Imatinib, a potent tyrosine kinase inhibitor, is effluxed from cells by the breast cancer resistance protein (BCRP/ABCG2), yet published studies to date fail to demonstrate resistance to imatinib cytotoxicity in BCRP-overexpressing cells in vitro. We investigated cellular resistance to imatinib in BCR-ABL–expressing cells transduced and selected to overexpress BCRP (K562/BCRP-MX10). These cells exhibited a 2- to 3-fold increase in resistance to imatinib (P < .05) and a 7- to 12-fold increase in resistance to mitoxantrone, a known BCRP substrate. Resistance to imatinib was completely abolished by the specific BCRP inhibitor fumitremorgin C. Studies of the mechanism of the diminished resistance to imatinib compared with mitoxantrone revealed that imatinib decreased the expression of BCRP in K562/BCRP-MX10 cells without affecting mRNA levels. BCRP levels in cells that do not express BCR-ABL were not affected by imatinib. Loss of BCRP expression was accompanied by imatinib-induced reduction of phosphorylated Akt in the BCRP-expressing K562 cells. The phosphoinositol-3 kinase (PI3K) inhibitor LY294002 also decreased BCRP levels in K562/BCRP-MX10 cells. These studies show that BCRP causes measurable imatinib resistance, but this effect is attenuated by imatinib-mediated inhibition of BCR-ABL, which in turn downregulates overall BCRP levels posttranscriptionally via the PI3K-Akt pathway.
Introduction
Chronic myelogenous leukemia (CML) is a clonal disorder that arises by the translocation of chromosomes 9 and 22 in an early hematopoietic stem cell (HSC) to produce the Philadelphia chromosome (Ph).1 This translocation results in production of the chimeric BCR-ABL protein, a constitutively active tyrosine kinase that drives hyperproliferation of stem and progenitor cells and the consequent panmyelosis associated with CML in the chronic phase of this disease.2 Imatinib mesylate (STI571; Gleevec) is a potent inhibitor of the BCR-ABL tyrosine kinase and is cytotoxic to cells that are dependent on BCR-ABL kinase activity for growth and survival. Accordingly, imatinib has proven to be highly effective in the treatment of the chronic phase of CML, with high rates of complete remission observed.3 Unfortunately, molecular complete remissions evidenced by negative polymerase chain reactions (PCRs) for BCR-ABL transcripts occur in only 35% of chronic-phase cases treated with imatinib,4 leading many to suspect that imatinib may not cure the majority of patients with chronic-phase CML.5
Primitive BCR-ABL–expressing stem cells have been described in chronic-phase CML that are resistant to the cytotoxic effects of imatinib,6 leading some to postulate that this resistant, self-renewing population could account for the failure of imatinib to cure the disease. Cellular resistance to imatinib can arise by a variety of mechanisms, including mutations in the BCR-ABL kinase7 and efflux by the multidrug resistance transporter P-glycoprotein (P-gp).8 Recently, imatinib was found to be effluxed by another multidrug resistance transporter, the breast cancer resistance protein (BCRP, ABCG2).9,10 Since BCRP is expressed in primitive normal HSCs,11 it is reasonable to posit the existence of counterpart BCR-ABL–expressing HSCs in CML that express BCRP. These cells may contribute to the inherent imatinib resistance of these primitive CML cells; hence, it is crucial to determine whether BCRP causes resistance of BCR-ABL–expressing CML cells to imatinib.
Current published studies indicate a clear interaction of imatinib and BCRP. Imatinib stimulates BCRP-specific ATPase activity9 and is both a substrate and an inhibitor of BCRP.10,12 Furthermore, imatinib bioavailability, pharmacokinetics, and disposition are influenced by BCRP, suggesting that BCRP functions as an imatinib transporter in vivo.13,14 Finally, chronic exposure of Caco2 cells to imatinib led to induction of BCRP expression.15 To date, however, the only examination of the consequences of BCRP on imatinib cytotoxicity revealed no difference in the survival of Saos2 cells transfected to overexpress BCRP compared with controls.12 Saos2 cells, however, are not growth or survival dependent on imatinib intracellular targets such as BCR-ABL, c-Kit kinase, or platelet-derived growth factor receptor tyrosine kinase, and relatively high concentrations (approximately 10 μM) of imatinib are required to cause 50% lethality. Hence, imatinib toxicity to Saos2 cells could be mediated by interaction with hitherto unknown cellular targets that are not blocked by efflux of intracellular drug. This prompted us to examine the effects of BCRP expression on cellular resistance to imatinib in CML cells that are dependent on BCR-ABL for growth and survival.
Here we report investigations of imatinib cytotoxicity in BCR-ABL–expressing K562 human CML cells that have been transduced and selected to express BCRP (K562/BCRP-MX10). We find that K562 cells are sensitive to imatinib at nanomolar concentrations. K562/BCRP-MX10 cells are measurably resistant to imatinib cytotoxicity; however, this effect is attenuated by imatinib inhibition of BCR-ABL, which we find exerts a posttranscriptional enhancing effect on BCRP expression mediated by the phosphoinositol-3 kinase (PI3K)–Akt signaling pathway. In contrast to previous studies in mice, we find that inhibition of PI3K-Akt signaling in these CML cells causes downregulation of total BCRP expression, not a shift in BCRP localization from plasma membrane to cytoplasm.
Materials and methods
Cells, media, and reagents
K562 and K562/BCRP cells were kindly obtained from Dr Yoshikazu Sugimoto of the Japanese Foundation for Cancer Research16 and were maintained in RPMI 1640 medium (Biofluids, Camarillo, CA) supplemented with 10% heat-inactivated fetal bovine serum (Gemini Bioproducts, Woodland, CA). In order to increase BCRP-mediated drug resistance, BCRP-transduced K562/BCRP cells were treated with 10 nM mitoxantrone for 1 hour prior to weekly passage (designated after mitoxantrone treatment as K562/BCRP-MX10). Human breast carcinoma MCF7/AdrVp cells, which overexpress BCRP following selection with doxorubicin and verapamil,17 were cultivated in improved minimal essential medium (IMEM; Biofluids) supplemented with 10% heat-inactivated fetal bovine serum in the presence of 1 μg/mL doxorubicin and 2.5 μg/mL verapamil. BCRP-overexpressing human ovarian carcinoma Igrov1/T8 cells (selected with topotecan) were provided by Dr Marc Maliepaard of the Netherlands Cancer Institute and were maintained in RPMI 1640 supplemented with 10% fetal bovine serum.18 P-gp–overexpressing human promyelocytic leukemia HL60/vinc cells19 (selected with vincristine) were cultivated in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum. Samples of gastrointestinal stromal tumors (GISTs) were obtained anonymously from the University of Maryland Marlene and Stewart Greenebaum Cancer Center (UMGCC) Tissue Bank shared service. Imatinib and mitoxantrone were from Novartis International AG (Basel, Switzerland) and Immunex (Seattle, WA), respectively. LY294002 was obtained from Sigma Chemicals (St Louis, MO). MG132 was purchased from Carbiochem (San Diego, CA). Fumitremorgin C (FTC) was provided by Dr Susan Bates of the Medicine Branch, National Cancer Institute.
Cytotoxicity assay
To determine the sensitivity of cells to mitoxantrone and imatinib, cell viability was assessed either by the fluorescein diacetate (FDA)/propidium iodide (PI) method or by the 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) method. For the FDA/PI method, viable cells were counted by flow cytometry (FACS Scan; Becton Dickinson, San Jose, CA) after staining with FDA and PI (Sigma-Aldrich, St Louis, MO) as described previously.20 In general, cells were washed with phosphate-buffered saline (PBS) and then resuspended in growth media at an initial cell density of 0.5 × 105 cells/mL. Cells were continuously incubated at 37°C in an atmosphere of 5% CO2 for 3 days. At the end of the experiment, an aliquot of each cell culture was stained with FDA (0.5 μg/mL) in a test tube at room temperature for 30 minutes and then PI (50 μg/mL) was added to each tube. Cells were placed on ice until flow cytometric analysis. Cells that were FDA+ and PI– were considered to be viable and were counted. The XTT assay was performed using the Cell Proliferation Kit II (Roche Applied Science, Indianapolis, IN). Cells were exposed continuously to mitoxantrone for 5 days or to imatinib for 3 days.
Cell proliferation assay
The effects of imatinib on cell proliferation were evaluated by counting cell numbers. Generally, K562 and K562/BCRP-MX10 cells were washed with PBS twice and then resuspended at initial density of 1.0 × 105 cells/mL in growth media or media including imatinib (0.1 μM) and/or FTC (5 μM). Cells were continuously exposed to imatinib and/or FTC for 5 days. Cell numbers were determined with a Coulter Counter (Beckman Coulter, Fullerton, CA) every day for up to 5 days.
Cellular surface expression of BCRP
Cells were stained with PE-conjugated 5D3 antibody to BCRP (R&D Systems, Minneapolis, MN). Generally cells were washed with PBS and then resuspended in PBS containing BSA (2%). Cells were stained directly with conjugated antibody at room temperature for 30 minutes and then washed twice with ice-cold PBS. Cellular fluorescence was determined by flow cytometry (FACScan; Becton Dickinson, Mountain View, CA).
Western blots
Cells were cultured at 37°C with imatinib or LY294002 at the indicated concentrations for varying exposure times. Following treatment, the cells were washed with ice-cold PBS and homogenized in RIPA buffer (50 mM Tris-HCl, pH 7.5; 1% NP-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EGTA; 1 mM sodium orthovanadate; 1 mM NaF; 10 mM sodium pyrophosphate; 1 μg/mL leupeptin; 1 μg/mL aprotinin; and 2 mM Pefabloc SC) by sonication. Debris was removed by centrifugation. The protein concentration of the resultant supernatant was determined by the method of Bradford.21 A 40-μg aliquot of cellular lysate was subjected to 10% SDS–polyacrylamide gel electrophoresis and then was electrotransferred onto a polyvinylidene difluoride membrane. The blots were then probed with one of the following antibodies and then followed by appropriate secondary antibodies conjugated to horseradish peroxidase (Amersham Pharmacia Biotech, Piscataway, NJ): the BXP-21 monoclonal antibody to BCRP,22 the F4 monoclonal antibody to P-gp (Kamiya Biomedical, Seattle, WA), a rabbit monoclonal antibody to Akt (Akt1, Akt2, and Akt3), phospho-Akt (Ser473), c-Kit (Cell Signaling Technology, Beverly, MA), a polyclonal antibody to ubiquitin (Sigma), or β-actin (Abcam, Cambridge, MA).
RT-PCR
RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA) and then treated with RNase-free DNase I (Roche Applied Science). PCR primers for the b3a2 isoform of BCR-ABL (p210) were prepared as described previously.23 Real-time, quantitative reverse transcriptase–PCR (RT-PCR) for BCRP, MDR1, or β-actin transcripts was performed with a Light Cycler (Roche Applied Science) using an RNA amplification kit from Sigma. Primers for BCRP and β-actin were prepared as described previously.24 The sequences of the primers for detecting MDR1 transcripts were as follows: sense, 5′-gtcacaatgcagacagc-3′; and antisense, 5′-ccaacaacaaaataaggcca-3′. Data were analyzed as described previously.24,25
Results
Selection of K562/BCRP with mitoxantrone
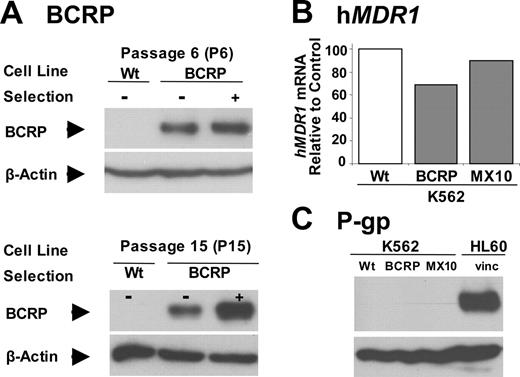
As received in our laboratory, K562/BCRP cells were only 5.5-fold resistant to mitoxantrone and 1.3-fold resistant to imatinib. Hence, K562/BCRP cells were exposed to 10 nM mitoxantrone for 1 hour prior to each weekly passage to select cells with higher levels of BCRP and drug resistance. As shown in Figure 1A, BCRP expression was slightly greater than that of unselected K562/BCRP cells after 6 weeks of selection (P6) and was substantially greater after 15 weeks (P15). At this point, we stopped the selection process and these cells were designated as K562/BCRP-MX10. During this selection process, no increase in hMDR1 mRNA (Figure 1B) or P-gp (Figure 1C) was observed.
Sensitivity of BCRP-expressing K562 cells to mitoxantrone and imatinib
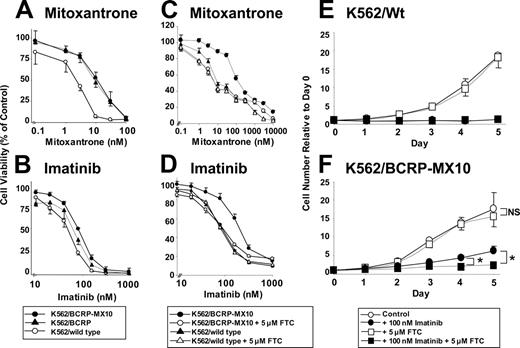
To test the sensitivity of K562, K562/BCRP, and K562/BCRP-MX10 cells to mitoxantrone and imatinib, cell viability was assessed by the FDA/PI method (Figure 2A-B) and by the XTT method (Figure 2C-D). The 50% inhibitory concentration (IC50) values obtained from these experiments are summarized in Tables 1 and 2. IC50 values obtained with the XTT method were somewhat higher than those obtained with the FDA/PI method, but the relative resistance ratios for mitoxantrone or imatinib were similar between the 2 methods. K562/BCRP cells were 5.5-fold resistant to mitoxantrone compared with wild-type K562 cells (Table 1), in agreement with the original report.16 Selection with mitoxantrone (K562/BCRP-MX10) increased mitoxantrone resistance to 7.2-fold by the FDA/PI assay (Figure 2A; Table 1), with comparable relative levels of resistance (11.8-fold) detected by the XTT assay (Figure 2C; Table 2). Similarly, K562/BCRP-MX10 cells were more resistant to imatinib than K562 wild-type cells determined by both cytotoxicity assays (Figure 2B,D; Tables 1, 2), with levels of resistance for K562/BCRP-MX10 cells ranging from 2- to 3-fold for the FDA/PI and XTT methods, respectively.
IC50 values (nM) of mitoxantrone and imatinib determined by FDA/PI assay in K562, K562/BCRP, and K562/BCRP-MX10 cells
. | K562 wild type . | K562/BCRP . | K562/BCRP-MX10 . |
|---|---|---|---|
| Mitoxantrone | 1.52 ± 0.38 (1.0) | 8.35 ± 1.6* (5.5) | 10.9 ± 3.8* (7.2) |
| Imatinib | 44.4 ± 5.4 (1.0) | 58.7 ± 9.4 (1.3) | 89.3 ± 4.7† (2.0) |
. | K562 wild type . | K562/BCRP . | K562/BCRP-MX10 . |
|---|---|---|---|
| Mitoxantrone | 1.52 ± 0.38 (1.0) | 8.35 ± 1.6* (5.5) | 10.9 ± 3.8* (7.2) |
| Imatinib | 44.4 ± 5.4 (1.0) | 58.7 ± 9.4 (1.3) | 89.3 ± 4.7† (2.0) |
Each value represents the mean value ± SEM (n = 4). Relative resistance is shown in parentheses as the ration of the IC50 of K562-expressing BCRP over the IC50 of K562 wild type.
Two-sample t test; P < .05.
Two-sample t test; P < .001.
IC50 values (nM) of mitoxantrone and imatinib determined by XTT assay in K562, and K562/BCRP-MX10 cells
. | K562 wild type . | . | K562/BCRP-MX10 . | . | ||
|---|---|---|---|---|---|---|
. | - FTC . | + FTC . | - FTC . | + FTC . | ||
| Mitoxantrone | 24.0 ± 4.6 (1.0) | 12.5 ± 3.7* | 283 ± 19.1 (11.8)†‡ | 21.3 ± 3.3* | ||
| Imatinib | 91.6 ± 10.8 (1.0) | 89.0 ± 30.3* | 274 ± 66.2 (3.0)§∥ | 94.5 ± 10.6* | ||
. | K562 wild type . | . | K562/BCRP-MX10 . | . | ||
|---|---|---|---|---|---|---|
. | - FTC . | + FTC . | - FTC . | + FTC . | ||
| Mitoxantrone | 24.0 ± 4.6 (1.0) | 12.5 ± 3.7* | 283 ± 19.1 (11.8)†‡ | 21.3 ± 3.3* | ||
| Imatinib | 91.6 ± 10.8 (1.0) | 89.0 ± 30.3* | 274 ± 66.2 (3.0)§∥ | 94.5 ± 10.6* | ||
Each value represents the mean value ± SEM (n = 8). Relative resistance is shown in parentheses as the ratio of the IC50 of K562-expressing BCRP over the IC50 of K562 wild type.
Two-sample t test versus K562 wild type without FTC; P = NS (not significant).
Two-sample t test versus K562 wild type without FTC; P < .001.
Two-sample t test versus K562/BCRP-MX10 with FTC; P < .001.
Two-sample t test versus K562 wild type without FTC; P < .05.
Two-sample t test versus K562/BCRP-MX10 with FTC; P < .05.
To determine whether the resistance to imatinib is due to only BCRP overexpression, the effect of a specific inhibitor of BCRP, FTC, on resistance was studied in K562/BCRP-MX10 cells, with cellular viability assessed by the XTT assay (Figure 2C-D; Table 2). FTC (5 μM) completely sensitized K562/BCRP-MX10 cells to mitoxantrone and to imatinib; in contrast, FTC had no effect on mitoxantrone or imatinib cytotoxicity to wild-type K562 cells.
Imatinib effects on the proliferation of K562 and K562/BCRP-MX10 cells
Proliferation assays were performed in the presence or absence of 5 μM FTC, with cell numbers monitored daily for 5 days by Coulter Counter (Figure 2E-F). K562 wild-type cells did not grow in the presence of 0.1 μM imatinib; furthermore, FTC had no protective effect on imatinib growth inhibition in wild-type cells (Figure 2E ▪). In contrast, K562/BCRP-MX10 cells did grow in 0.1 μM imatinib, reaching numbers equivalent to approximately 30% that of control by day 5 (Figure 2F •). Addition of 5 μM FTC sensitized these cells to imatinib growth inhibition (Figure 2F ▪). FTC alone had no effect on the growth of either wild-type K562 or K562/BCRP-MX10 cells (Figure 2E-F □).
Effect of selection with mitoxantrone on BCRP and MDR1 expression. (A) The effect of mitoxantrone selection on BCRP expression. K562/BCRP cells were selected with 10 nM of mitoxantrone for 1 hour immediately prior to each passage. BCRP expression was determined by Western blots. Forty micrograms of total protein lysates was loaded onto each lane. Blots were probed with the BXP-21 antibody to BCRP. Wt indicates wild type. (B-C) The effect of mitoxantrone selection on MDR1 mRNA and P-gp expression was examined by quantitative RT-PCR (qRT-PCR) (B) or Western blot, using the F4 antibody to P-gp (C), respectively. For panel B, each bar represents the mean value (n = 2). HL60/vinc cells, which are known to overexpress P-gp, were used as positive controls.
Effect of selection with mitoxantrone on BCRP and MDR1 expression. (A) The effect of mitoxantrone selection on BCRP expression. K562/BCRP cells were selected with 10 nM of mitoxantrone for 1 hour immediately prior to each passage. BCRP expression was determined by Western blots. Forty micrograms of total protein lysates was loaded onto each lane. Blots were probed with the BXP-21 antibody to BCRP. Wt indicates wild type. (B-C) The effect of mitoxantrone selection on MDR1 mRNA and P-gp expression was examined by quantitative RT-PCR (qRT-PCR) (B) or Western blot, using the F4 antibody to P-gp (C), respectively. For panel B, each bar represents the mean value (n = 2). HL60/vinc cells, which are known to overexpress P-gp, were used as positive controls.
Cytotoxicity of mitoxantrone and imatinib assessed by FDA/PI and XTT methods. (A-B) FDA/PI assay. The cytotoxicity of mitoxantrone (A) or imatinib (B) was examined in K562/wild-type (○), K562/BCRP (▴), and K562/BCRP-MX10 cells (•). Cells were exposed to either drug at 37°C for 3 days and then stained with FDA and PI. Cellular fluorescence was measured by FACScan. Each point represents the mean value of 4 individual assays with SEM. (C-D) XTT assay and effects of FTC on resistance to mitoxantrone or imatinib. The cytotoxicity of mitoxantrone (C) or imatinib (D) was examined in K562/wild-type (triangles) and K562/BCRP-MX10 cells (circles) in the presence (open symbols) or absence (closed symbols) of 5 μM FTC, using the XTT assay. Cells were exposed to mitoxantrone for 5 days or to imatinib for 3 days at 37°C. Each point represents the mean value of 8 individual assays with SEM. (E-F) Effects of imatinib ± FTC on cell growth. Wild-type (Wt) K562 (E) or K562/BCRP-MX10 cells (F) were cultivated without any drugs (○) or with 5 μM FTC (□), 100 nM imatinib (•), or both drugs (▪). Cell proliferation was determined by counting cell number by Coulter Counter for up to 5 days. Cell numbers relative to day 0 are plotted. Each point represents the mean value of 4 to 5 individual assays with SEM. *P < .05, Student t test. NS indicates no significant difference.
Cytotoxicity of mitoxantrone and imatinib assessed by FDA/PI and XTT methods. (A-B) FDA/PI assay. The cytotoxicity of mitoxantrone (A) or imatinib (B) was examined in K562/wild-type (○), K562/BCRP (▴), and K562/BCRP-MX10 cells (•). Cells were exposed to either drug at 37°C for 3 days and then stained with FDA and PI. Cellular fluorescence was measured by FACScan. Each point represents the mean value of 4 individual assays with SEM. (C-D) XTT assay and effects of FTC on resistance to mitoxantrone or imatinib. The cytotoxicity of mitoxantrone (C) or imatinib (D) was examined in K562/wild-type (triangles) and K562/BCRP-MX10 cells (circles) in the presence (open symbols) or absence (closed symbols) of 5 μM FTC, using the XTT assay. Cells were exposed to mitoxantrone for 5 days or to imatinib for 3 days at 37°C. Each point represents the mean value of 8 individual assays with SEM. (E-F) Effects of imatinib ± FTC on cell growth. Wild-type (Wt) K562 (E) or K562/BCRP-MX10 cells (F) were cultivated without any drugs (○) or with 5 μM FTC (□), 100 nM imatinib (•), or both drugs (▪). Cell proliferation was determined by counting cell number by Coulter Counter for up to 5 days. Cell numbers relative to day 0 are plotted. Each point represents the mean value of 4 to 5 individual assays with SEM. *P < .05, Student t test. NS indicates no significant difference.
Evaluation of other possible imatinib resistance mechanisms in K562/BCRP-MX10 cells
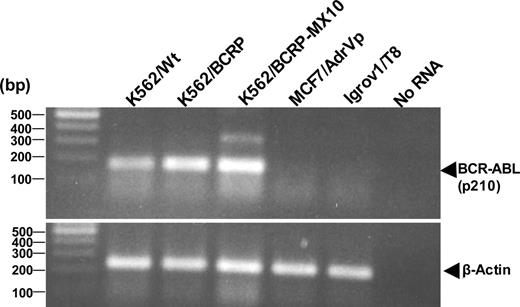
To rule out upregulation of BCR-ABL as a possible cause of imatinib resistance in this system, we measured BCR-ABL mRNA transcripts in wild-type K562 and K562/BCRP-MX10 cells. Both cell types expressed comparable amounts of the p210 form of BCR-ABL (b3a2 form), as measured by a 168-bp PCR product from RT-PCR analysis (Figure 3). The sizes of faint PCR products observed in MCF-7/AdrVp and Igrov1/T8 cells did not correspond to those of another form of p210 (b2a2, 68 bp) and p230 (b19a2) so that we considered these products as nonrelated to BCR-ABL.
To rule out the possibility of upregulation of another molecular target of imatinib contributing to imatinib resistance in the K562/BCRP-MX10 cells, we measured the expression of c-Kit, a receptor tyrosine kinase that is known to activate the PI3K pathway and also a known target of imatinib. K562 wild-type cells have been reported to have very low expression of c-Kit protein.26 Using Western blots, we were unable to detect any c-Kit protein in wild-type K562 or K562/BCRP-MX10 cells (data not shown). In contrast, strong and distinct immunoreactive bands of the characteristic molecular mass for c-Kit (120 and 145 kDa) were observed in 2 samples of human GIST tissue, which were studied as positive controls.
Taken together, the data in Figures 2 and 3 and the c-Kit measurements clearly illustrate that the expression of BCRP/ABCG2 enables K562/BCRP-MX10 cells to resist imatinib-induced cytotoxicity and growth inhibition. However, BCRP overexpression appears to provide more protection from mitoxantrone toxicity than from imatinib toxicity. To investigate this further, we examined the effects of imatinib on the expression of transduced BCRP in K562/BCRP-MX10 cells.
Effect of imatinib on BCRP expression
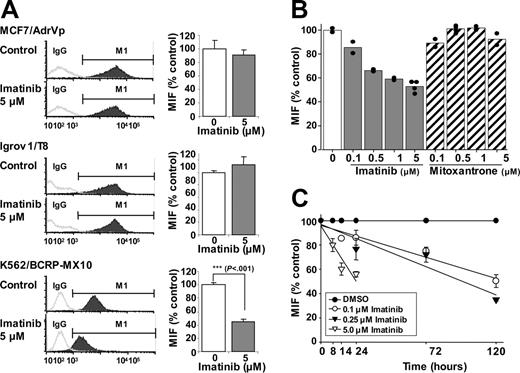
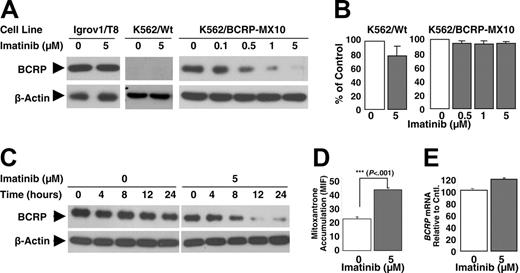
Cellular transformation by BCR-ABL is associated with high PI3K activity.27,28 Furthermore, it is known that murine Bcrp1 membrane localization and hence Bcrp1 function is regulated by PI3K-Akt signaling.29 We therefore hypothesized that imatinib inhibition of BCR-ABL diminishes PI3K-Akt signaling, which may thereby diminish BCRP function in these K562 cells, resulting in less relative resistance to imatinib than to mitoxantrone. Indeed, 14-hour exposure to 5 μM imatinib caused a 60% reduction in the expression of BCRP on the surface membranes of K562/BCRP-MX10 cells, as measured by flow cytometric quantification of 5D3 monoclonal antibody binding, which recognizes a surface epitope of BCRP in intact cells (Figure 4A, K562/BCRP-MX10). In contrast, BCRP expression in MCF7/AdrVp cells and Igrov1/T8 cells, which do not express BCR-ABL, was unaffected by imatinib (Figure 4A). Figure 4A shows typical histograms of cellular fluorescence along with averaged values of mean intensity of fluorescence, expressed as percentage of control.
RT-PCR for p210 BCR-ABL. RT-PCR products from the indicated cells were visualized in 2% agarose gel, along with a 100-bp DNA ladder.
RT-PCR for p210 BCR-ABL. RT-PCR products from the indicated cells were visualized in 2% agarose gel, along with a 100-bp DNA ladder.
Cell surface BCRP expression in K562/BCRP-MX10 cells measured by the 5D3 antibody displayed a time- and dose-dependent decrease in response to imatinib concentrations in the IC50 range (0.1 and 0.25 μM, Figure 4B-C). Imatinib (0.1 or 0.25 μM) reduced surface BCRP expression to 50% and 35% of control, respectively, after 5 days of exposure (Figure 4C). Imatinib at 5 μM reduced BCRP surface expression to approximately 50% of control by 24 hours; for cells exposed to 5 μM imatinib for longer than 24 hours, we were unable to collect reliable BCRP expression data because of loss of cells due to cytotoxicity (Figure 4C). In contrast to imatinib, exposure of K562/BCRP-MX10 cells to a concentration range of mitoxantrone for 14 hours did not alter the cell-surface expression of BCRP (Figure 4B).
Characterization of BCRP downregulation by imatinib
To determine whether the observed reduction in cell-surface BCRP expression is caused by internalization of protein or a decrease in total BCRP, the expression of total BCRP was examined by Western blot following 14-hour exposure to imatinib (Figure 5A). Total cellular BCRP expression, like surface BCRP expression (Figure 4B), was decreased in K562/BCRP-MX10 cells in a dose-dependent fashion (Figure 5A) following 14-hour treatment with imatinib, but the same imatinib treatment did not affect BCRP expression in BCR-ABL– Igrov1/T8 cells. The 14-hour imatinib treatment did not cause a decrease in K562 wild-type or K562/BCRP-MX10 cell number, suggesting that the observed decrease in BCRP expression is not the result of death of cells (Figure 5B). A decrease in total BCRP in K562/BCRP-MX10 cells was observed with this relatively short exposure time even at 0.1 μM imatinib, which is close to the IC50 for these cells determined by the FDA/PI assay and well below the IC50 observed with the XTT assay.
Effect of imatinib on BCRP expression in plasma membrane. (A) MCF-7/AdrVp, Igrov1/T8, or K562/BCRP-MX10 cells were exposed to 5 μM of imatinib for 14 hours at 37°C and then stained with PE-conjugated 5D3 antibody to BCRP. Cellular fluorescence was assessed by flow cytometry. Mean intensities of fluorescence (MIF) obtained from control experiments were normalized to 100%. These experiments were performed twice on different days, with duplicate studies for each experimental condition on a given day. Each bar represents the mean value of 4 individual assays. ***P < .001, Student t test. M1 indicates the channels that exclude 98% of the isotype (IgG) control. (B) K562/BCRP-MX10 cells were cultured for 14 hours with a concentration range of imatinib or mitoxantrone, then BCRP expression on the cell surface was measured as described for panel A. Each bar represents the mean MIF value from at least 2 individual experiments; each dot represents an individual data point. (C) K562/BCRP-MX10 cells were cultured for periods of time up to 120 hours with 0, 0.1, 0.25, or 5 μM imatinib. BCRP expression on the cell surface was then measured as described for panel A. Each point represents the mean value ± SEM of MIFs relative to controls (0 μM imatinib) done at the same time point. Experiments were repeated 2 to 3 times. The lines drawn for the experimental points were obtained by linear regression analysis. The correlation coefficients (R2) were 0.94, 0.91, and 0.89 for cells treated with 0.1, 0.25, and 5 μM imatinib, respectively.
Effect of imatinib on BCRP expression in plasma membrane. (A) MCF-7/AdrVp, Igrov1/T8, or K562/BCRP-MX10 cells were exposed to 5 μM of imatinib for 14 hours at 37°C and then stained with PE-conjugated 5D3 antibody to BCRP. Cellular fluorescence was assessed by flow cytometry. Mean intensities of fluorescence (MIF) obtained from control experiments were normalized to 100%. These experiments were performed twice on different days, with duplicate studies for each experimental condition on a given day. Each bar represents the mean value of 4 individual assays. ***P < .001, Student t test. M1 indicates the channels that exclude 98% of the isotype (IgG) control. (B) K562/BCRP-MX10 cells were cultured for 14 hours with a concentration range of imatinib or mitoxantrone, then BCRP expression on the cell surface was measured as described for panel A. Each bar represents the mean MIF value from at least 2 individual experiments; each dot represents an individual data point. (C) K562/BCRP-MX10 cells were cultured for periods of time up to 120 hours with 0, 0.1, 0.25, or 5 μM imatinib. BCRP expression on the cell surface was then measured as described for panel A. Each point represents the mean value ± SEM of MIFs relative to controls (0 μM imatinib) done at the same time point. Experiments were repeated 2 to 3 times. The lines drawn for the experimental points were obtained by linear regression analysis. The correlation coefficients (R2) were 0.94, 0.91, and 0.89 for cells treated with 0.1, 0.25, and 5 μM imatinib, respectively.
Like surface expression of BCRP (Figure 4C), the imatinib-induced reduction in total BCRP expression was also time dependent, with lowest levels observed after 12- to 24-hour treatment with 5 μM (Figure 5C); BCRP expression levels were reduced by approximately 50% after 8 hours (Figure 5C).
The downregulation of BCRP induced by 14-hour exposure to 5 μM imatinib was accompanied by a statistically significant increase in the intracellular accumulation of mitoxantrone, suggesting loss of BCRP functional activity at the cell surface (Figure 5D). No significant change was observed in the level of BCRP mRNA transduced from the retroviral vector in the K562/BCRP-MX10 cells under these conditions (Figure 5E).
Taken together, these data indicate that the imatinib-induced decrease of functional BCRP in the plasma membrane of BCR-ABL+ cells (Figure 4) is due to loss of overall BCRP expression (Figure 5) without any alteration in the production of BCRP mRNA. This suggests that BCR-ABL is involved in a posttranscriptional process that regulates the synthesis of BCRP or modulates expression at a posttranslational level such as degradation of BCRP.
Effect of LY294002 on BCRP expression
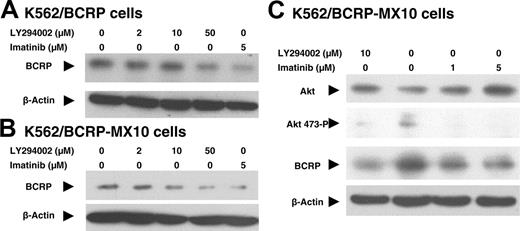
If BCRP expression in K562/BCRP-MX10 cells is enhanced by PI3K-Akt signaling stimulated by the action of BCR-ABL, then BCRP expression in these cells should also be downregulated by direct inhibition of the PI3K-Akt signaling pathway. To test this hypothesis, we studied the effects of the PI3K inhibitor LY294002 on total BCRP expression in K562/BCRP and K562/BCRP-MX10 cells, using Western blots (Figure 6A-B). Total BCRP expression was reduced in a dose-dependent manner by treatment with LY294002. Treatment with 10 to 50 μM LY294002 for 14 hours reduced overall BCRP expression to a level similar to that obtained with 5 μM imatinib, suggesting that an active PI3K-Akt pathway in these cells is responsible, at least in part, for maintaining BCRP expression.
Characterization of BCRP downregulation by imatinib. (A,C) Cells were exposed to imatinib under the concentrations indicated at 37°C for 14 hours (A) or the times indicated (C). Total cell lysates were then extracted and subjected to Western blotting. These Western blots represent 3 individual experiments done on different days. (B) Cell growth relative to control (without imatinib) was determined by counting cell number with a Coulter Counter (n > 3). (D) Effects of pre-exposure to imatinib on BCRP function. K562/BCRP-MX10 cells were cultured with 0 or 5 μM imatinib for 14 hours, and then the cells were washed free of drug and cultured for an additional 80 minutes in fresh medium containing 10 μM mitoxantrone. At this point, intracellular mitoxantrone content was determined using flow cytometry. Each bar represents the mean value ± SEM for 4 individual experiments. The difference between imatinib-treated and control cells was evaluated by Student t test. (E) Relative BCRP mRNA expression was quantified by 4 individual qRT-PCR experiments. Each vertical bar represents the mean value ± SEM.
Characterization of BCRP downregulation by imatinib. (A,C) Cells were exposed to imatinib under the concentrations indicated at 37°C for 14 hours (A) or the times indicated (C). Total cell lysates were then extracted and subjected to Western blotting. These Western blots represent 3 individual experiments done on different days. (B) Cell growth relative to control (without imatinib) was determined by counting cell number with a Coulter Counter (n > 3). (D) Effects of pre-exposure to imatinib on BCRP function. K562/BCRP-MX10 cells were cultured with 0 or 5 μM imatinib for 14 hours, and then the cells were washed free of drug and cultured for an additional 80 minutes in fresh medium containing 10 μM mitoxantrone. At this point, intracellular mitoxantrone content was determined using flow cytometry. Each bar represents the mean value ± SEM for 4 individual experiments. The difference between imatinib-treated and control cells was evaluated by Student t test. (E) Relative BCRP mRNA expression was quantified by 4 individual qRT-PCR experiments. Each vertical bar represents the mean value ± SEM.
Imatinib effects on phosphorylation of Ser473 of Akt
Based on these results, we tested whether imatinib downregulates the phosphorylation of Akt. When K562/BCRP-MX10 cells were treated for 14 hours with 1 or 5 μM imatinib, or 10 μM LY294002, the expression of phosphorylated Akt (Ser473) and BCRP were clearly decreased (Figure 6C). Overall Akt protein (Akt1, Akt2, and Akt3) level was essentially unaltered by LY294002 or imatinib. Similar results of LY294002 downregulation of phosphorylated Akt were observed in K562/BCRP cells and in K562 (wild-type) cells (data not shown). These data suggest that phosphorylation of Akt is involved in sustaining BCRP expression in these CML-derived cells.
Investigation of posttranslational mechanisms: effect of imatinib on the proteasome
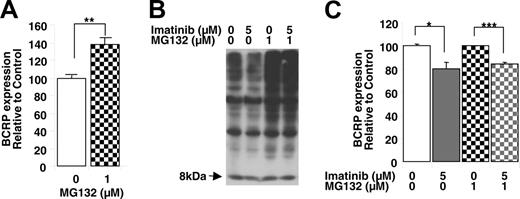
Exposure of K562/BCRP-MX10 cells to the proteasome inhibitor MG132 (1 μM, 8 hours) resulted in an approximately 40% increase in the expression of BCRP on the cell surface (Figure 7A), suggesting that the proteasome is responsible, at least in part, for the posttranslational regulation of BCRP. To our knowledge, this is the first report of proteasomal regulation of BCRP levels. The exposure to MG132 used caused proteasomal inhibition, evidenced by the increase in ubiquitinated proteins in the K562/BCRP-MX 10 cells (Figure 7B). Imatinib (5 μM, 8 hours) did not appear to affect ubiquitinated proteins in the presence or absence of MG132 (Figure 7B). Proteasomal inhibition, however, could not block the imatinib-induced downregulation of BCRP (Figure 7C), suggesting that the BCRP-lowering effect of imatinib is not the result of stimulation of proteasomal activity either directly (by imatinib itself) or indirectly (via inhibition of the PI3K-Akt pathway).
Discussion
In the present work, we demonstrate that BCRP protects BCR-ABL+ cells from the cytotoxicity of imatinib. However, we also find that BCR-ABL, by activation of the PI3K-Akt pathway, enhances BCRP expression via a posttranscriptional mechanism and that imatinib inhibition of BCR-ABL decreases Akt phosphorylation (Ser473) and reduces BCRP expression. These 2 opposing responses to imatinib in a BCRP-expressing, BCR-ABL+ cell line are novel findings. A key question relevant to clinical practice is whether BCRP is functionally expressed in self-renewing CML cells and which of the opposing effects (cytotoxic resistance caused by BCRP efflux of imatinib or imatinib-induced downregulation of BCRP expression) predominates in clinical cases. Although the system we used employed an “artificial” means of enforcing BCRP expression, the effects we observed of imatinib on BCRP levels appear to be posttranscriptional and should apply to naturally occurring expression of BCRP in BCR-ABL+ cancer cells as well.
A practical question to ask at this point is whether BCRP is functionally expressed in clinical cases of CML, particularly since the CML-derived wild-type K562 cells used express very low levels of BCRP. CML is regarded to be a clonal proliferative disease, with self-renewing populations of stem and progenitor cells.30,31 In normal HSCs, BCRP is functionally expressed in a cell-cycle–quiescent, self-renewing population characterized by its ability to efflux Hoechst 33342 dye.11 This population is termed the side population (SP) because of the characteristic appearance of the population on plots of blue versus red Hoechst cellular fluorescence. It is reasonable to believe that a Ph+ counterpart to SP cells exists in cases of chronic-phase CML. Remarkably, quiescent self-renewing Ph+ cells with stem-cell characteristics have been described in patients with chronic-phase CML, and these cells were found to be resistant to imatinib.6 Initially, this resistance was attributed to these cells being in a quiescent state; however, a more recent study finds no correlation between the cytotoxic activity of imatinib and proliferative status.32 Hence, it is reasonable to postulate that these imatinib-resistant quiescent cells are BCRP-expressing Ph+ SP cells. Further research into this question is underway in our laboratory. K562 cells, on the other hand, are derived from a CML patient in blastic phase33 and may have evolved additional mutations enabling self-renewal capacity in the absence of an SP phenotype.
Effect of imatinib or LY294002 on BCRP expression and Akt expression or phosphorylation. (A-B) Effects on BCRP expression. K562/BCRP (A) and K562/BCRP-MX10 cells (B) were exposed to LY294002 or imatinib for 14 hours at 37°C. Total cell lysates were extracted and then 40 μg of total cell lysates was subjected to Western blotting. These Western blots represent 3 individual experiments. (C) Effects on Akt expression and phosphorylation. K562/BCRP-MX10 cells were exposed to imatinib or LY294002 for 14 hours at 37°C. Total cell lysates were then extracted and subjected to Western blotting for Akt or phospho-Akt (S473) protein. Forty micrograms of cell lysate was loaded onto each lane. These Western blots are representative of 3 individual experiments done on different days.
Effect of imatinib or LY294002 on BCRP expression and Akt expression or phosphorylation. (A-B) Effects on BCRP expression. K562/BCRP (A) and K562/BCRP-MX10 cells (B) were exposed to LY294002 or imatinib for 14 hours at 37°C. Total cell lysates were extracted and then 40 μg of total cell lysates was subjected to Western blotting. These Western blots represent 3 individual experiments. (C) Effects on Akt expression and phosphorylation. K562/BCRP-MX10 cells were exposed to imatinib or LY294002 for 14 hours at 37°C. Total cell lysates were then extracted and subjected to Western blotting for Akt or phospho-Akt (S473) protein. Forty micrograms of cell lysate was loaded onto each lane. These Western blots are representative of 3 individual experiments done on different days.
The interaction of imatinib with BCRP has been characterized by several groups9,10,12,34 ; however, to date no published studies have produced evidence that BCRP expression actually causes cytotoxic resistance to imatinib.10,12,34 Our results clearly demonstrate that BCRP overexpression causes resistance to imatinib in cells where growth and survival are dependent on BCR-ABL signaling (Figures 1, 2), consistent with a recent report that BCRP transports imatinib in HEK-293 with enforced BCRP expression.10 To date, the only published study of the effects of BCRP on imatinib cytotoxicity to cancer cells examined Saos2 osteosarcoma cells,12 which do not overexpress cellular targets of imatinib. BCRP-transfected Saos2 cells did not demonstrate resistance to imatinib compared with wild-type or vector control cells. For BCR-ABL–expressing K562 cells (wild type), the IC50 of 40 to 90 nM that we observed was 80- to 180-fold lower than that reported for imatinib in Saos2 cells (IC50, vector control cells 7300 nM).12 The higher doses of imatinib required to kill Saos2 cells may have overcome the effects of BCRP efflux or may have killed the cells by a mechanism unaffected by efflux of intracellular drug, resulting in no apparent protective effect of BCRP in these cells.
A potential concern with our methods is that the mitoxantrone selection we used to increase BCRP expression and drug resistance might provoke other mechanisms of resistance to imatinib, such as an increase in P-gp, which is reported to transport imatinib.8 In the original description, K562/BCRP cells did not have high P-gp expression.16 During the course of selection we did not observe any induction of MDR1 mRNA or P-gp as shown in Figure 1B-C. Furthermore, we did not observe an increase in BCR-ABL mRNA or c-Kit protein as the result of mitoxantrone selection. Finally, the ability of the BCRP-specific inhibitor FTC to completely sensitize K562/BCRP-MX cells to imatinib as well as mitoxantrone, a key substrate for BCRP (Figure 2C-D), clearly indicates that BCRP is the cause of cytotoxic resistance to imatinib in these cells.
The BCR-ABL tyrosine kinase activates signal transduction pathways that stimulate growth and prevent apoptosis, such as JAK/STAT, Raf/MEK/ERK, and PI3K/Akt, and confers cytokine-independent survival to hematopoietic cells.28 Therefore, it is reasonable to find the PI3K-Akt pathway activated in K562 cells and that phosphorylation of Ser473 of Akt is diminished by imatinib treatment.35 A number of reports indicate that the PI3K-Akt pathway is involved in posttranscriptional/posttranslational modulation of BCRP. Bcrp1 protein expressed in murine hematopoietic SP cells was found to be translocated from the plasma membrane to the cytoplasm in Akt1–/– mice or after exposure of wild-type SP cells to LY294002, without a decrease in total Bcrp1 expression.29,36 In another study, BCRP expressed on the plasma membrane of guinea pig LLC-PK1 cells was internalized after treatment with LY294002 or wortmannin.36 In our present study, overall BCRP was reduced significantly when K562/BCRP-MX10 cells were treated with imatinib or LY294002, both of which reduced phosphorylation of Akt (Figure 6), suggesting strongly that overall BCRP expression is regulated through the PI3K-Akt pathway. However, our findings in human leukemia cells clearly differ from the previous studies in murine or guinea pig cells, which show that PI3K-Akt signaling causes translocation of BCRP without loss of overall protein expression.
In summary, we find that BCRP confers resistance to imatinib in a Ph+ CML cell line. Additionally we find that the PI3K-Akt signaling pathway, which is activated in K562 cells by BCR-ABL, controls total BCRP expression posttranscriptionally/translationally by a mechanism that is independent of the proteasome. Imatinib inhibition of BCR-ABL hence downregulates BCRP expression in BCR-ABL+ cells. These observations may have profound implications for the treatment of CML, where it is likely that a BCR-ABL+ stem-cell population analogous to normal hematopoietic SP cells exists, which expresses BCRP. Further studies of clinical cases of CML to determine the presence of such BCRP+ CML cells is warranted, and if such cells do exist, it should be determined whether these cells are resistant to imatinib and whether imatinib downregulates their BCRP expression. Even if it is found that BCRP is downregulated by imatinib in clinical CML, it may be possible that the initial protection offered by BCRP will enable the cells to develop other mechanisms of resistance to imatinib, such as mutations of BCR-ABL, during the period that BCRP expression levels are declining. The protective effects of BCRP could be overcome by combining imatinib treatment with one of the many commercially available drugs that are BCRP inhibitors, which would also serve to enhance the bioavailability of imatinib by inhibition of BCRP expression in the intestine and bile canaliculi.
Role of proteasome on posttranscriptional downregulation of BCRP by imatinib. (A) Effects of the proteasomal inhibitor MG132 on cell surface BCRP expression. K562/BCRP-MX10 cells were treated with 1 μM MG132 for 8 hours and then BCRP expression was measured by binding of the 5D3 antibody, as determined by flow cytometry. Each bar represents the mean value with SEM of at least 3 individual experiments. **P < .01, Student t test. (B) Effectiveness of MG132 in causing proteasomal inhibition; effects of imatinib. K562/BCRP-MX10 cells in culture were exposed to the indicated concentrations of MG132 or imatinib for 8 hours. Then, the ubiquitination of proteins in total cell lysates was determined by Western blot, using rabbit polyclonal antibody to ubiquitin. The 8-kDa band represents free, unconjugated ubiquitin ligand. (C) Proteasomal inhibition does not block the down-regulation of BCRP by imatinib. K562/BCRP-MX10 cells in culture were exposed to the indicated concentrations of MG132 or imatinib for 8 hours. Then, BCRP expression was measured by flow cytometry as in panel A. Each bar represents the mean value with SEM of at least 3 individual experiments. *P < .05, ***P < .001 by Student t test.
Role of proteasome on posttranscriptional downregulation of BCRP by imatinib. (A) Effects of the proteasomal inhibitor MG132 on cell surface BCRP expression. K562/BCRP-MX10 cells were treated with 1 μM MG132 for 8 hours and then BCRP expression was measured by binding of the 5D3 antibody, as determined by flow cytometry. Each bar represents the mean value with SEM of at least 3 individual experiments. **P < .01, Student t test. (B) Effectiveness of MG132 in causing proteasomal inhibition; effects of imatinib. K562/BCRP-MX10 cells in culture were exposed to the indicated concentrations of MG132 or imatinib for 8 hours. Then, the ubiquitination of proteins in total cell lysates was determined by Western blot, using rabbit polyclonal antibody to ubiquitin. The 8-kDa band represents free, unconjugated ubiquitin ligand. (C) Proteasomal inhibition does not block the down-regulation of BCRP by imatinib. K562/BCRP-MX10 cells in culture were exposed to the indicated concentrations of MG132 or imatinib for 8 hours. Then, BCRP expression was measured by flow cytometry as in panel A. Each bar represents the mean value with SEM of at least 3 individual experiments. *P < .05, ***P < .001 by Student t test.
Prepublished online as Blood First Edition Paper, March 16, 2006; DOI 10.1182/blood-2005-10-4020.
Supported in part by a Department of Veterans Affairs Merit Review Grant (D.D.R.).
One of the authors (D.D.R.) holds a patent related to the work that is described in the present study (US Patent 6313277, breast cancer resistance protein [BCRP] and the DNA which encodes it).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Victoria S. Yang for her assistance in performing the initial cytotoxicity assays done in this work. Thanks also to Dr Susan Bates of the National Cancer Institute for providing our laboratory with fumitremorgin C. We are grateful to Drs Raymond Jones and William Rodgers and to Mr John Cottrell of the UMGCC Tissue Bank shared service for providing, in an anonymous fashion, 2 samples of human GIST tissue. All specimens distributed by the UMGCC Tissue Bank are collected after informed consent, according to a protocol approved by the University of Maryland, Baltimore institutional review board.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal