Gimap4, a member of the newly identified GTPase of the immunity-associated protein family (Gimap), is strongly induced by the pre–T-cell receptor in precursor T lymphocytes, transiently shut off in double-positive thymocytes, and reappears after TCR-mediated positive selection. Here, we show that Gimap4 remains expressed constitutively in the cytosol of mature T cells. A C-terminal IQ domain binds calmodulin in the absence of calcium, and conserved PKC phosphorylation motifs are targets of concanavalin A (ConA)– or PMA/ionomycin-induced PKC activation. To address the role of Gimap4 in T-cell physiology, we completed the genomic organization of the gimap4 locus and generated a Gimap4-null mutant mouse. Studies in these mice revealed no critical role of Gimap4 in T-cell development but in the regulation of apoptosis. We have found that Gimap4 accelerates the execution of programmed cell death induced by intrinsic stimuli downstream of caspase-3 activation and phosphatidylserine exposure. Apoptosis directly correlates with the phosphorylation status of Gimap4.
Introduction
T-lymphocyte development proceeds through several genetically controlled stages. It is characterized by drastic changes in gene expression profiles resulting in the generation of highly specialized lymphocyte populations. Three critical checkpoints have to be passed to guarantee that only qualified precursors continue their development (reviewed by Rodewald and Fehling1 ). (1) The IL-7 receptor and c-Kit expressed on the earliest thymocyte progenitors control their expansion into the developmental stages double-negative (DN) (CD4–CD8–), DNI (CD25–CD44+), DNII (CD25+ CD44+), and DNIII (CD25+CD44–). (2) The pre–T-cell receptor (pre-TCR), expressed upon successful rearrangement of a TCRβ allele, controls the developmental transition of DNIIIE (expected size) into DNIIIL (large) cells. Because the expression of a pre-TCR is controlled by the rearrangement and onset of TCRβ expression, this checkpoint is referred to as β-selection.2 β-selection ensures that only those cells equipped with a functional pre-TCR can proliferate and progress via a DNIV (CD25–CD44–) and an immature single-positive (ISP) (CD4–CD8+) stage into immature CD4+CD8+ double-positive (DP) thymocytes.2,3 Simultaneously, further TCRβ gene rearrangements are prohibited, a process known as allelic exclusion. Further development requires functional rearrangement(s) at the TCRα locus and expression of a clonotypic αβTCR. (3) Finally, the αβTCR controls whether a given thymocyte fulfills requirements for self-tolerance on the basis of TCR–self-peptide major histocompatibility complex (pMHC) interactions. Positively selected thymocytes differentiate into single-positive (SP) mature T cells.
During development most lymphoid precursors do not fulfill the necessary requirements and die by apoptosis. The interaction of the TCR with pMHC ligands in the thymus determines the outcome for positive and negative selection. Only thymocytes with the right affinity strength for pMHC receive a survival signal from the TCR, whereas thymocytes with no or too high affinity for pMHC undergo apoptosis. Considering the high cellular turnover within the immune system, effective apoptosis is a critical event, not only during formation of the primary lymphocyte repertoire but also in controlling the size of the antigen-selected lymphocyte pool during an infection, thus ensuring cellular homeostasis during lymphocyte development and immunity. An effective death program is therefore a prerequisite for the rapid clearance of cells from the immune system (reviewed by Danial and Korsmeyer4 and Marsden and Strasser5 ).
Two pathways can activate the death machinery. The extrinsic pathway is triggered by binding of ligands to so-called death receptors such as TNF receptor-1, Fas, and TRAIL receptor-1 and -2. The intrinsic mitochondrion-mediated pathway is stress induced and triggered by lack of survival factors or by DNA damage. The decision to undergo apoptosis involves the integration of multiple proapoptotic and antiapoptotic signals collectively controlling the integrity of the outer mitochondrial membrane.6 The execution of mitochondrion-mediated apoptosis is initiated by the release of cytochrome c and other proapoptotic proteins that synergize to activate effector caspases.7 The release of cytochrome c in the stress pathways seems to be generally caspase independent.8 By cleaving various substrates, active effector caspases, such as caspase-3, trigger a series of events that are part of the apoptotic execution. These include exposure of phosphatidylserine (PS) in the outer lipid layer of the plasma membrane and a subsequent loss of plasma membrane integrity, such as permeabilization and blebbing of the plasma membrane as well as cell and organelle condensation, chromatin condensation, DNA cleavage, nuclear segmentation and, finally, cell fragmentation into apoptotic bodies.
To gain further insight into the molecular processes of TCR-mediated development and survival, the identification and characterization of novel differentially expressed genes that are directly or indirectly controlled by the onset of TCR expression is of special interest. By anti-CD3–induced thymocyte differentiation in Rag knockout (ko) mice,9 developmental subsets were isolated and, by differential mRNA display, a series of tightly regulated genes was identified. Here, we report on our strategy to identify developmentally regulated genes in early thymocytes and the identification of Gimap4, a small GTPase that has been identified independently,10,11 whose function in lymphocyte physiology, however, remains ill defined.
Given the interesting role of another family member, Gimap5, in controlling T-cell homeostasis and diabetes mellitus in BioBreeding (BB) rats,12-18 we have set out to define the function of Gimap4 in vitro and in vivo. Besides a GTPase domain, characteristic for all members within the gimap gene cluster,19 other conserved protein motifs are proven to be functional. The generation and characterization of Gimap4-deficient mice revealed a role of Gimap4 in the execution of apoptosis.
Materials and methods
Mice
Mice deficient for Rag2,20 cγ/Rag (mixed), TCRα and TCRβ, the TCR transgenic strains HA, HY, LCMV, F5 (BL10), OT-I, and OT-II, as well as ΔVTCRβ and Ly5.1 C57BL/6 were available at The Netherlands Cancer Institute. PKCθ-deficient mice were kindly provided by Dirk Schlueter and Dan Littman (New York University School of Medicine, NY).21 If not stated otherwise, mice were backcrossed to C57BL/6 for at least 6 generations. All animal experiments were performed according to institutional and national guidelines.
Generation of Gimap4-deficient mice
A commercially available 129SVJ mouse genomic library (Phage FTX*II Vector; Stratagene, La Jolla, CA) was used to isolate genomic clones that hybridize with a Gimap4 cDNA probe. From one phage a genomic NotI fragment (12.8 kb) was subcloned into the cosmid vector pWE15, which was used for physical mapping of the Gimap4 locus. From this cosmid a blunted SphI fragment (7.7 kb) containing the exon 2 was subcloned into the EcoRV site of the pSK+ vector (GenBank). Murine Gimap4 was inactivated by gene targeting in the mouse germ line. To derive the targeting vector, an EcoRV fragment (1.1 kb) was replaced with a blunted Asp718/BamHI fragment containing the floxed neocassette (1.4 kb) from the pGL2 neovector (kind gift from Raul Torres, National Jewish Medical and Research Center, Denver, CO).
A SalI-NotI fragment (8.5 kb) was isolated from the pSK-Gimap4 vector to target E14 embryonic stem (ES) cells (129 derived). After electroporation and selection, ES-cell clones were screened by Southern blot using EcoRI-digested DNA in combination with a genomic Sph/NotI probe from cosmid pWE15. ES clones that underwent homologous recombination were used to introduce the mutation into the mouse germ line by blastocyst injection and transfer into pseudopregant foster mothers.22 To facilitate the phenotyping of Gimap4 mutant mice, a polymerase chain reaction (PCR) screening was established including primers that locate outside of the EcoRV site: forward primer (GCTGGCTAATTATGGTGTAG) and reverse primer (CTGGATCACACACTCGGCAC). Gimap4 ko mice were bred on the C57BL/6 background for at least 12 generations and used for experiments at 6 to 10 weeks of age.
In vitro transcription/translation
For protein synthesis, Gimap4 cDNA was cloned into the pBluescript II SK vector and used in an in vitro transcription/translation system (TNT Quick coupled TS/TL System; Promega, Utrecht, The Netherlands).
Differential mRNA display and full-length cDNA cloning of Gimap4
For the differential mRNA display, the anti-CD3ϵ–inducible T-cell differentiation system was used as previously described.9 In short, 120 Rag2 ko mice were injected intraperitoneally with 50 μg anti-CD3ϵ (clone 145-2C11), and the thymi were isolated up to 4 days after injection. Thymocytes were labeled with anti-CD25, anti-CD44, anti-CD4, and anti-CD8 monoclonal antibodies. Viable (propidium iodide–negative) DNIV, ISP, and DP T-cell subsets were sorted independently to high purity (more than 98%) by fluorescence-activated cell sorting (FACS) MoFlo (Cytomation, Fort Collins, CO) and FACSstar (Becton Dickinson, San Jose, CA). To identify genes that are specifically induced by signaling through the pre-TCR–CD3 complex, differentiation-arrested DNIIIE thymocytes from nontreated Rag2 ko mice were isolated following the same procedure. All 4 subsets were sorted in doublet.
From these 8 populations total RNA was isolated using the RNeasy Mini Kit (Qiagen, Cologne, Germany) and treated with DNAse1. Single-stranded T11G-, T11C-, and T11A-primed cDNA libraries were generated and used for differential display, as described by GeneHunter (Nashville, TN).
Gimap4 was identified independently with 2 arbitrary primers (APs) (AP03, AAGCTTTGGTCAG; and AP04, AAGCTTCTCAACG; underlined nucleotides are not homologous to genomic sequence) in combination with the T11G primers. Differential expression was further confirmed using gene-specific primer sets and Western blotting.
The full-length cDNA was derived by 5′ rapid amplification of cDNA ends (5′RACE) (Clontech, Mountain View, CA).
Derivation of rabbit anti-Gimap4 antibodies
Three peptides encompassing 20 to 30 amino acids (aa's) of the Gimap4 protein were synthesized and are listed in the order in which they occur within the protein: MEVQCGGAGFIPESSRSSHEL, KDDLEDTDIHEYLEKAPKFFQ, and IQKETLRMQELYREELEREKARIRREYEEQ. The synthetic peptides were conjugated to the carrier protein KLH. For each peptide, one rabbit (NZW strain) was primed with the conjugated peptide (1 mg) in complete Freund adjuvant and boosted on days 14 (0.3 mg), 21 (0.2 mg), and 28 (0.1 mg) with the conjugated peptide in incomplete Freund adjuvant. Sera were isolated on days 0, 35, 77, 112, and 119 to check for presence of peptide-specific antibodies by Western blot. To isolate secondary IgG antibodies from sera, Gamma Bind Plus Sepharose column (Bio-Rad, Munich, Germany) was used.
Phosphorylation studies by 2D gels
T cells were purified from wild-type splenocytes by depletion of B cells with anti-CD19 beads (AutoMACS; Miltenyi Biotec, Utrecht, The Netherlands), stimulated with 2 μg/μL concanavalin A (ConA) for 3 days or with 10 ng/mL PMA and 1 ng/mL ionomycin for 10, 20, and 60 minutes, harvested, and lysed in extraction buffer. The 2D gels were performed according to the Bio-Rad ReadyStrip IPG Strip instruction manual using 7 cm, pH 3-10 strips.
Calmodulin-binding assay
The Gimap4 protein was produced using the in vitro Transcription/Translation Kit (Promega). The pulldown was performed according to Seraphin Lap Tap protocol at EMBL (European Molecular Biology Laboratory) Heidelberg. In short, a 2 μL 35S-radioactive aliquot was washed in binding buffer and then incubated with 50 μL calmodulin (CaM)–bead suspension (Stratagene) either in the presence or absence (EGTA) of calcium for 1 hour at 4°C under rotation. Beads were isolated by centrifugation and washed 3 times. The bound protein was released from the beads by boiling the samples for 3 minutes at 95°C in reducing Laemmli sample buffer. Calmodulin (CaM)–bound proteins were resolved by gel electrophoresis and detected by Fluorescent image analyzer 3000 series (Fujifilm, Dublin, Ireland).
Isolation of T cells
T cells were isolated from mouse spleens using Becton Dickinson Mouse T lymphocyte Enrichment Set or FACS with anti-CD3 or anti-CD90. Cells prepared in this manner were typically more than 98% pure by FACS analysis.
Flow cytometry
Single cells from thymus or spleen were obtained by mincing through cell strainers, and red blood cells were lysed. Cells were incubated for 15 minutes on ice with specific antibodies conjugated to FITC, PE, or APC or biotin as indicated; biotinylated monoclonal antibodies were revealed with streptavidin-PerCP. All antibodies were purchased from BD PharMingen (San Diego, CA). Analysis was performed on a FACS Calibur using CellQuest software. Viable cells were gated on the basis of PI exclusion. For intracellular staining of Gimap4, a mixture of 3 antibodies was conjugated to FITC, which was incubated with permeabilized splenocytes of C57BL/6 mice.
Apoptosis assays
Splenic T cells (0.2 106 cells per well from Gimap4 ko and heterozygous Gimap4 littermates were incubated with various apoptotic stimuli for up to 3 days in 96-well plates. For inhibition studies cells were incubated in the presence or absence of DEVD (Calbiochem, Darmstadt, Germany). Apoptotic cells were identified by annexin V and PI staining according to the apoptosis detection kit from Becton Dickinson. Active capase-3 was measured according to the recommended protocol using rabbit anticaspase-3 and anti–rabbit Ig-PE The mitochondrial membrane potential was determined by incubation of cells with 1 μM tetramethylrhodamine ethyl ester (TMRE; Molecular Probes, Leiden, The Netherlands) for 30 minutes at 37°C. Cytochrome c release was measured as described elsewhere.23
Quantitative real-time PCR
Total RNA was used for cDNA synthesis. Sequences for primers and fluorescence-labeled probe (5′FAM, 3′TAMRA) for all 9 Gimap genes were selected using the primer sequence software (PE Biosystems, Foster City, CA) and ordered at Sigma (Haverhill, United Kingdom). As housekeeping gene, GAPDH was used, for which primers and probes were commercially available (Applied Biosystems, Foster City, CA). Standard curves for control and marker gene expression were generated using serially diluted cDNA. Runs and analyses were performed according to TaqMan Universal PCR Master Mix Protocol (Applied Biosystems) using ABI PRISM 7700 Sequence Detector. Fluorescence was measured from 0 to 40 PCR cycles and resulted in the threshold cycle (CT) value for each dilution and target. The quantities found for GAPDH control and target genes were used to calculate the relative quantity of gene expression.
Confocal microscopy
Images were obtained by using a Leica TCS NT confocal laser-scanning microscope equipped with a 63 ×/1.32 oil-immersion objective lens (Leica Microsystems, Heidelberg, Germany). Leica Confocal 2.4 software (Leica Microsystems) and Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA) were used for image processing.
Results
Identification and cloning of Gimap4, a gene that is differentially expressed during T-cell development
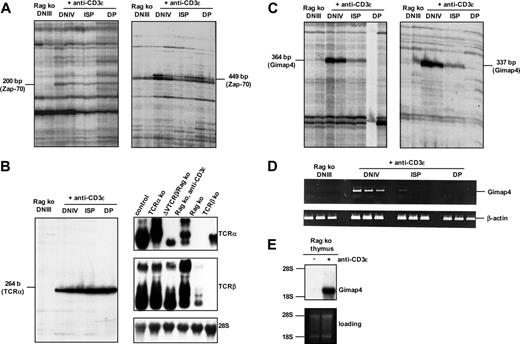
The development of thymocytes from recombination-activating gene-deficient mice (Rag ko) is blocked at the DNIIIE stage. Crosslinking of CD3 molecules, occurring at very low levels on the surface of these thymocytes, allows development up to the DP stage in vivo.9 This antibody-mediated induction of thymocyte differentiation provides a unique possibility to enrich in vivo the low frequent subsets DNIII, DNIV, ISP, as well as DP and sort these efficiently to high purity (not shown). In combination with differential mRNA display or microarray analysis, this system provides an effective strategy to identify genes regulated by the pre-TCR checkpoint. Gene loci, known to be transcriptionally activated upon pre-TCR–induced differentiation, the protein tyrosine kinase Zap-70 (Figure 1A), and germ-line TCRα (Figure 1B), were identified by cloning and sequencing. Both transcripts were indeed found in DNIV, ISP, and DP but not in untreated, differentiation-arrested DNIII thymocytes of Rag ko mice.
Besides Zap-70, another message was identified independently with 2 arbitrary primers in T11G-primed cDNA libraries. Based on the differential display analysis, this message appears absent in Rag ko thymocytes, present at high levels in DNIV, decreased in ISP, and completely absent in DP thymocytes (Figure 1C). Because our differential mRNA display limits sequence information to the 3′ end of the message, the lacking 5′ part was identified by rapid amplification of 5′ cDNA ends (5′RACE). The full-length cDNA covers 1.8 kb, and its open reading frame (ORF) (984 bp) encodes a 328-aa residue polypeptide, initially described as immune-associated nucleotide-1 (IAN-1), a marker for positive selected thymocytes.11 It is a member of a novel GTPase family,10 expressed in mammals, and recently renamed Gimap4 (GTPases of the immunity-associated protein 4).19,24 Gimap4 is a member of 8 gimap genes that are encoded within a single gimap gene cluster on mouse chromosome 6B (about 120 kb in length) and on human chromosome 7q36.1 (about 300 kb in length).19
To verify the transient expression of Gimap4 during early thymocyte development, semiquantitative reverse transcriptase (RT)–PCR was performed by making use of gene-specific forward and reverse primers located in exons 2 and 3 of the Gimap4 locus, respectively (Figure 1D). The data confirmed the differential expression deduced from the mRNA display analysis. The critical dependence of Gimap4 expression on CD3-mediated T-cell differentiation was further revealed by Northern blot analysis of total RNA extracted from whole thymi of anti-CD3ϵ–treated and nontreated Rag2 ko mice. In agreement with our 5′RACE analysis, a single message of 1.8 kb was found that was absent in the thymus of nontreated Rag ko mice (Figure 1E). These data indicate that Gimap4 is directly or indirectly induced by CD3 signals and is lacking in TCR-deficient T-cell progenitors as well as thymic epithelial cells and other minor cell types present in the thymus.
The Gimap4 cDNA encodes a lymphoid signaling protein
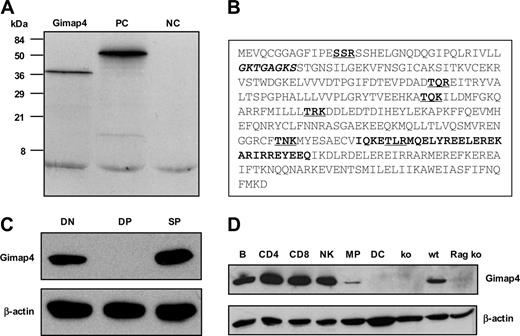
The ORF of the full-length Gimap4 cDNA predicts a hydrophilic protein of 38 kDa, where on average every third residue is charged. Consistent with the predicted molecular weight, a single protein product of 38 kDa was detected in an in vitro transcription/translation system using full-length cDNA as a template (Figure 2A). A Prosite motif search revealed several interesting motifs (Figure 2B): an amino-terminal ATP/GTP-binding P loop,25 a consensus sequence consisting of a glycine-rich sequence, followed by a conserved lysine and a serine or threonine (GxxxxGKT/S). Furthermore, a carboxy-terminal IQ domain, known to bind the second messenger molecule CaM.26 In most proteins, the complete IQ motif consists of about 23 residues. The consensus sequence comprises a hydrophobic aa (not necessarily an isoleucine) followed by a glutamine, and 3 arginines wherefrom the last one can be substituted by a lysine (hydrophobQxxxRxxxxRxxxR/K). Interestingly, this motif has not been earlier defined for Gimap4 and appears to be absent in other family members (Prosite; Swiss Institute of Bioinformatics, Basel, Switzerland). Moreover, 6 dispersed PKC phosphorylation sites are predicted, the inner 4 of which are conserved between the mouse and human. The diverse motifs strongly point toward a role of Gimap4 in signal transduction.
Differential mRNA display of T-cell subsets derived from the anti-CD3–inducible T-cell differentiation system. (A) Arrested double-negative DNIII thymocytes from untreated as well as DNIV immature single-positive (ISP) and double-positive (DP) thymocytes from anti-CD3ϵ–treated Rag ko mice were sorted in doublet, and total RNA was used for differential mRNA display. Zap-70 mRNA was identified twice with the 2 arbitrary 5′ primers AP02 (200 bp, left) and AP11 (449 bp, right) in combination with T11G primers. (B) Germ-line TCRα transcripts were found with the primer set AP16 and T11G (264 bp). Northern blot of total RNA from thymi of the indicated mice (right panels) confirmed the onset of germ-line TCRα transcription in anti-CD3ϵ–induced Rag ko thymocytes. (C) With 2 different primer sets, AP03/T11G (left) and AP04/T11G (right), a single differentially expressed cDNA species was identified independently. (D) Semiquantitative PCR with gene-specific primers verified the differential expression of Gimap4; depicted are 40, 30, and 20 cycles from left to right. (E) Northern blot of RNA from thymi of anti-CD3ϵ–treated or nontreated Rag ko mice confirmed quantitatively the expression and the CD3-dependent induction of Gimap4. A labeled cDNA probe revealed a single message of 1.8 kb.
Differential mRNA display of T-cell subsets derived from the anti-CD3–inducible T-cell differentiation system. (A) Arrested double-negative DNIII thymocytes from untreated as well as DNIV immature single-positive (ISP) and double-positive (DP) thymocytes from anti-CD3ϵ–treated Rag ko mice were sorted in doublet, and total RNA was used for differential mRNA display. Zap-70 mRNA was identified twice with the 2 arbitrary 5′ primers AP02 (200 bp, left) and AP11 (449 bp, right) in combination with T11G primers. (B) Germ-line TCRα transcripts were found with the primer set AP16 and T11G (264 bp). Northern blot of total RNA from thymi of the indicated mice (right panels) confirmed the onset of germ-line TCRα transcription in anti-CD3ϵ–induced Rag ko thymocytes. (C) With 2 different primer sets, AP03/T11G (left) and AP04/T11G (right), a single differentially expressed cDNA species was identified independently. (D) Semiquantitative PCR with gene-specific primers verified the differential expression of Gimap4; depicted are 40, 30, and 20 cycles from left to right. (E) Northern blot of RNA from thymi of anti-CD3ϵ–treated or nontreated Rag ko mice confirmed quantitatively the expression and the CD3-dependent induction of Gimap4. A labeled cDNA probe revealed a single message of 1.8 kb.
To characterize Gimap4 at the protein level, rabbit polyclonal antibodies were raised against Gimap4-specific peptide sequences. Pooled antibodies specifically detected one protein of the expected size in total splenocytes and no expression in splenocytes of the Gimap4 ko mice (Figures 2D, 5). Western blot analysis of sorted DN (comprising DNI, DNII, DNIIIE, as well as pre-TCR–positive DNIIIL and DNIV), DP, and SP thymocytes confirmed the absence of Gimap4 in DP thymocytes. The presence of Gimap4 in the DN fraction is in line with the presence of β-selected DNIIIL and DNIV subsets in this fraction. Interestingly, Gimap4, while absent in DP thymocytes, is reexpressed in SP thymocytes (Figure 2C).
Characterization of the GTPase family member Gimap4. (A) In vitro transcription/translation of Gimap4 cDNA revealed a 38 kDa protein. Positive control (PC) (51 kDa luciferase) and negative control (NC) (the empty vector) are included. (B) A motif search revealed a P loop (bold, italic), an IQ domain (bold), and several PKC phosphorylation sites (bold, underlined). (C) The protein expression pattern of Gimap4 was determined by Western blotting on sorted DN, DP, and SP thymocytes using polyclonal rabbit antibodies directed against various parts of mouse Gimap4. Blots were reprobed with anti–β-actin as a loading control. (D) Western blot of sorted hematopoietic subsets: B cells (B), CD4+ T cells, CD8+ T cells, natural killer cells (NK), macrophages (MP), and dendritic cells (DC). Thymocytes from a Rag ko mouse and splenocytes from Gimap4 ko (see Figure 5) served as a negative control; splenocytes from a wild-type (wt) mouse, as a positive control.
Characterization of the GTPase family member Gimap4. (A) In vitro transcription/translation of Gimap4 cDNA revealed a 38 kDa protein. Positive control (PC) (51 kDa luciferase) and negative control (NC) (the empty vector) are included. (B) A motif search revealed a P loop (bold, italic), an IQ domain (bold), and several PKC phosphorylation sites (bold, underlined). (C) The protein expression pattern of Gimap4 was determined by Western blotting on sorted DN, DP, and SP thymocytes using polyclonal rabbit antibodies directed against various parts of mouse Gimap4. Blots were reprobed with anti–β-actin as a loading control. (D) Western blot of sorted hematopoietic subsets: B cells (B), CD4+ T cells, CD8+ T cells, natural killer cells (NK), macrophages (MP), and dendritic cells (DC). Thymocytes from a Rag ko mouse and splenocytes from Gimap4 ko (see Figure 5) served as a negative control; splenocytes from a wild-type (wt) mouse, as a positive control.
To examine the expression of Gimap4 in the hematopoietic system, various cell types (B cells, CD4 T cells, CD8 T cells, natural killer cells, macrophages, and dendritic cells) were sorted and analyzed by RT-PCR (not shown) and Western blotting (Figure 2D). From this analysis, expression of Gimap4 appeared to be restricted to the lymphoid lineage. The marginal detection in macrophages is likely due to a contamination of lymphocytes (about 2%).
In summary, Gimap4 mRNA and protein expression follow 2 critical TCR-mediated checkpoints of T-cell development: β-selection and positive selection. Additionally, Gimap4 appears to be a lymphoid-specific signaling molecule.
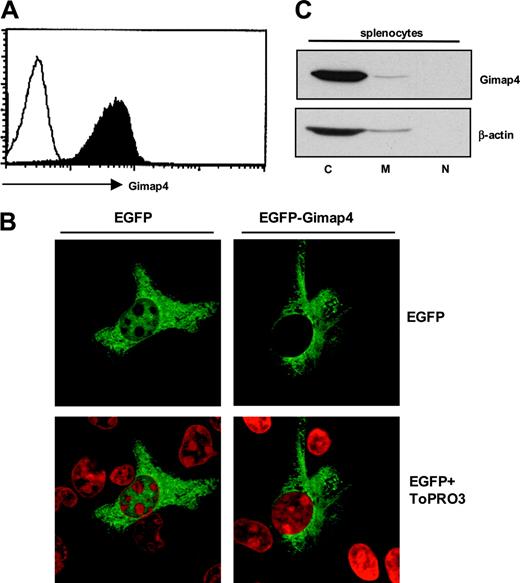
Distribution and subcellular localization of Gimap4. (A) Flow cytometric detection of Gimap4. Splenocytes from a wild-type mouse were intracellularly stained with Gimap4-fluorochrome (black), and nonstained cells were used as comparison (white), showing that expression of Gimap4 on lymphocytes is ubiquitous. (B) HeLa cells were transiently transfected with an N-terminal Gimap4-EGFP fusion protein. Cells were incubated with ToPRO3 for nuclear staining (red), and confocal microscopy was used to visualize the localization of Gimap4. HeLa cells transfected with the empty vector served as a control. (C) Different cellular fractions (cytosol, C; membranes, M; nucleus, N) were isolated from splenocytes,27 and Western blot analysis was performed with anti-Gimap4.
Distribution and subcellular localization of Gimap4. (A) Flow cytometric detection of Gimap4. Splenocytes from a wild-type mouse were intracellularly stained with Gimap4-fluorochrome (black), and nonstained cells were used as comparison (white), showing that expression of Gimap4 on lymphocytes is ubiquitous. (B) HeLa cells were transiently transfected with an N-terminal Gimap4-EGFP fusion protein. Cells were incubated with ToPRO3 for nuclear staining (red), and confocal microscopy was used to visualize the localization of Gimap4. HeLa cells transfected with the empty vector served as a control. (C) Different cellular fractions (cytosol, C; membranes, M; nucleus, N) were isolated from splenocytes,27 and Western blot analysis was performed with anti-Gimap4.
Gimap4 resides in the cytosol in resting cells
To address the question of whether Gimap4 is expressed constitutively in all lymphocytes or differentially in subsets, an intracellular staining of splenocytes with fluorochrome-conjugated Gimap4-specific polyclonal antibodies was performed. Flow cytometry demonstrates a positive staining of all lymphocytes (Figure 3A).
To localize Gimap4 at the subcellular level, we transfected HeLa cells with a Gimap4-EGFP fusion protein. Confocal microscopy analysis showed that Gimap4-EGFP resides in the cytoplasm and, compared with the EGFP control, is excluded from the nucleus (Figure 3B). No colocalization with mitochondria could be found in HeLa cells under nonstimulating conditions (not shown). The subcellular distribution was further addressed by Western blot analysis of cytosolic, membrane, and nuclear fractions of lymphocytes. Because Gimap4 is not processed proteolytically, the 2 putative myristoylation motifs at residue 40-45 and 88-93 (Scan-Prosite) are likely nonfunctional. In accordance with the lack of a functional myristoylation site, a transmembrane domain, and a nuclear localization sequence, most Gimap4 resides in the cytosol, excluded from the nucleus. The minor signal in the membrane fraction either relates to a specific translocation or a technical problem in isolating pure membrane fractions (Figure 3C).
Functional characterization of the Gimap4 protein motifs
Besides a functional GTPase domain10 characteristic for the Gimap protein family,19 Gimap4 contains other conserved protein motifs. To investigate their functional relevance, several assays were performed.
First, we investigated whether the C-terminal IQ domain of Gimap4 is capable of binding CaM. An IQ domain mutant was derived by site-directed mutagenesis; 3 critical arginine residues within the IQ consensus sequence were substituted by glutamines, which is sufficient to prevent CaM binding.28 35S-radiolabeled wild-type and mutant Gimap4 proteins were produced in vitro, and equal amounts were used in a pulldown with agarose CaM beads in the presence or absence of calcium ions. Wild-type Gimap4 was found to bind CaM only in the absence of Ca2+ (Figure 4A). The interaction between CaM and Gimap4 is likely to be mediated by the IQ domain because mutations in this domain abolish interaction. Thus, the C-terminal IQ domain of Gimap4 appears to be functional and, like most IQ-domain containing proteins, Gimap4 is found to bind CaM only at low Ca2+ levels.
Murine Gimap4 contains 6 putative PKC phosphorylation sites; the central 4 are conserved between the mouse and human (Figure 2B). To determine if these sites are targets of PKC-mediated phosphorylation, T cells were stimulated with PMA/ionomycin in the absence or presence of the PKC inhibitor rottlerin for the indicated time points (Figure 4B). Equal amounts of protein from NP-40 lysates were separated by 2D gel electrophoresis followed by immunoblotting with Gimap4 polyclonal antibodies. In this assay, phospho-Gimap4 species are identified by a shift in their isoelectric point. The isoelectric point shift found after PMA/ionomycin stimulation suggests phosphorylation of Gimap4. While phosphorylation occurs rapidly in the presence of PMA/ionomycin, phospho-Gimap4 is not found in rottlerin-treated samples, indicating that Gimap4 is a PKC target. Similar results were found with the Roche (Almere, The Netherlands) inhibitor Ro 318220 (not shown). A critical role for PKCθ has been excluded because phosphorylation of Gimap4 appears normal in PMA/ionomycin–stimulated PKCθ-deficient T cells (not shown).
To further examine Gimap4 phosphorylation upon stimulation, freshly isolated peripheral splenocytes were incubated in the absence or presence of the pan–T-cell activator ConA. While only low steady state levels of phospho-Gimap4 were found in nonactivated lymphocytes, phospho-Gimap4 levels increase dramatically in ConA-stimulated splenocytes (Figure 4C). To determine the duration of Gimap4 phosphorylation, primary splenocytes were stimulated with PMA/ionomycin for the indicated time points (Figure 4D). While again a low steady state phosphorylation is found at time point zero, phospho-Gimap4 levels increased significantly within 10 minutes of PMA/ionomycin stimulation and diminished completely after 40 to 60 minutes. The rapid phosphorylation and dephosphorylation/degradation suggests a tight post-translational control of Gimap4. This would be in accordance with our finding that the mRNA expression level appears unaltered regardless of the activation status, as examined by semiquantitative RT-PCR, whereas protein levels decrease in T cells cultured for 3 days with anti-CD3/28 (not shown and Cambot et al10 ).
Functional analysis of Gimap4 motifs. (A) Gimap4 protein was produced in an in vitro TS/TL system. A pulldown of the protein with CaM beads in the absence (–) or presence (+) of Ca2+ was used to determine the binding capacity of CaM to Gimap4. A functional mutant (IQ*), where the 3 arginine residues (R) within the IQ domain were substituted for 3 glutamine residues (Q), failed to bind CaM. Furthermore, the empty vector was used as a negative control (NC). (B) Phosphorylation level of Gimap4 in the presence or absence of ConA (2 mg/mL). (C) Equal amounts of cell lysates from T cells treated with 10 ng/mL PMA plus 1 ng/mL ionomycin in the absence or presence of the PKC inhibitor rottlerin (1 mM) for 20 minutes were used for 2D gel analysis, and immunoblots were performed using anti-Gimap4 polyclonal antibody. Phospho-Gimap4 species are identified by a shift in the isoelectric point. In vitro inhibition of PKC activity abrogates phosphorylation of Gimap4. (D) Gimap4 phosphorylation diminishes after 60 minutes of PMA/ionomycin stimulation.
Functional analysis of Gimap4 motifs. (A) Gimap4 protein was produced in an in vitro TS/TL system. A pulldown of the protein with CaM beads in the absence (–) or presence (+) of Ca2+ was used to determine the binding capacity of CaM to Gimap4. A functional mutant (IQ*), where the 3 arginine residues (R) within the IQ domain were substituted for 3 glutamine residues (Q), failed to bind CaM. Furthermore, the empty vector was used as a negative control (NC). (B) Phosphorylation level of Gimap4 in the presence or absence of ConA (2 mg/mL). (C) Equal amounts of cell lysates from T cells treated with 10 ng/mL PMA plus 1 ng/mL ionomycin in the absence or presence of the PKC inhibitor rottlerin (1 mM) for 20 minutes were used for 2D gel analysis, and immunoblots were performed using anti-Gimap4 polyclonal antibody. Phospho-Gimap4 species are identified by a shift in the isoelectric point. In vitro inhibition of PKC activity abrogates phosphorylation of Gimap4. (D) Gimap4 phosphorylation diminishes after 60 minutes of PMA/ionomycin stimulation.
Genomic organization of the gimap4 locus and its inactivation in the mouse germ line
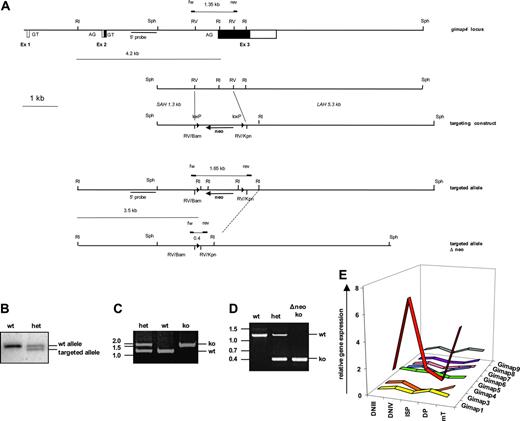
To determine the precise genomic structure of the gimap4 locus, a 129SVJ liver DNA–derived mouse genomic library was screened with a Gimap4 cDNA probe. Two independent lambda phages were identified, one of which contained the complete Gimap4 locus, and were partially sequenced to complete the organization of the mouse Gimap4 locus (Figure 5A). The full-length Gimap4 cDNA is encoded by 3 exons separated by 2 introns of 2215 and 3460 bp. Exon 1 is 28 bp in length, and noncoding, exon 2 is 72 bp in length, and the last 58 bp are coding. Most (309 residues of the 328) of the mouse Gimap4 protein is encoded by the last exon. Exon 3 is 1690 bp in length; the first 929 bp encode the N-terminal GTPase domain, 5 of the 6 putative PKC phosphorylation sites, as well as the C-terminal CaM-binding region. The last 761 bp of the Gimap4 message are untranslated (3′UTR).
Targeting of the Gimap4 locus. (A) An internal 1.1 kb EcoRV fragment (comprising the 3′ end of the intron and most of exon 3 encoding the GTPase domain) was replaced by a blunted 1.4 kb BamHI/KpnI selection cassette containing a floxed neomycin gene. (B) Gimap4-targeted mice were screened by Southern blot of EcoRI-digested tail DNA using a 3′-specific probe and (C) by PCR; a 1.35 kb and a 1.65 kb band identified the wt and the targeted allele, respectively. (D) Targeted Gimap4 ko's were also crossed with Cre deleter, and PCR analysis was performed; the wt allele is 1.35 kb, the ko allele 0.4 kb. (E) Total RNA from DNIII, DNIV, ISP, and DP thymic subsets, obtained from Rag-deficient mice triggered with or without anti-CD3, as well as RNA from peripheral T cells (mT) have been used for quantitative PCR for Gimap family members. GAPDH was used as an endogenous control for normalization of targets.
Targeting of the Gimap4 locus. (A) An internal 1.1 kb EcoRV fragment (comprising the 3′ end of the intron and most of exon 3 encoding the GTPase domain) was replaced by a blunted 1.4 kb BamHI/KpnI selection cassette containing a floxed neomycin gene. (B) Gimap4-targeted mice were screened by Southern blot of EcoRI-digested tail DNA using a 3′-specific probe and (C) by PCR; a 1.35 kb and a 1.65 kb band identified the wt and the targeted allele, respectively. (D) Targeted Gimap4 ko's were also crossed with Cre deleter, and PCR analysis was performed; the wt allele is 1.35 kb, the ko allele 0.4 kb. (E) Total RNA from DNIII, DNIV, ISP, and DP thymic subsets, obtained from Rag-deficient mice triggered with or without anti-CD3, as well as RNA from peripheral T cells (mT) have been used for quantitative PCR for Gimap family members. GAPDH was used as an endogenous control for normalization of targets.
To study the role of Gimap4 in vivo, we generated a Gimap4-null mutant mouse. To inactivate Gimap4 in the mouse germ line, a 7.7 kb SphI fragment containing exon 3 was subcloned. From this subclone, the targeting vector was derived by replacing an internal 1.1 kb EcoRV fragment comprising the 3′ end of intron 2, the splice-acceptor site, and the first 431 coding bases of exon 3, including the GTPase domain, by a 1.4 kb fragment containing a neomycin gene flanked by loxP sites (Figure 5A). An 8 kb SalI/NotI fragment was released and used to inactivate Gimap4 in E14 ES cells.22 Southern blot analysis with EcoRI-digested DNA and an external Sph probe revealed 2 independent clones that contained besides a wild-type a correctly targeted Gimap4 allele (4191 and 3523 bp). These clones were used to introduce the mutation into the mouse germ line and derive Gimap4-deficient mice (Figure 5B). While crossing the targeted Gimap4 allele onto the C57BL/6 background, we also deleted the floxed neomycin cassette by intercrossing the Cre deleter allele.29 A PCR with gene-specific primers located just outside the deleted EcoRV fragment was established to simplify the genotyping of the mice before (Figure 5C) and after deletion of the neomycin cassette (Figure 5D).
Southern blot, PCR, and RT-PCR analysis indicated the proper targeting of the Gimap4 allele. Western blot analysis of total lysates from Gimap4-proficient and -deficient lymphocytes further revealed a complete inactivation of gimap4 (Figure 2D).
Characterization of Gimap4-deficient mice
Despite the interesting expression pattern of Gimap4, in Gimap-deficient mice the lymphoid development, selection, as well as activation in vivo appear to occur normally as based on steady state numbers of cells in lymphoid organs, thymic selection, cytokine profiling, induction of an influenza virus-specific CD8+ T-cell response in vivo (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article), and competitive bone marrow reconstitution. Taken together, T-cell development, selection, and activation in vivo appear to occur normally in Gimap4-targeted mice.
When examining mRNA expression of other Gimap family members in T-cell progenitors, only Gimap4 was found to be distinctly regulated during development (Figure 5E). Furthermore, no significant differences were found comparing wild-type and Gimap4-deficient peripheral T cells (S.S., unpublished data, November 2005). These data argue for a nonredundant role of Gimap4 in lymphocyte biology, at least on a transcription level. However, at present we cannot exclude compensatory activation of other Gimap family members at the protein level.
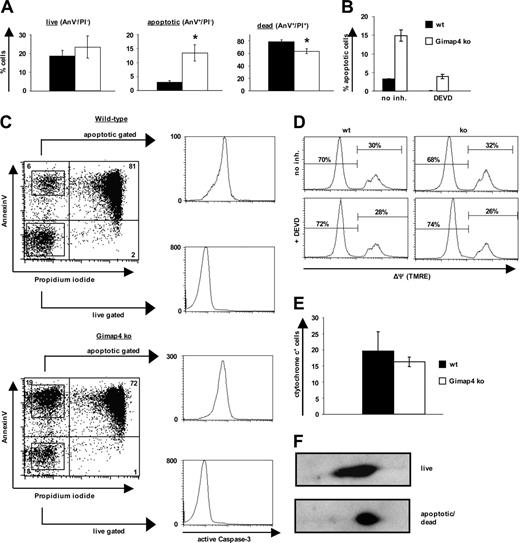
Gimap4 accelerates stress-induced death in T cells. Sorted T cells from Gimap ko mice and heterozygous littermates were incubated for 1 day in serumfree medium. Data are representative of at least 3 independent experiments using 3 mice in each experiment. Error bars indicate standard deviation. (A) Live, apoptotic, or dead cells were distinguished by fluorochrome–annexin V and PI labeling. The apoptotic population was always 2- to 4-fold higher in Gimap4-deficient T cells. Student t test revealed significant differences; asterisk represents P value of less than .005. (B) Sorted T cells were incubated in the presence or absence of the effector-caspase inhibitor DEVD (100 μM) and subsequently stained with fluorochrome–annexin V and PI. (C) After 1 day of serum deprivation, live and apoptotic T cells were sorted based on their annexin V and PI staining pattern and stained for intracellular active caspase-3. (D) For the mitochondrial membrane potential (ΔΨ) cells were stained with the voltage-dependent membrane permeable dye TMRE. (E) Cells were stained with fluorochrome-labeled cytochrome c antibody. (F) Equal amounts of proteins from lysates from purified live and dying T lymphocytes after 1 day of serum starvation were used for 2D analysis.
Gimap4 accelerates stress-induced death in T cells. Sorted T cells from Gimap ko mice and heterozygous littermates were incubated for 1 day in serumfree medium. Data are representative of at least 3 independent experiments using 3 mice in each experiment. Error bars indicate standard deviation. (A) Live, apoptotic, or dead cells were distinguished by fluorochrome–annexin V and PI labeling. The apoptotic population was always 2- to 4-fold higher in Gimap4-deficient T cells. Student t test revealed significant differences; asterisk represents P value of less than .005. (B) Sorted T cells were incubated in the presence or absence of the effector-caspase inhibitor DEVD (100 μM) and subsequently stained with fluorochrome–annexin V and PI. (C) After 1 day of serum deprivation, live and apoptotic T cells were sorted based on their annexin V and PI staining pattern and stained for intracellular active caspase-3. (D) For the mitochondrial membrane potential (ΔΨ) cells were stained with the voltage-dependent membrane permeable dye TMRE. (E) Cells were stained with fluorochrome-labeled cytochrome c antibody. (F) Equal amounts of proteins from lysates from purified live and dying T lymphocytes after 1 day of serum starvation were used for 2D analysis.
Gimap4 accelerates T-cell death
Given the high cellular turnover within the immune system, effective apoptosis plays an essential role not only during formation of the primary immune repertoire but also in controlling the size of antigen-selected lymphocyte pool during an infection. A characteristic feature of apoptosis is PS exposure followed by plasma membrane permeabilization; both can be readily detected by flow cytometry using fluorochrome-conjugated annexin V and the accumulation of PI in the nucleus, respectively.30
Because present data suggest a function of the Gimap family members in survival and proliferation, a possible role of Gimap4 in programmed cell death of T cells was examined by comparing wild-type versus Gimap4 ko cells. For this purpose, mature splenic T cells were positively sorted to ensure that apoptotic cells were not removed by macrophages. Very similar results were obtained using negatively enriched T cells to exclude the possibility of a receptor-triggered phenotype. To induce cell death, T cells were incubated for up to 2 days either in serumfree medium or exposed to γ-irradiation, etoposide, and dexamethasone under conventional conditions. PS exposure and PI uptake were measured by flow cytometry. These intrinsic stimuli caused significant cell death in both wild-type and Gimap4-deficient T cells. Interestingly, while the number of live cells (AnV–/PI–) was indistinguishable between ko and wild-type T cells, the number of apoptotic cells (AnV+/PI–) was 2- to 4-fold higher in Gimap4-deficient T cells, and dead cells (AnV+/PI+) were reduced in accordance with the increase in the apoptotic population. Exemplarily, the effect of serum starvation is shown in Figure 6A; a kinetic analysis of this phenotype is shown in Figure S2.
The increased frequency of apoptotic T cells (AnV+/PI–) from Gimap4 ko mice could be inhibited by the effector-caspase inhibitor DEVD (Figure 6B). Similar results were found with the pancaspase inhibitor zVAD (not shown). These data indicate that the Gimap4 phenotype is caspase dependent and thus apoptosis related.
To determine whether the delay in membrane permeabilization relates to inappropriate caspase-3 activation, live and apoptotic cells were sorted on the basis of annexin V/PI pattern and consecutively stained intracellularly for active caspase-3 (Figure 6C). Nonapoptotic T cells stained negatively, and apoptotic T cells stained strongly positive for active caspase-3. Because all apoptotic T cells, independent of the genotype, have active caspase-3, the induction phase of apoptosis seems to be unaffected in the absence of Gimap4. Based on these considerations, we propose a role of Gimap4 in the execution rather than the induction phase of apoptosis.
Consistent with the assumption that Gimap4 acts downstream of caspase-3 activation and PS exposure, the frequency of cells with mitochondrial depolarization (ΔΨm) is not impaired as measured by TMRE (Figure 6D), and the frequency of cells with high cytochrome c levels was comparable between wild-type and ko T cells (Figure 6E).
Because our data suggested that Gimap4 is a target of PKC phosphorylation, we compared the phosphorylation status of wild-type Gimap4 in live versus dying cells. The AnV–/PI– population as well as a mixture of AnV+/PI– and AnV+/PI+ populations were sorted, and equal amounts of protein from NP-40 lysates were separated by 2D gel electrophoresis followed by immunoblotting (Figure 6F). A higher level of phosphorylation was found in apoptotic/dead cells in comparison with live cells, suggesting a necessity of Gimap4 phosphorylation to accelerate T-cell death. Second messengers could lead to modulation of the activity of Gimap4 through modification of its phosphorylation status.
In summary, these data indicate a role for Gimap4 in the execution phase of programmed cell death to accelerate T-cell death.
Discussion
By using the inducible anti-CD3 system in vivo9 we identified Gimap4. Its developmentally regulated expression pattern during T-cell development is similar in B cells (C.D. and H.J., unpublished data, May 1998). Despite previous data on the cloning, expression, and GTPase activity of Gimap4, the role of Gimap4 in lymphocyte physiology remains ill defined.10,11 The ATP/GTP-binding P loop of Gimap4 is part of the prototype AIG1 domain found in the pathogen-induced plant protein Aig1,31 which was suggested to be associated with programmed cell death.32 The AIG1 domain contains 5 motifs (G1 to G5) characteristic for the GTP-binding sites of most GTPases and is common to all Gimap proteins.19 We here report on the existence of a functional C-terminal CaM-binding IQ domain and conserved target sites of PKC-mediated phosphorylation in Gimap4 implicating a role in lymphocyte signaling. The lack of a transmembrane domain and a functional N-myristoylation site in the human and mouse Gimap4 protein is in agreement with our finding that most endogenous Gimap4 resides in the water-soluble fraction of the cytosol (ie, not associated with cytoplasmic organelles) and outside the nucleus. These data contrast with a previous report suggesting that Gimap4 has a transmembrane region and resides at the endoplasmic reticulum (ER) and Golgi apparatus, as deduced in CHO-K1 cells overexpressing a V5-tagged hGimap4.19
Gimap4 protein is lacking in any established lymphoid cell line (not shown and Poirier et al11 ) and shut off gradually in T cells cultured ex vivo for several days (not shown and Cambot et al10 ), hampering a proper functional analysis of Gimap4 in vitro. We therefore decided to address the function of Gimap4 in vivo. To inactivate the gimap4 locus in the mouse germ line, we first completed the analysis of the mouse Gimap locus by sequencing gimap4-specific inserts of a 129SVJ fetal liver–derived lambda phage library. When verifying the exon/intron organization of mGimap4 it became clear that the 1.8 kb transcript of Gimap4 is encoded in 3 rather than 2 exons (see “Results” and Figure 5), identical to the exon/intron organization of the rat and human gimap4 loci.
The best-studied Gimap member is Gimap5. The functional analysis of Gimap5 has been greatly advanced by the identification of the BB diabetes-prone rat, which carries a spontaneous deletion in the Gimap5 locus. In fact, the functional analysis of Gimap5 preceded its identification. The recessive mutation is associated with severe lymphopenia (lyp mutation) and the development of insulin-dependent diabetes mellitus (Iddm1 mutation). Recently, rat Ian5 (Gimap5) was cloned and proven to be responsible for severe lymphopenia (lyp mutation) and the development of insulin-dependent diabetes mellitus (iddm1) in the BB diabetes-prone rat.15,16,18 In addition, there are several indications for a function of Gimap family members in survival and proliferation in the mouse,33-36 rat,14-17,37,38 as well as human.10,34,39,40 Specifically, there are indications for an antiapoptotic activity of rat37 and human34 Gimap5, mouse Gimap8,35 as well as a survival function of mouse Gimap136 and Gimap3.33
These data strongly argue for a role of Gimap proteins in controlling apoptotic processes and thereby the homeostasis within the lymphoid lineage. Because Gimap4 is expressed transiently in β- as well as positively-selected thymocytes, but not in the major apoptosis-prone DP subset (more than 85%), we focused our functional analysis of Gimap4 on mature T cells. Significant differences were found regarding the sensitivity of T cells to several intrinsic stimuli in mature Gimap4-deficient T cells. In the absence of Gimap4, apoptosis appears to be delayed at the transition from apoptotic to dead cells. While the number of viable T cells (AnV–/PI–) remained unaltered, the number of apoptotic cells (AnV+/PI–) was 2- to 4-fold increased, and dead cells (AnV+/PI+) were decreased relative to the increase of apoptotic cells in the Gimap4-deficient samples. Exposure of PS is a direct consequence of a loss of phopholipid asymmetry in the plasma membrane.41 The cell-surface exposure of PS is dependent on the activation of a Ca2+-induced flip-flop activity of a so-called phospholipid scramblase.42 The finding that PS exposure is not impaired in the absence of Gimap4 indicates that Gimap4 is likely not to be involved in controlling the scramblase. Rather, Gimap4 might be involved in a consecutive step, controlling directly or indirectly the permeabilization of the plasma membrane of dying cells, a phenomenon that is mechanistically poorly understood.
The fact that Gimap4 protein expression is lost after 2 to 3 days in culture makes it difficult to perform standard experiments testing extrinsic stimuli or activation-induced cell death, because these assays require several days of culturing. Therefore, at present no statement can be made in regard to extrinsic stimuli.
In vivo, apoptotic cells are readily detected by the presence of PS in the outer layer of the plasma membrane.43 PS-specific receptors on phagocytes trigger the uptake of apoptotic cells. The early uptake of apoptotic cells, prior to membrane permeabilization, could explain the absence of a phenotype regarding the cellularity and development of lymphocytes in Gimap4 ko mice. We speculate that Gimap4 accelerates the formation and clearance of apoptotic bodies to help phagocytes cope with the lymphoid remnants generated during lymphocyte development in primary lymphatic organs as well as in secondary lymphatic organs during the contraction phase of an immune response. Taking the enormous overlays and shared signaling pathways among the different death programs (ie, apoptosis- or necrosis-like programmed cell death),44-46 a role of Gimap4 in the latter process cannot be excluded.
Interestingly, ectopic expression of Gimap4 in mice causes growth retardation and premature graying (S.S., C.D., and H.J., December 1998). This phenotype is similar to the one found in Bcl-2 ko mice, arguing independently for an apoptotic function of Gimap4 in vivo, besides the apoptotic phenotype in vitro.
In summary, our data provide a role for Gimap4 in the execution of programmed cell death in T cells.
Prepublished online as Blood First Edition Paper, March 28, 2006; DOI 10.1182/blood-2005-11-4616.
Supported by The Netherlands Cancer Institute (grant SFN SFR 2.1.29).
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mark Dessing, Anita Pfauth, and Frank van Diepen from The Netherlands Cancer Institute for help with the cell sorting. We thank the caretaker teams of the animal facility and furthermore thank Jannie Borst, Stephen Tait, and Ton Schumacher for scientific discussions and comments.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal