a result of BCR-ABL kinase activity. DNA microarray analysis was performed on FDCP-Mix control cells and cells expressing BCR-ABL kinase activity. The microarray identified genes differentially expressed as a result of the BCR-ABL kinase activity, a small subset including CDK4, MIP 1α, CCN3 is shown (A). Northern blotting was used to confirm expression of these genes in control cells (-) and BCR-ABL kinase-active cells (+) after 24 hours in culture. Fold changes in gene expression are shown (B).
We also used confocal microscopy to examine CCN3 expression (Figure 2B). CCN3 was clearly detected in most control cells grown at 39°C and 32°C (Figure 2Bi-ii) and in ts-BCR-ABL FDCP-Mix cells grown at the restrictive temperature (Figure 2Biii). CCN3 expression was significantly reduced in ts-BCR-ABL FDCP-Mix cells grown at the permissive temperature (kinase active; Figure 2Biv). Most cells were negative, although a small number of positively staining vesicles could still be observed in occasional cells (mean fluorescent intensity, 2.6 ± 0.65 × 105; n = 5). The decreased CCN3 fluorescence in the kinase active cells was significantly reduced compared with the control or kinase inactive cells (mean fluorescence intensity, 4.6 ± 0.87 × 105; P = .043). The fluorescence intensity in both the BCR-ABL kinase-active or -inactive cells was strongest at the cell periphery, where it was concentrated into globular or vesicular-like structures resembling those for exportation from the cell, a phenomenon known to occur with CCN3.
Because there was a suggestion that export vesicles contained CCN3, we prepared the medium in which these cells had been grown from kinase-active or control FDCP-Mix cells and subjected these to Western blotting (Figure 2C). This showed that the level of secreted CCN3 was increased in BCR-ABL kinase-active cells (BCR-ABL, 32°C) compared with control cells grown at 39°C and 32°C and in ts-BCR-ABL FDCP-Mix cells grown at the restrictive temperature (BCR-ABL, 39°C). The increased secretion of CCN3 in kinase-active cells compared with control cells (mean percentage IOD, 163.8 ± 20.8 and 116.3 ± 1.0, respectively; n = 3, P = .034) suggests that regulation of CCN3 expression within cells may be regulated by posttranslational processing as well as transcription. Critically, protein stability does not appear to play a part in the decreased cellular protein levels observed, as we found no evidence of protein degradation over a period of 72 hours.
We next examined the phosphorylation status of intracellular CCN3 in control and BCR-ABL active FDCP-Mix by immunoprecipitating CCN3 and Western blotting with antiphosphotyrosine antibodies (Figure 2D). CCN3 was strongly phosphorylated in control cells and weakly phosphorylated in BCR-ABL kinase active cells (Figure 2Di). The reduction of phosphorylation detected in kinase active cells is probably due to the overall decrease in CCN3 expression in BCR-ABL kinase active cells (Figure 2Dii). The cell lysates were tested for actin expression (Figure 2Diii) to ensure equal protein loading.
CCN3 is down-regulated in human CML cell lines
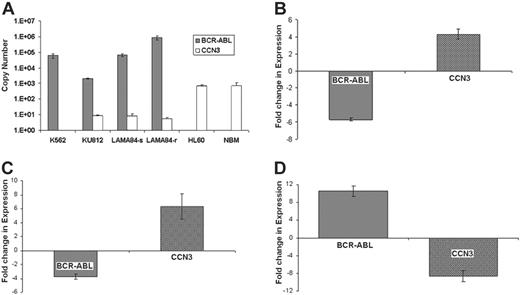
In order to confirm the effect of BCR-ABL expression on CCN3 in human cells, we used real-time PCR, which showed strong expression of BCR-ABL and weak or absent expression of CCN3 in CML cell lines (Figure 3A). In K562, KU812, and LAMA84-s cell lines, BCR-ABL expression was high, while CCN3 expression was low. CCN3 expression in the BCR-ABL-negative cell line, HL-60, and normal bone marrow (NBM) was high (n = 3). Treatment of the CML cell lines with imatinib or siRNA against BCR-ABL significantly reduced BCR-ABL and increased CCN3 expression. For example, K562 cells treated with 1 μM imatinib for 96 hours showed a 5.9-fold decrease in BCR-ABL expression and a 4.2-fold increase in CCN3 expression, as shown in Figure 3B (mean Ct change, 2.5 ± 0.1 and 2.1 ± 0.2 for BCR-ABL and CCN3, respectively; n = 3, P = .001). Treatment of K562 cells with siRNA directed against BCR-ABL resulted in a 3.7-fold decrease in BCR-ABL and a 6.1-fold increase in CCN3, as shown in Figure 3C (mean Ct change, 1.9 ± 0.2 and 2.6 ± 0.5 for BCR-ABL and CCN3, respectively; n = 3, P = .001). The difference in BCR-ABL and CCN3 expression in imatinib-resistant cells, LAMA84-r, compared with the sensitive LAMA84-s cells is shown (Figure 3D). Cumulative resistance to imatinib in vitro due to a 10-fold increase in BCR-ABL expression showed a corresponding 9-fold decrease in CCN3 expression (mean Ct change, 3.4 ± 0.1 and 3.1 ± 0.3 for BCR-ABL and CCN3, respectively; n = 3, P = .001). Furthermore, in order to analyze whether there was a relationship between BCR-ABL expression and the secretion of CCN3 in a human cell line (K562), we used siRNA to decrease expression of BCR-ABL and measured the extracellular CCN3 levels. We found that there was a significant decrease of 33% ± 5% in secreted levels of CCN3 protein resulting from siRNA expression (P = .038, n = 3). This demonstrates that down-regulation of CCN3 is a direct consequence of BCR-ABL activation in human cells as well as murine cells.
CCN3 protein expression is down-regulated as a result of BCR-ABL kinase activity. (A) FDCP-Mix control and ts-BCR-ABL cells were grown at the restrictive temperature (39°C) and permissive temperature for BCR-ABL activity (32°C) for 24 hours. Protein lysates were extracted and probed for CCN3 and actin expression; levels of expression corrected for protein loading are shown as integrated optical densitometry units (IODs). *P = .026; n = 3. (B) Confocal microscopy was also used to identify CCN3 protein (green) in FDCP-Mix control cells at 39°C (i) and 32°C (ii) and ts-BCR-ABL cells at 39°C (iii) and 32°C (iv); propidium iodide nuclear staining (red) is also shown. Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 160). (C) Medium in which FDCP-Mix control and ts BCR-ABL cells were grown for 24 hours was collected and probed for secreted CCN3; levels of secreted CCN3 are shown as IODs for comparison. *P = .014; n = 3. (D) Lysates from control cells and BCR-ABL kinase-active cells grown at the permissive temperature (32°C) were immunoprecipitated using CCN3 antibody and probed for (i) phosphotyrosine, (ii) CCN3, and (iii) actin expression. Levels of CCN3 and phosphorylated CCN3 detected by Western blot analysis in FDCP-Mix control and BCR-ABL kinase-active cells were compared using densitometry as shown.
CCN3 protein expression is down-regulated as a result of BCR-ABL kinase activity. (A) FDCP-Mix control and ts-BCR-ABL cells were grown at the restrictive temperature (39°C) and permissive temperature for BCR-ABL activity (32°C) for 24 hours. Protein lysates were extracted and probed for CCN3 and actin expression; levels of expression corrected for protein loading are shown as integrated optical densitometry units (IODs). *P = .026; n = 3. (B) Confocal microscopy was also used to identify CCN3 protein (green) in FDCP-Mix control cells at 39°C (i) and 32°C (ii) and ts-BCR-ABL cells at 39°C (iii) and 32°C (iv); propidium iodide nuclear staining (red) is also shown. Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 160). (C) Medium in which FDCP-Mix control and ts BCR-ABL cells were grown for 24 hours was collected and probed for secreted CCN3; levels of secreted CCN3 are shown as IODs for comparison. *P = .014; n = 3. (D) Lysates from control cells and BCR-ABL kinase-active cells grown at the permissive temperature (32°C) were immunoprecipitated using CCN3 antibody and probed for (i) phosphotyrosine, (ii) CCN3, and (iii) actin expression. Levels of CCN3 and phosphorylated CCN3 detected by Western blot analysis in FDCP-Mix control and BCR-ABL kinase-active cells were compared using densitometry as shown.
CCN3 expression is reduced in human BCR-ABL+ cell lines and can be reversed in K562 cells treated with imatinib or siRNA to BCR-ABL. (A) RNA was extracted from human CML cell lines (K562, KU812, LAMA), BCR-ABL- cells (HL60), and NBM, and subjected to real-time PCR to identify the level of BCR-ABL ( ) and CCN3 (□) expression. Level of gene expression is shown as Ct values (n = 3). (B) K562 cells were treated with imatinib (1 μM) for 96 hours, and gene expression was analyzed by real-time PCR. Fold changes in BCR-ABL (
) and CCN3 (□) expression. Level of gene expression is shown as Ct values (n = 3). (B) K562 cells were treated with imatinib (1 μM) for 96 hours, and gene expression was analyzed by real-time PCR. Fold changes in BCR-ABL ( ) and CCN3 (□) expression as a result of imatinib treatment are shown (n = 3; P = .001). (C) K562 cells were treated with siRNA directed to BCR-ABL (0.5 μg/106 cells) for 24 hours. Real-time PCR was used to identify changes in BCR-ABL (
) and CCN3 (□) expression as a result of imatinib treatment are shown (n = 3; P = .001). (C) K562 cells were treated with siRNA directed to BCR-ABL (0.5 μg/106 cells) for 24 hours. Real-time PCR was used to identify changes in BCR-ABL ( ) and CCN3 (□) transcripts, which are shown as fold change in expression (n = 3; P = .001). (D) Levels of BCR-ABL (
) and CCN3 (□) transcripts, which are shown as fold change in expression (n = 3; P = .001). (D) Levels of BCR-ABL ( ) and CCN3 (□) transcripts were compared using real-time PCR in human CML cells that are sensitive (LAMA84-s) and resistant (LAMA84-r) to imatinib. Alterations in BCR-ABL and CCN3 expression as a result of imatinib resistance are shown as fold changes (n = 3; P = .001).
) and CCN3 (□) transcripts were compared using real-time PCR in human CML cells that are sensitive (LAMA84-s) and resistant (LAMA84-r) to imatinib. Alterations in BCR-ABL and CCN3 expression as a result of imatinib resistance are shown as fold changes (n = 3; P = .001).
CCN3 expression is reduced in human BCR-ABL+ cell lines and can be reversed in K562 cells treated with imatinib or siRNA to BCR-ABL. (A) RNA was extracted from human CML cell lines (K562, KU812, LAMA), BCR-ABL- cells (HL60), and NBM, and subjected to real-time PCR to identify the level of BCR-ABL ( ) and CCN3 (□) expression. Level of gene expression is shown as Ct values (n = 3). (B) K562 cells were treated with imatinib (1 μM) for 96 hours, and gene expression was analyzed by real-time PCR. Fold changes in BCR-ABL (
) and CCN3 (□) expression. Level of gene expression is shown as Ct values (n = 3). (B) K562 cells were treated with imatinib (1 μM) for 96 hours, and gene expression was analyzed by real-time PCR. Fold changes in BCR-ABL ( ) and CCN3 (□) expression as a result of imatinib treatment are shown (n = 3; P = .001). (C) K562 cells were treated with siRNA directed to BCR-ABL (0.5 μg/106 cells) for 24 hours. Real-time PCR was used to identify changes in BCR-ABL (
) and CCN3 (□) expression as a result of imatinib treatment are shown (n = 3; P = .001). (C) K562 cells were treated with siRNA directed to BCR-ABL (0.5 μg/106 cells) for 24 hours. Real-time PCR was used to identify changes in BCR-ABL ( ) and CCN3 (□) transcripts, which are shown as fold change in expression (n = 3; P = .001). (D) Levels of BCR-ABL (
) and CCN3 (□) transcripts, which are shown as fold change in expression (n = 3; P = .001). (D) Levels of BCR-ABL ( ) and CCN3 (□) transcripts were compared using real-time PCR in human CML cells that are sensitive (LAMA84-s) and resistant (LAMA84-r) to imatinib. Alterations in BCR-ABL and CCN3 expression as a result of imatinib resistance are shown as fold changes (n = 3; P = .001).
) and CCN3 (□) transcripts were compared using real-time PCR in human CML cells that are sensitive (LAMA84-s) and resistant (LAMA84-r) to imatinib. Alterations in BCR-ABL and CCN3 expression as a result of imatinib resistance are shown as fold changes (n = 3; P = .001).
The expression of CCN3 protein is decreased in primary human CML cells
We next determined whether CCN3 expression was altered in primary CML cells. Real-time PCR was used to measure CCN3 and BCR-ABL expression in 3 patients with CML at diagnosis and following treatment with imatinib; 1 patient had a hematopoietic response, and 2 patients had a complete cytogenetic response. In each of the patients, a fall in BCR-ABL expression was associated with a reciprocal rise in CCN3 (Figure 4A). Since CCN3 expression is highest in BCR-ABL-negative cells, it reflects hematologic response. There does not appear to be a direct correlation between the decrease in BCR-ABL expression and increased CCN3 transcripts, possibly reflecting the heterogeneity of the cell types present.
CCN3 protein expression was then examined using Western blotting of whole-cell lysates of normal bone marrow and bone marrow from 3 patients with CML taken at diagnosis and following response to therapy. Expression of full-length CCN3 was detected in all 3 normal bone marrow samples. A lower-molecular-weight form of CCN3 (45 kDa) was weakly detectable in 2 of the patients at diagnosis, returning to a normal expression pattern on response to treatment (Figure 4B). Optical densitometry of the blot from Figure 4B shows patients responding to treatment have levels of CCN3 comparable with NBM (Figure 4D). CD34+ cells taken from 3 patients with CML at diagnosis were treated with imatinib for 72 hours in vitro, and a significant increase in both cellular and secreted CCN3 expression was demonstrated in all 3 samples (Figure 4C). Optical densitometry of the blot from Figure 4C shows that treating CD34+ CML cells with imatinib increases CCN3 protein expression (Figure 4D).
CCN3 expression was also examined in primary human cells by confocal microscopy. Mononuclear cells from a patient with CML at diagnosis showed only occasional weakly staining cells (Figure 4Ei), but on entering complete cytogenetic remission, most cells stained positively for CCN3 (Figure 4Eii). Similarly, CD34+ cells from a patient with CML at diagnosis (Figure 4Fi) showed a modest amount of staining compared with CD34+ cells from a normal marrow (Figure 4Fii). CD34+ cells from a patient with thrombocythemia (Figure 4Fiii) exhibited a normal expression pattern for CCN3 consistent with the changes in CCN3 being dependent on BCR-ABL activity rather than a reflection of myeloproliferation.
Increasing CCN3 expression in BCR-ABL+ cells
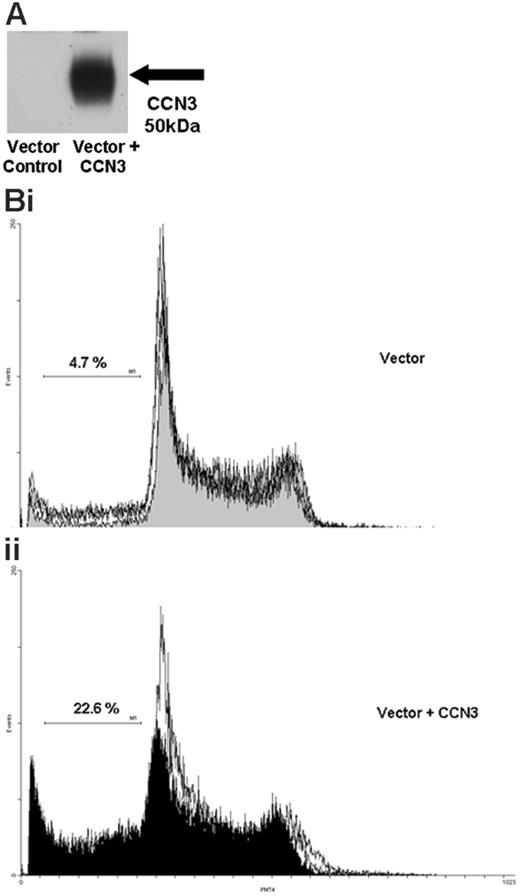
To further investigate the role of CCN3 expression we transfected human K562 cells with a full-length CCN3 construct. Western blot analysis confirmed a strong increase in CCN3 protein in cells transfected with the vector containing the CCN3 construct compared with cells transfected with vector alone (Figure 5A). Flow cytometry was used to perform cell-cycle analysis. K562 cells transfected with vector containing the full-length CCN3 construct (Figure 5Bii) showed significantly more cells accumulating in the sub-G0 area of the cell cycle than cells that had been transfected with vector alone (Figure 5Bi; mean sub-G0 21.8% ± 0.7% and 9.9% ± 4.6%, respectively; P = .028, n = 3). The ability of transfected cells to form colonies on methyl cellulose was also assessed. Colony formation of K562 cells transfected with vector containing the CCN3 construct was significantly reduced by one third compared with cells transfected with vector alone (P = .027, n = 3).
Next we assessed whether BCR-ABL+ cells could respond to exogenous CCN3 by incubating K562 cells with recombinant CCN3 for 48 hours. Flow cytometry demonstrated a significant increase in the sub-G0 peak when cells were treated with 100 ng/mL CCN3 (increased from 9.3% ± 3.9% to 23.7% ± 6.9%; P = .014).
Discussion
CCN3 (Nov) is a member of the CCN family of regulatory proteins.30 This family was named using the initial letter of the first 3 family members: CYR61 (cysteine rich), CTGF (connective tissue growth factor), and NOV (nephroblastoma overexpressed). With the other family members, ELM1, rCOP1, and WISPs, all CCN proteins share significant structural homology and play key regulatory roles in fundamental biological processes. CCN3 is composed of 5 distinct structural modules. The signal peptide is responsible for secretion of the full-length protein either into the extracellular matrix or to the cell membrane. The remaining 4 modules show significant homology with the other CCN family members and have motifs associated with insulin-like growth factor-binding protein (IGFBP), von Willebrand type C repeat (VWC), thrombospondin type 1 repeat (TSP1), and a cysteine knot (CT) involved in dimerization of several matrix proteins and growth factors.
CCN3 mRNA and protein is down-regulated in primary cells from patients with CML, and expression increases upon entering remission. (A) RNA was extracted from bone marrow samples taken from patients with CML at diagnosis and following treatment (patient 1 hematopoietic response, HR; patients 2 and 3 complete cytogenetic response, CCR), and real-time PCR was performed to determine CCN3 and BCR-ABL mRNA levels. Results are presented as the fold change in BCR-ABL ( ) and CCN3 (□) expression following treatment (mean of 3 determinations ± SD). (B) Protein lysates extracted from CML patient bone marrow samples taken at diagnosis (“D”) and following response to treatment (“R”) were subjected to Western blot analysis to identify CCN3 protein expression. Three normal bone marrow samples are included for comparison. Patient 1, HR; patients 2 and 3, CCR. (C) CD34+ cells were extracted from the bone marrow of 3 patients with CML at diagnosis and treated for 72 hours in vitro with imatinib (1 μM). Western blot analysis of the medium in which the cells were grown and protein cell lysates was performed to identify CCN3 protein. Optical densitometry was performed on the Western blot. The mean signal from NBM was assigned 100% and measurements were expressed as a percentage compared to the signal from NBM. (D) Optical densitometry was performed on the Western blot from panel C. CCN3 expression in CD34+ cells grown without imatinib were assigned 100%. CCN3 expression in CD34+ cells treated with imatinib for 72 hours was expressed as a percentage compared with cells that were not treated. (E) Confocal microscopy was used to detect CCN3 expression (green fluorescence) in mononuclear cells from a patient with CML at diagnosis (i) and following a complete cytogenetic response (ii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 80). Propidium iodide was used to stain the nuclei. (F) Confocal microscopy was used to detect CCN3 protein (green fluorescence) in CD34+ cells from a patient with CML at diagnosis (i), normal bone marrow (ii), and a patient with thrombocythemia (iii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 110). Propidium iodide was used to stain the nuclei.
) and CCN3 (□) expression following treatment (mean of 3 determinations ± SD). (B) Protein lysates extracted from CML patient bone marrow samples taken at diagnosis (“D”) and following response to treatment (“R”) were subjected to Western blot analysis to identify CCN3 protein expression. Three normal bone marrow samples are included for comparison. Patient 1, HR; patients 2 and 3, CCR. (C) CD34+ cells were extracted from the bone marrow of 3 patients with CML at diagnosis and treated for 72 hours in vitro with imatinib (1 μM). Western blot analysis of the medium in which the cells were grown and protein cell lysates was performed to identify CCN3 protein. Optical densitometry was performed on the Western blot. The mean signal from NBM was assigned 100% and measurements were expressed as a percentage compared to the signal from NBM. (D) Optical densitometry was performed on the Western blot from panel C. CCN3 expression in CD34+ cells grown without imatinib were assigned 100%. CCN3 expression in CD34+ cells treated with imatinib for 72 hours was expressed as a percentage compared with cells that were not treated. (E) Confocal microscopy was used to detect CCN3 expression (green fluorescence) in mononuclear cells from a patient with CML at diagnosis (i) and following a complete cytogenetic response (ii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 80). Propidium iodide was used to stain the nuclei. (F) Confocal microscopy was used to detect CCN3 protein (green fluorescence) in CD34+ cells from a patient with CML at diagnosis (i), normal bone marrow (ii), and a patient with thrombocythemia (iii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 110). Propidium iodide was used to stain the nuclei.
CCN3 mRNA and protein is down-regulated in primary cells from patients with CML, and expression increases upon entering remission. (A) RNA was extracted from bone marrow samples taken from patients with CML at diagnosis and following treatment (patient 1 hematopoietic response, HR; patients 2 and 3 complete cytogenetic response, CCR), and real-time PCR was performed to determine CCN3 and BCR-ABL mRNA levels. Results are presented as the fold change in BCR-ABL ( ) and CCN3 (□) expression following treatment (mean of 3 determinations ± SD). (B) Protein lysates extracted from CML patient bone marrow samples taken at diagnosis (“D”) and following response to treatment (“R”) were subjected to Western blot analysis to identify CCN3 protein expression. Three normal bone marrow samples are included for comparison. Patient 1, HR; patients 2 and 3, CCR. (C) CD34+ cells were extracted from the bone marrow of 3 patients with CML at diagnosis and treated for 72 hours in vitro with imatinib (1 μM). Western blot analysis of the medium in which the cells were grown and protein cell lysates was performed to identify CCN3 protein. Optical densitometry was performed on the Western blot. The mean signal from NBM was assigned 100% and measurements were expressed as a percentage compared to the signal from NBM. (D) Optical densitometry was performed on the Western blot from panel C. CCN3 expression in CD34+ cells grown without imatinib were assigned 100%. CCN3 expression in CD34+ cells treated with imatinib for 72 hours was expressed as a percentage compared with cells that were not treated. (E) Confocal microscopy was used to detect CCN3 expression (green fluorescence) in mononuclear cells from a patient with CML at diagnosis (i) and following a complete cytogenetic response (ii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 80). Propidium iodide was used to stain the nuclei. (F) Confocal microscopy was used to detect CCN3 protein (green fluorescence) in CD34+ cells from a patient with CML at diagnosis (i), normal bone marrow (ii), and a patient with thrombocythemia (iii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 110). Propidium iodide was used to stain the nuclei.
) and CCN3 (□) expression following treatment (mean of 3 determinations ± SD). (B) Protein lysates extracted from CML patient bone marrow samples taken at diagnosis (“D”) and following response to treatment (“R”) were subjected to Western blot analysis to identify CCN3 protein expression. Three normal bone marrow samples are included for comparison. Patient 1, HR; patients 2 and 3, CCR. (C) CD34+ cells were extracted from the bone marrow of 3 patients with CML at diagnosis and treated for 72 hours in vitro with imatinib (1 μM). Western blot analysis of the medium in which the cells were grown and protein cell lysates was performed to identify CCN3 protein. Optical densitometry was performed on the Western blot. The mean signal from NBM was assigned 100% and measurements were expressed as a percentage compared to the signal from NBM. (D) Optical densitometry was performed on the Western blot from panel C. CCN3 expression in CD34+ cells grown without imatinib were assigned 100%. CCN3 expression in CD34+ cells treated with imatinib for 72 hours was expressed as a percentage compared with cells that were not treated. (E) Confocal microscopy was used to detect CCN3 expression (green fluorescence) in mononuclear cells from a patient with CML at diagnosis (i) and following a complete cytogenetic response (ii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 80). Propidium iodide was used to stain the nuclei. (F) Confocal microscopy was used to detect CCN3 protein (green fluorescence) in CD34+ cells from a patient with CML at diagnosis (i), normal bone marrow (ii), and a patient with thrombocythemia (iii). Images were collected using a Bio-Rad Microradiance confocal laser scanning microscope with an oil immersion lens (magnification, × 110). Propidium iodide was used to stain the nuclei.
There have now been several reports of the existence of CCN3 isoforms, lacking 1 or 2 of the basic modules.31 High concentrations of an amino-truncated form of the molecule missing the IGFBP and VWC modules have been detected in the nucleus of malignant cells. In addition, expression of the truncated form has the potential to translocate to the nucleus and interact with the RPB7 subunit of RNA polymerase II, suggesting transcriptional activity.32 Expression of CCN3 is abnormal in tumor cells and CCN3 may be a proto-oncogene, the expression of which is disrupted in malignancy.33,34 Expression of full-length CCN3 has previously been associated with differentiation during chondrogenesis, development of the central nervous system, and muscular differentiation.21,35,36 The “full-length” form is inhibitory to fibroblastic cell growth, and has been associated with cell quiescence and increased adhesion, therefore suggesting negative growth regulatory properties.37,38 In contrast, expression of the N-terminal-truncated form promotes cell transformation and exhibits oncogenic properties.33 Matricellular regulatory proteins are involved in internal and external cell signaling.39 As such, the CCN proteins play a key role in controlling cell proliferation and differentiation, and aberrant expression has been demonstrated in many solid tumors.34 We have shown that CCN3 is down-regulated as a consequence of BCR-ABL expression in both murine and human hematopoietic progenitor cells. CCN3 has not been associated previously with hematologic malignancy.
Northern blotting clearly demonstrated down-regulation of CCN3 expression in kinase-active FDCP-Mix cells, and there was no evidence of alternative gene splicing. Real-time PCR confirmed that high expression of BCR-ABL correlated with low expression of CCN3 in both human CML cell lines and in primary cells from patients with CML at diagnosis. In vitro treatment of CML cell lines with imatinib or siRNA directed against BCR-ABL significantly reduced BCR-ABL expression with a concomitant increase in CCN3 expression. Similarly, patients responding to treatment with imatinib showed a decrease in BCR-ABL and increase in CCN3 expression. LAMA84-r cells, which are resistant to imatinib due to increased expression of BCR-ABL had a significantly lower level of CCN3 than their drug-sensitive counterparts. Furthermore, expression of CCN3 in the BCR-ABL+ cell line, K562, resulted in inhibition of cell proliferation and decreased clonogenicity, and exogenous CCN3 also decreased the rate of cell proliferation and increased cell death. We are now in the process of analyzing the effects of CCN3 on normal and BCR-ABL+ cell populations. These experiments demonstrate that CCN3 expression is directly suppressed by BCR-ABL kinase, and that expression of CCN3 in BCR-ABL+ cells can partially reverse the malignant phenotype. Loss of this protein may therefore be associated with the molecular pathogenesis of CML.
Increasing CCN3 expression in Bcr-Abl+ cells causes accumulation in the sub-G0 area of the cell cycle. (A) Protein lysates were extracted from K562 cells 24 hours after transfection with vector alone or vector containing the full-length CCN3 construct. Western blotting was performed to detect CCN3 protein. (B) K562 cells transfected with vector alone or vector containing the full-length CCN3 construct were harvested at 24 hours, fixed, and stained with propidium iodide. Cell-cycle profiles were generated and compared for cells containing the vector alone (i) and vector with CCN3 (ii).
Increasing CCN3 expression in Bcr-Abl+ cells causes accumulation in the sub-G0 area of the cell cycle. (A) Protein lysates were extracted from K562 cells 24 hours after transfection with vector alone or vector containing the full-length CCN3 construct. Western blotting was performed to detect CCN3 protein. (B) K562 cells transfected with vector alone or vector containing the full-length CCN3 construct were harvested at 24 hours, fixed, and stained with propidium iodide. Cell-cycle profiles were generated and compared for cells containing the vector alone (i) and vector with CCN3 (ii).
CCN3 contains a signal peptide responsible for secretion of the protein from the cell and 4 functional domains. It is not yet clear whether CCN3 acts mainly at the site of production or if it is transported to a remote site as yet undefined.34 Our observations (Figure 2C) demonstrate that BCR-ABL-transformed cells show a decrease in cellular CCN3 protein and an associated increase in the protein secreted by the cell. Crucially, we demonstrated that protein stability does not appear to play a part in the decreased cellular protein levels observed. BCR-ABL PTK clearly affects posttranslational processing of CCN3 protein as well as gene transcription. Potentially, CCN3 phosphorylation status is regulated by BCR-ABL to potentiate secretory rates of this protein. We are currently mapping CCN3 phosphorylation sites to investigate this further. CCN3 potentially plays a role in both external and internal cell-signaling pathways.
It has been shown that p210 BCR-ABL expression in cell lines and bone marrow progenitor cells leads to increased binding to stromal cells and fibronectin, although the mechanism by which this occurs is poorly understood.40-45 Several CCN family proteins support cell adhesion and stimulate adhesive signaling.31,39,46,47 CCN1 and CCN2 are direct ligands of multiple integrins, which mediate many of their functions.46-50 Purified recombinant CCN3 interacts with multiple integrin receptors and binds directly to integrins αvβ3 and α5β1.51 Studies have also shown that the C-terminal of CCN3 interacts with fibulin 1C, which is consistent with a role in cell adhesion.52 Increased secretion and decreased cellular expression of CCN3 in BCR-ABL+ cells may be linked with the changes in cellular adhesion properties associated with CML.
CCN3 has been shown to play a key role in mesenchymal cell differentiation. CCN3 associates with the Notch1 extracellular domain and inhibits myoblast differentiation via the Notch signaling pathway.53 The Notch pathway has also been implicated in regulating myeloid differentiation.54,55 In contrast to mesenchymal cells, activation of the Notch1 transmembrane receptor induces myeloid differentiation of multipotent hemopoietic progenitor cells. Loss of CCN3 expression would be consistent with the accumulation of immature myeloid progenitors characteristic of CML.
Our results strongly suggest that BCR-ABL PTK regulates transcription and posttranslational processing of CCN3. CCN3 has been shown to act as a tumor suppressor gene and to play a role in cell adhesion and differentiation in other tumor types. Down-regulation of CCN3 may contribute to the complex phenotype observed in CML.
Prepublished online as Blood First Edition Paper, May 2, 2006; DOI 10.1182/blood-2006-04-016113.
Supported by the Leukaemia Research Fund, Northern Ireland Leukaemia Research Fund, Elimination of Leukaemia Fund, and Ministere de l'Education Nationale, de la Recherche et de la Technologie.
L.M. carried out the majority of the practical work, data analysis, and interpretation, and contributed to preparation of the manuscript; S.P. played a major part in experimental design and execution; N.P. and B.P. provided essential biological material and made a major contribution to the interpretation of data; A.P. and A.D.W. provided essential biological reagents and contributed to experimental design and writing of the manuscript; A.E.I. was responsible for the overall conception and design of the project, interpretation of data, and writing most of the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank the consultant hematologists at Belfast City Hospital; Ken Arthur, Tom Gardiner, Alexandra Kwasniewska, and Wanhua Lu of Queen's University Belfast; and Vincent Martinez of Universite de Paris for technical assistance; and Prof Junia Melo of Imperial College London for provision of the LAMA84-s and LAMA84-r cell lines.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal