Abstract
The translocation t(12;22) involves MN1 and TEL and is rarely found in acute myeloid leukemia (AML). Recently, it has been shown in a mouse model that the fusion protein MN1-TEL can promote growth of primitive hematopoietic progenitor cells (HPCs) and, in cooperation with HOXA9, induce AML. We quantified MN1 expression by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) in 142 adult patients with AML with normal cytogenetics treated uniformly in trial AML-SHG 01/99. AML samples were dichotomized at the median MN1 expression. High MN1 expression was significantly correlated with unmutated NPM1 (P < .001), poor response to the first course of induction treatment (P = .02), a higher relapse rate (P = .03), and shorter relapse-free (P = .002) and overall survivals (P = .03). In multivariate analysis, MN1 expression was an independent prognostic marker (P = .02) in addition to age and Eastern Cooperative Oncology Group (ECOG) performance status. Excluding patients with NPM1mutated/FLT3ITDnegative, high MN1 expression was associated with shorter relapse-free survival (P = .057). MN1 was highly expressed in some patients with acute lymphoblastic but not chronic lymphocytic or myeloid leukemia. MN1 was highly expressed in HPCs compared with differentiated cells and was down-regulated during in vitro differentiation of CD34+ cells, suggesting a functional role in HPCs. In conclusion, our data suggest MN1 overexpression as a new prognostic marker in AML with normal cytogenetics.
Introduction
The meningioma (disrupted in balanced translocation) 1 (MN1) gene, localized on human chromosome 22, was first cloned in 1995 from a patient with meningioma with translocation t(4;22)(p16;q11), which disrupts MN1 in its first exon.1 At the same time it was shown that TEL, an ETS transcription factor, was fused to MN1 in patients with myeloid leukemia or myelodysplastic syndrome containing the translocation t(12; 22)(p13;q11).2 The fusion protein MN1-TEL has transforming activity on NIH 3T3 cells and most likely acts as a disregulated transcription factor.3 Recently, the MN1-TEL fusion protein was studied in a mouse model, in which it was expressed under the control of the AML1 regulatory sequences. After long latency 30% of the MN1-TEL–expressing mice developed T-lymphoid tumors, whereas 10% of these mice developed severe anemia with altered myelopoiesis but not myeloid leukemia.4,5 However, MN1-TEL in cooperation with HOXA9, which is overexpressed in patients with acute myeloid leukemia (AML),6 induced AML in mice within 3 to 6 months after transplantation compared with 10 months if HOXA9-transduced cells were transplanted alone.5 In addition, MN1-TEL promoted the proliferation of normal myeloid and lymphoid progenitors and partially blocked their differentiation in vivo.4 Whereas an oncogenic function of the fusion protein MN1-TEL is now established in hematopoiesis, the role of MN1 alone remains to be defined.
van Wely et al7 showed that MN1 acts on the MSV-LTR as a transcription coactivator in retinoic acid receptor (RAR)–retinoic X receptor (RXR)–mediated transcription leading to a synergistic induction of expression when MN1 and the RAR-RXR ligand retinoic acid are combined. Furthermore, it has been shown that transcription is activated by recruitment of RAC3 and P300, both known coactivators of retinoic acid receptors.7 In osteoblasts, MN1 is a target of vitamin D3, which stimulates transcriptional activity of the vitamin D receptor and inhibits proliferation of osteoblasts.8 A mouse knock-out model of Mn1 showed that Mn1 plays a role in cranial bone development, because Mn1 null mice lack several cranial bones, exhibit cleft palate defects, and, because of these defects, die shortly after birth.9 Previously, we compared gene expression profiles of patients with AML with either good or poor response with the first course of induction chemotherapy. There we identified a predictive treatment-response signature containing MN1, which was associated with poor response to the first course of induction therapy.10 To investigate the role of MN1 overexpression in AML, we quantified MN1 expression in a uniformly treated cohort of adult patients with AML with normal karyotype and compared the prognostic relevance of MN1 with other prognostic factors, notably FLT3 internal tandem duplication (ITD), MLL, and the newly identified mutations of the NPM1 gene.11
We show that high MN1 expression is an independent prognostic marker in patients with AML with normal cytogenetics associated with a significantly worse day 15 response rate, shorter relapse-free survival (RFS), and shorter overall survival (OS). Moreover, MN1 is highly expressed in cell populations enriched for primitive hematopoietic cells and in patients with acute lymphoblastic leukemia but it is down-regulated on differentiation, thus suggesting roles both in normal hematopoiesis and leukemia.
Patients, materials, and methods
Patients and treatment
Diagnostic bone marrow (BM) or peripheral blood (PB) samples were analyzed from 142 adult patients (aged 16-60 years) with de novo or secondary AML (French-American British [FAB] classification M0-M2, M4-M7)12 and normal cytogenetics who had been entered into the multicenter treatment trial AML-SHG 01/99 (June 1999 to September 2004). For this molecular study, inclusion criteria were normal cytogenetics, the availability of a BM or PB sample from diagnosis, and at least one course of induction chemotherapy. Median percentage of blasts in diagnostic samples was 80% for BM (n = 104; data missing, n = 7) and 65% for PB (n = 30; data missing, n = 1). Written informed consent was obtained prior to therapy according to the Declaration of Helsinki, and the study was approved by the institutional review board of Hannover Medical School.
All patients received intensive, response-adapted double induction and consolidation therapy. Double induction therapy consisted of a course of IVA (idarubicin 12 mg/m2 on days 2, 4, and 6; etoposide 100 mg/m2 on days 3 through 7; and cytosine-arabinoside 100 mg/m2 continuously on days 1 through 7), followed by a second course of IVA (idarubicin 10 mg/m2 on days 2 and 4, etoposide 100 mg/m2 on days 2 through 6, and cytosinearabinoside 100 mg/m2 continuously on days 1 through 6) on day 21 in patients with a good response to the first course of induction therapy. Patients with a poor response to the first course of induction therapy were randomly assigned between a second course of IVA and a course of FLAG-Ida (fludarabin 30 mg/m2 days 1 through 4, cytosine-arabinoside 1 g/m2 days 1 through 4, granulocyte colony–stimulating factor (G-CSF) 5 μg/kg starting day 0, and idarubicin 8 mg/m2 days 1 and 3). Response to induction cycle 1 was assessed cytomorphologically on day 15 by the initial blast cell reduction: good response was defined as no blasts in peripheral blood, less than 5% blasts in bone marrow, and no extramedullary manifestation. Poor response was defined as residual blasts in peripheral blood, 5% or more blasts in bone marrow, or extramedullary AML manifestation. First consolidation therapy consisted of cytosine-arabinoside 1 g/m2 on days 1 through 4 and daunorubicin 45 mg/m2 days 5 and 6 or a second course of FLAG-Ida in patients who responded to the previous course of FLAG-Ida.
For second consolidation therapy, patients with normal karyotype were randomly assigned to either high-dose AraC/DNR (cytosine-arabinoside 3 g/m2 days 1 through 6 and daunorubicin 45 mg/m2 days 7 through 9) or myeloablative therapy (total body irradiation/cyclophosphamide or busulfan/cyclophosphamide), followed by autologous stem cell transplantation (SCT). Alternatively, patients were assigned to allogeneic SCT if an HLA-compatible family donor was available. In 129 patients information about the availability status of an HLA-compatible family donor was present at the time of diagnosis. In 36 (27.9%) of the 129 patients and in 29 (25.2%) of 115 patients achieving a complete remission (CR) after induction therapy, respectively, an HLA-compatible family donor was available.
In addition, diagnostic specimens were obtained from patients with chronic lymphocytic leukemia (CLL; n = 12; 3 BM and 9 PB specimens), chronic myeloid leukemia in chronic phase (n = 10; 4 BM and 6 PB specimens) or in blast crisis (n = 2 specimens from PB), acute lymphoblastic leukemia (ALL; n = 13; 6 BM and 7 PB specimens), and from healthy volunteers (n = 6 specimens from PB) after obtaining written informed consent. G-CSF–mobilized peripheral blood aphereses were obtained from 4 healthy donors after written informed consent.
Cytogenetic and molecular genetic analysis
Pretreatment samples from all patients were studied centrally by G-banding and fluorescence R-banding analysis and fluorescence in situ hybridization (FISH).13,14 Conventional cytogenetic studies were performed using standard techniques, and chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature.15 All specimens were also analyzed by FISH using a comprehensive DNA probe set allowing for the detection of the most relevant AML-associated genomic aberrations.14 In addition, diagnostic samples from all patients with AML were analyzed for mutations in FLT3 (ITDs and activation loop mutations at D835),16 MLL (partial tandem duplications [PTDs]),17 and NPM1 genes (NPM1, exon 12 mutations).11
Real-time RT-PCR
Total RNA from stored, frozen, Ficoll-separated mononuclear AML cell pellets was isolated using TRIZOL reagent (Invitrogen, Paisley, United Kingdom) and subsequently purified by a Qiagen RNeasy column (Qiagen, Hilden, Germany). Random hexamer priming and MuLV reverse transcriptase (Fermentas, Hanover, MD) were used to generate cDNA. Real-time reverse transcriptase–polymerase chain reaction (RT-PCR) was carried out on a LightCycler (Roche Diagnostics, Mannheim, Germany) using the QuantiTect SYBR Green PCR kit as described in the manufacturer's instructions (Qiagen). The following primers were used to measure RNA abundance (5′ to 3′): MN1 forward primer, GACGACGACAAGACGT-TGG, and MN1 reverse primer, GACAGACAGGCACTGCAAG; CD34 forward primer, AGGTATGCTCCCTGCTCCTT, and CD34 reverse primer, ATCCCCAGCTTTTTCAGGTC. Primers for MN1 and CD34 are intron spanning and do not function on genomic DNA. The expression of ABL was used as an endogenous control.18 The reactions were carried out in duplicate under the following conditions: 95°C for 15 minutes, then 45 cycles of 94°C for 15 seconds, 53°C (MN1) or 54°C (CD34) for 25 seconds, and 72°C for 15 seconds. Melting curve analyses were performed to verify amplification specificity. First, quantitative PCR reactions were carried out with 6-fold dilutions (each with 5 replicates) covering the expected detection range with a reference cDNA (ME1 cell line) to obtain standard curves for the target genes and ABL. Amplification efficencies were 1.91 for MN1, 1.9 for CD34, and 1.95 for ABL. On the basis of these curves, the relative concentrations (defined as 1.00 in ME1 cells) were calculated, based on the Second Derivative Maximum method using the LightCycler Relative Quantification Software. The intra-assay and interassay variation of 5 independent replicates was less than 0.1% for MN1, CD34, and ABL transcripts.
Cell sorting
Previously, MN1 has been identified as part of a gene signature of hematopoietic stem cells and stem cell-like AML cells.10,19 Therefore, we investigated MN1 expression levels in different hematopoietic cell subsets and during in vitro differentiation of CD34+ cells. CD34+ cells were enriched from G-CSF–mobilized peripheral blood mononuclear cells (PB MNCs) with an anti–human CD34 monoclonal antibody (mAb) labeled with magnetic beads on an affinity column (Miltenyi Biotec, Bergisch-Gladbach, Germany). The flow-through constituted the CD34– population. CD34+-enriched cells were either used for RNA extraction or incubated with phycoerythrin-conjugated anti–human CD34 and allophycocyanin-conjugated anti–human CD38 mAb (both Becton Dickinson, San Jose, CA). Finally, the cells were washed in phosphate-buffered saline and 10% fetal calf serum (FCS) and stained with propidium iodide (PI) to identify dead cells. The CD34+/CD38– population was sorted using the automatic cell-depositing unit on a fluorescence-activated cell sorting (FACS) flow cytometry system (FACSAria; Becton Dickinson). Cells positive for PI were excluded. The purity of cell populations was 80% to 97%.
In vitro differentiation of CD34+ cells
Immunomagnetically purified CD34+ cells were cultured in IMDM (Biochrom, Berlin, Germany) with 10% FCS (Biochrom) or in X-vivo 10 medium (Cambrex Bio Science Verviers, Verviers, Belgium) without addition of serum, 1% penicillin/streptomycin, and various cytokines, including stem cell factor (SCF), interleukin 3 (IL-3), granulocytemacrophage colony-stimulating factor (GM-CSF), FLT3-ligand (FL), thrombopoietin (TPO), interleukin 6 (IL-6), G-CSF, macrophage colony-stimulating factor (M-CSF; all PAN-Biotech, Aidenbach, Germany), and erythropoietin (EPO; R&D Systems, Wiesbaden-Nordenstadt, Germany). Cells were cultured in 6-well tissue culture plates seeded at an initial density of 0.5 × 105/mL in 2 mL/well. Cultures were semidepleted every 4 days. On days 4, 8, 12, and 16, medium and cytokines were replaced, and harvested cells were processed for RNA isolation. At various time points, cells were harvested for slide preparation, which were subsequently stained with Giemsa for morphologic analysis. Images were visualized using a Zeiss Axiovert 40 microscope (Carl Zeiss, Oberkochen, Germany) and a 100×/1.4 numerical aperture objective, with Zeiss Immersol medium. A Canon Powershot G2 camera and Canon ZoomBrowser EX 2.0 software (Canon, Krefeld, Germany) were used to capture images, which were subsequently processed using Adobe Photoshop version 6.0 (Adobe Systems, San Jose, CA).
Statistical analysis
To evaluate the effect of MN1 expression values on clinical outcome without seeking an optimal cutpoint, AML samples were dichotomized at the median value (0.0646; range, 0-6.85; fold-change in expression between highest and lowest measurable value, 2900-fold) and divided into 2 expression groups: a low MN1 group with MN1 values below the median value and a high MN1 group with MN1 values above the median value. Similarly, CD34 expression values were dichotomized at the median value (0.273; range, 0-47.5; fold-change in expression between highest and lowest measurable value, 23 750-fold). Survival analysis was repeated using maximally selected log-rank statistics20 to identify the optimal cutpoint of MN1 expression, and results are shown in part in Table S1 and Figure S1 (available at the Blood website; see the Supplemental Materials link at the top of the online article). The definition of CR followed the recommended criteria.21 Overall survival end points, measured from entry into the prospective study, were death (failure) and alive at last follow-up (censored).21 Relapse-free survival end points, measured from the date of documented CR, were relapse (failure), death in CR (failure), and alive in CR at last follow-up (censored).21 Pairwise comparisons between patient characteristics were performed by Student t test for continuous variables and by chi-square test for categorical variables. The Kaplan-Meier method and log-rank test were used to estimate the distribution of RFS and OS and to compare differences between survival curves, respectively. To further investigate the effect of increasing expression of MN1, a Cox proportional hazards model was constructed, adjusting for potential confounding covariates, using the forward selection method.22 To provide quantitative information on the relevance of results, 95% confidence intervals (95% CIs) of hazard ratios were computed.
Expression levels of MN1 and CD34 in CD34+ cells were similar. To compare expression levels of MN1 and CD34 in CD34– and CD34+/CD38– cell populations, MN1 expression levels in all 3 cell populations were adjusted by multiplication with a correction factor, so that mean MN1 expression in CD34+ cells equaled mean CD34 expression in CD34+ cells. To compare data of the in vitro differentiation assays, MN1 and CD34 expression levels in CD34+ cells at day 0 were arbitrarily set to 1000, and subsequent expression values were adjusted accordingly. The 2-sided level of significance was set at P values less than .05. The statistical analyses were performed with the statistical software package SPSS 13.0 (SPSS Science, Chicago, IL).
Results
Patient characteristics
Adult patients with newly diagnosed AML with normal cytogenetics were evaluated for MN1 transcript expression by real-time RT-PCR. All 142 patients evaluated were treated according to the AML-SHG 01/99 multicenter trial protocol. Patients with normal cytogenetics, treated within the AML-SHG 01/99 trial but not included in this analysis because of a lack of frozen cell samples (n = 86), did not differ from included patients for CR rate (P = .62, chi-square test), RFS (P = .78, log-rank test), and OS (P = .2, log-rank test). There were no significant differences in presenting clinical characteristics between patients with high (n = 71) or low (n = 71) MN1 expression, including secondary or post-MDS AML, FLT3-ITD or MLL mutations except a strong association of low MN1 expression with mutated NPM1, and the mutation status NPM1mutated/FLT3ITDnegative (P < .001 and P = .001, respectively, chi-square test; Table 1). MN1 expression was independent of the blast percentage in diagnostic bone marrow or peripheral blood specimens (correlation coefficient R2 = .002). MN1 and CD34 transcript expression were positively correlated (correlation coefficient R2 = 0.2, P < .001). Of 29 patients with poor response to the first course of induction treatment, 4 were randomly assigned to receive a second course of IVA, and 25 were randomly assigned to receive FLAG-Ida. Patients with high MN1 expression are overrepresented in the FLAG-Ida group compared with patients with low MN1 expression (23.9% versus 11.3%, P = .047, chi-square test). The small sample size of the IVA group prevented a more detailed analysis of the influence of FLAG-Ida compared with IVA on the clinical outcome of patients randomly assigned to treatment. Late consolidation treatment was similar for patients with high versus low MN1 expression (chemotherapy in 21 versus 24 patients, P =NS; autologous transplantation in 13 versus 22 patients, P = .08; allogeneic transplantation in 21 versus 18 patients, P = NS; high versus low MN1 expression, respectively). The median follow-up time for surviving patients (n = 78) was 30 months (range, 3-61 months).
Patient characteristics according to MN1 expression levels
Characteristic . | MN1 low . | MN1 high . | P . |
|---|---|---|---|
| Age, y | |||
| Mean | 45.3 | 46.3 | .58 |
| Range | 18-60 | 22-60 | |
| Sex, n (%) | .13 | ||
| Male | 34 (48) | 43 (61) | |
| Female | 37 (52) | 28 (39) | |
| FAB subtype, n (%) | .43 | ||
| M0 | 0 (0) | 3 (4) | |
| M1 | 5 (7) | 7 (10) | |
| M2 | 17 (24) | 21 (30) | |
| M4 | 30 (42) | 26 (37) | |
| M5 | 13 (19) | 9 (12) | |
| M6 | 3 (4) | 1 (1) | |
| M7 | 0 (0) | 0 (0) | |
| Not classified | 3 (4) | 4 (6) | |
| Type of specimen, n (%) | .16 | ||
| Bone marrow | 59 (83) | 52 (73) | |
| Peripheral blood | 12 (17) | 19 (27) | |
| % Blasts | |||
| Mean, % | 73.3 | 67.9 | .19 |
| Missing, n (%) | 3 (4) | 5 (7) | |
| Diagnosis, n (%) | .13 | ||
| De novo | 65 (92) | 59 (83) | |
| Post-MDS/secondary | 6 (8) | 12 (17) | |
| WBC count, × 109/L | |||
| Mean | 35.2 | 48.2 | .16 |
| Range | 1-226 | 1-328 | |
| ECOG performance status, n (%) | .73 | ||
| 0 | 16 (23) | 20 (28) | |
| 1 | 49 (69) | 46 (65) | |
| 2 | 6 (8) | 5 (7) | |
| FLT3-ITD, n (%) | 21 (30) | 20 (28) | .85 |
| MLL PTD, n (%) | 3 (4) | 5 (7) | .47 |
| NPM1, n (%) | |||
| Mutated | 38/48 (79) | 13/37 (35) | < .001 |
| NPM1 mutated/ FLT3ITD negative | 28/48 (58) | 8/37 (22) | .001 |
| Missing | 23 (32) | 34 (48) | |
| CD34 expression, n (%) | < .001 | ||
| CD34 low | 49 (69) | 12 (17) | |
| CD34 high | 12 (17) | 49 (69) | |
| Missing | 10 (14) | 10 (14) | |
| Induction treatment, n (%) | .047 | ||
| IVA/IVA | 63 (89) | 54 (76) | |
| IVA/FLAG-Ida | 8 (11) | 17 (24) | |
| First consolidation treatment,*n (%) | .11 | ||
| AraC/DNR | 63 (89) | 55 (77) | |
| FLAG-Ida | 6 (8) | 12 (17) | |
| Second consolidation treatment,†n (%) | .26 | ||
| HD-AraC/DNR | 27 (38) | 33 (46) | |
| Auto SCT | 23 (32) | 13 (18) | |
| Allo SCT | 18 (25) | 21 (30) |
Characteristic . | MN1 low . | MN1 high . | P . |
|---|---|---|---|
| Age, y | |||
| Mean | 45.3 | 46.3 | .58 |
| Range | 18-60 | 22-60 | |
| Sex, n (%) | .13 | ||
| Male | 34 (48) | 43 (61) | |
| Female | 37 (52) | 28 (39) | |
| FAB subtype, n (%) | .43 | ||
| M0 | 0 (0) | 3 (4) | |
| M1 | 5 (7) | 7 (10) | |
| M2 | 17 (24) | 21 (30) | |
| M4 | 30 (42) | 26 (37) | |
| M5 | 13 (19) | 9 (12) | |
| M6 | 3 (4) | 1 (1) | |
| M7 | 0 (0) | 0 (0) | |
| Not classified | 3 (4) | 4 (6) | |
| Type of specimen, n (%) | .16 | ||
| Bone marrow | 59 (83) | 52 (73) | |
| Peripheral blood | 12 (17) | 19 (27) | |
| % Blasts | |||
| Mean, % | 73.3 | 67.9 | .19 |
| Missing, n (%) | 3 (4) | 5 (7) | |
| Diagnosis, n (%) | .13 | ||
| De novo | 65 (92) | 59 (83) | |
| Post-MDS/secondary | 6 (8) | 12 (17) | |
| WBC count, × 109/L | |||
| Mean | 35.2 | 48.2 | .16 |
| Range | 1-226 | 1-328 | |
| ECOG performance status, n (%) | .73 | ||
| 0 | 16 (23) | 20 (28) | |
| 1 | 49 (69) | 46 (65) | |
| 2 | 6 (8) | 5 (7) | |
| FLT3-ITD, n (%) | 21 (30) | 20 (28) | .85 |
| MLL PTD, n (%) | 3 (4) | 5 (7) | .47 |
| NPM1, n (%) | |||
| Mutated | 38/48 (79) | 13/37 (35) | < .001 |
| NPM1 mutated/ FLT3ITD negative | 28/48 (58) | 8/37 (22) | .001 |
| Missing | 23 (32) | 34 (48) | |
| CD34 expression, n (%) | < .001 | ||
| CD34 low | 49 (69) | 12 (17) | |
| CD34 high | 12 (17) | 49 (69) | |
| Missing | 10 (14) | 10 (14) | |
| Induction treatment, n (%) | .047 | ||
| IVA/IVA | 63 (89) | 54 (76) | |
| IVA/FLAG-Ida | 8 (11) | 17 (24) | |
| First consolidation treatment,*n (%) | .11 | ||
| AraC/DNR | 63 (89) | 55 (77) | |
| FLAG-Ida | 6 (8) | 12 (17) | |
| Second consolidation treatment,†n (%) | .26 | ||
| HD-AraC/DNR | 27 (38) | 33 (46) | |
| Auto SCT | 23 (32) | 13 (18) | |
| Allo SCT | 18 (25) | 21 (30) |
For MN1 low and MN1 high groups, total n = 71 each.
n = 136.
n = 135.
Response to induction therapy
The response rate on day 15 after the first course of induction therapy was significantly different according to MN1 expression levels (Table 2). Only 71.8% of patients with high MN1 expression achieved a good response on day 15 compared with 87.3% of patients with low MN1 expression (P = .02, chi-square test). The complete remission rate did not significantly differ between MN1 groups (80.3% in MN1 high group versus 88.7% in MN1 low group, P = .16, chi-square test). However, when the optimal cutoff for MN1 expression was applied, patients with high MN1 expression (n = 82) had a significantly lower CR rate compared with patients with low MN1 expression (n = 60, 79.3% versus 91.7%, respectively, P = .02, chi-square test; Table S1).
Response to induction therapy according to MN1 expression levels
Remission . | MN1 low, n (%) . | MN1 high, n (%) . | P . |
|---|---|---|---|
| Good response day 15 | 62 (87.3) | 51 (71.8) | .02 |
| Complete remission | 63 (88.7) | 57 (80.3) | .16 |
| Early death | 3 (4.2) | 4 (5.6) | NS |
Remission . | MN1 low, n (%) . | MN1 high, n (%) . | P . |
|---|---|---|---|
| Good response day 15 | 62 (87.3) | 51 (71.8) | .02 |
| Complete remission | 63 (88.7) | 57 (80.3) | .16 |
| Early death | 3 (4.2) | 4 (5.6) | NS |
For MN1 low and MN1 high groups, total n = 71 each.
In a subgroup analysis differences in treatment response between the high and low MN1 groups were evaluated separately for BM and PB samples. Patients with a PB sample and high MN1 expression (n = 31) had a significantly worse CR rate (P = .037) but did not differ significantly for response to first induction treatment (P = .57) compared with patients with low MN1 expression. Patients with BM samples and high MN1 expression (n = 111) had significantly more often a poor response to the first induction treatment (30% versus 13%, P = .02), but the CR rate did not differ significantly compared with patients with low MN1 expression.
Relapse-free survival
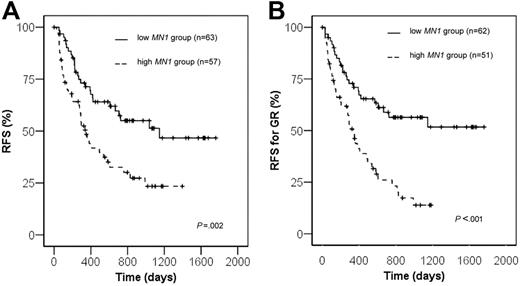
Fifty-seven patients with high MN1 expression and 63 with low MN1 expression achieved a complete remission and were evaluated for RFS. The relapse rate for patients with high MN1 expression was significantly higher compared with patients with low MN1 expression (56.1% versus 36.5%, respectively, P = .03, chi-square test). RFS was significantly shorter in the MN1 high group compared with the MN1 low group (23.4% versus 51.4% at 3 years, P = .002, log-rank test; Figure 1A). To rule out that the difference in RFS was due to the different early response rates at day 15 in MN1 high and low groups, RFS was determined for patients who achieved a good response at day 15. RFS was again significantly shorter in patients with high MN1 expression compared with low MN1 expression (13.9% versus 56.4% at 3 years, respectively, P = .0001 log-rank test; Figure 1B). Because of the higher rate of relapses, patients with high MN1 expression more often received an allogeneic transplant as salvage treatment compared with patients with low MN1 expression (26.3% versus 7.9%, P = .02, chi-square test). In a subgroup analysis patients with a PB sample and high MN1 expression showed a trend toward shorter RFS (P = .055, log-rank test), and patients with a BM sample and high MN1 expression had significantly shorter RFS (P = .01, log-rank test) compared with patients with low MN1 expression.
Treatment results according to theMN1expression levels. (A) Relapse-free survival. (B) Relapse-free survival of patients achieving a good treatment response (GR) after the first course of induction chemotherapy.
Treatment results according to theMN1expression levels. (A) Relapse-free survival. (B) Relapse-free survival of patients achieving a good treatment response (GR) after the first course of induction chemotherapy.
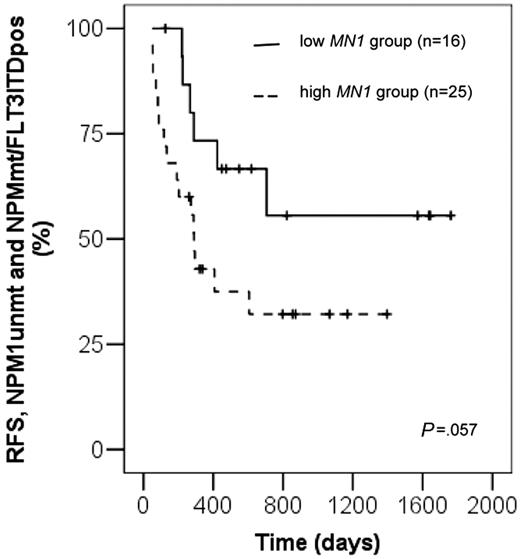
As low MN1 expression was strongly associated with the mutation status NPM1mutated/FLT3ITDnegative, which has been described as a favorable prognostic marker in patients with normal karyotype,11,23 the prognostic significance of MN1 was evaluated in patients with NPM1mutated/FLT3ITDnegative or all remaining patients. In the favorable risk group (NPM1mutated/FLT3ITDnegative) there were only 5 patients with high MN1 expression, which did not allow reasonable statistical analysis. However, in the remaining patient group (NPM1mutated/FLT3ITDpositive or NPM1unmutated) we found a trend toward shorter RFS for patients with high MN1 expression compared with patients with low MN1 expression (P = .057, log-rank test; Figure 2). Univariate analysis of other potential prognostic markers (response to first induction chemotherapy, age, de novo versus secondary or post-MDS AML, white blood cell [WBC] count, MLL-PTD or FLT3-ITD mutation status, CD34 expression) only revealed a significant difference in RFS for CD34 expression comparing low versus high expression (P = .01, log-rank test; data not shown). In bivariate analysis only MN1 was an independent unfavorable prognostic factor (P = .005; hazard ratio, 2.17; 95% CI, 1.26-3.74; CD34, P = .38).
Overall survival
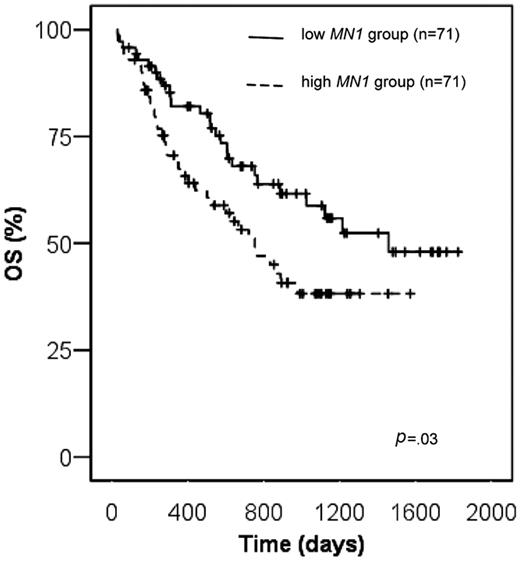
Overall survival for patients with high MN1 expression was significantly shorter compared with patients with low MN1 expression (38.1% versus 58.8% at 3 years, respectively, P = .03, log-rank test; Figure 3). The survival difference was even more pronounced, when the optimal cutoff for MN1 expression was applied (32.9% versus 71% at 3 years, respectively, P = .002, log-rank test; Figure S1). This finding was confirmed when only patients with BM samples were analyzed (P = .03, log-rank test), whereas the difference in OS of patients with PB samples did not reach statistical significance most likely because of the small patient number. Of other patient characteristics shown in Table 1, including NPM1mutated/FLT3-ITDnegative versus all others and CD34 low versus high expressers, only age (dichotomized at the median), Eastern Cooperative Oncology Group (ECOG) performance status (0 or 1 compared with 2), and FLT3-ITD were predictive for overall survival in univariate analysis (data not shown). Multivariable analysis for OS was performed, including the variables MN1 expression level, ECOG performance status, age, and FLT3-ITD. Of those, high MN1 expression (P = .02), ECOG performance status of 2 (P = .003), and age above the median (P = .007) were unfavorable prognostic factors in multivariable analysis (Table 3).
Results of Cox regression analysis of overall survival
Variable . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|
| MN1 expression above median | 1.87 | 1.12-3.13 | .02 |
| ECOG 2 | 2.96 | 1.44-6.11 | .003 |
| Age above median | 2.01 | 1.21-3.33 | .007 |
Variable . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|
| MN1 expression above median | 1.87 | 1.12-3.13 | .02 |
| ECOG 2 | 2.96 | 1.44-6.11 | .003 |
| Age above median | 2.01 | 1.21-3.33 | .007 |
Relapse-free survival according to MN1 expression levels in patients with mutation status NPM1unmutated and NPM1mutated/FLT3ITDnegative.
Relapse-free survival according to MN1 expression levels in patients with mutation status NPM1unmutated and NPM1mutated/FLT3ITDnegative.
The effect of the availability of an HLA-compatible family donor on survival was analyzed in patients with low and high MN1 expression. For 13 patients the availability of an HLA-compatible family donor was not known, mostly because of an early death of the patient. In patients with low MN1 expression, the availability of an HLA-compatible family donor did not influence RFS (P = .12, log-rank test; data not shown) or OS (P = .38, log-rank test; Figure S2A). Similarly, in patients with high MN1 expression, the availability of an HLA-compatible family donor did not influence RFS (P = .55, log-rank test; data not shown) or OS (P = .19, log-rank test; Figure S2B). There was no significant difference in OS when the analysis was restricted to younger patients (younger than 40 years of age) with high MN1 expression with or without an HLA-compatible family donor (n = 17; data not shown).
MN1 expression in different hematopoietic tissues and during in vitro differentiation of CD34+ cells
MN1 expression was evaluated in normal and leukemic cell samples (Figures S3-S4). There was no significant difference of mean MN1 expression comparing PB MNCs (n = 6) with CLL samples (n = 12; P = NS, t test), CML in chronic phase (n = 10; P = NS, t test), or CML in blast crisis (n = 2; P = NS, t test). There was a trend to higher mean MN1 expression in ALL (n = 13) compared with PB MNCs (P = .066, t test), and mean MN1 expression was significantly higher in AML samples (n = 142) compared with PB MNCs (P < .001, t test; Figure S3).
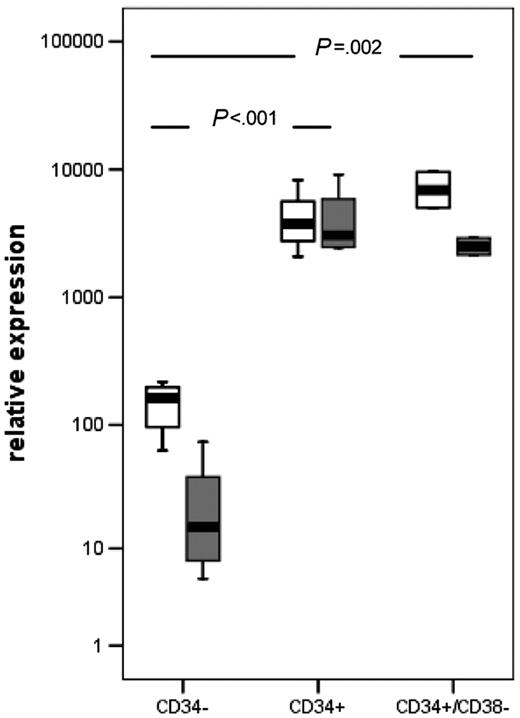
Expression of CD34 transcripts was evaluated in purified human CD34+, CD34–, and CD34+/CD38– bone marrow cells. In parallel, MN1 expression was evaluated in these 3 cell populations. MN1 and CD34 expression was significantly higher in CD34+ cells compared with CD34– cells (P < .001) and in CD34+/CD38– cells compared with CD34– cells (P = .002). Expression of MN1 was 1.64-fold higher in CD34+/CD38– cells compared with CD34+ cells, whereas expression of CD34 was 1.76-fold lower in CD34+/CD38– cells compared with CD34+ cells (Figure 4).
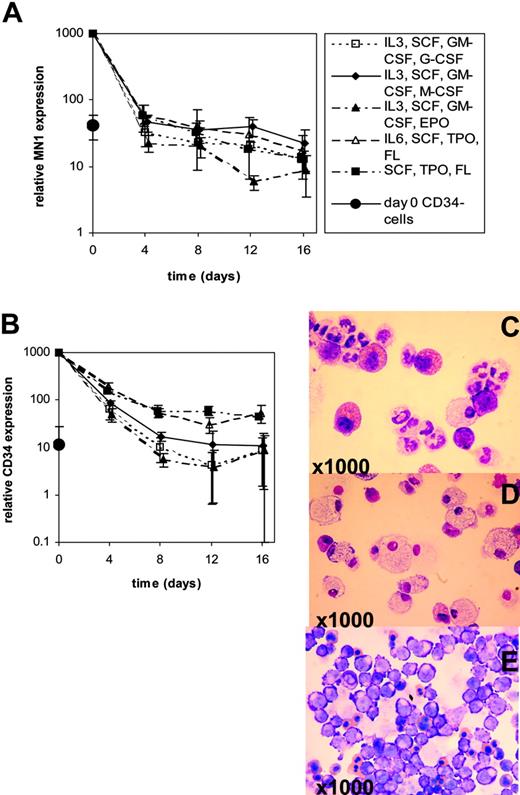
MN1 and CD34 transcript expressions were evaluated during in vitro differentiation of human CD34+ cells purified from G-CSF–mobilized PB MNCs. CD34+ cells were maintained in suspension culture in X-vivo 10 medium supplemented with lineage-unspecific cytokines supporting proliferation of early-stage progenitors (SCF [100 ng/mL], TPO [20 ng/mL], FLT3-ligand [100 ng/mL] ± IL-6 [20 ng/mL]). In parallel, for induction of differentiation cells were cultured in IMDM medium supplemented with cytokine combinations supporting differentiation (SCF [50 ng/mL], IL-3 [20 ng/mL], GM-CSF [20 ng/mL], and one of the lineage-specific cytokines G-CSF [50 ng/mL], M-CSF [50 ng/mL], or EPO [3 U/mL]). Morphologic analysis on day 10 (EPO) or day 18 (G-CSF, M-CSF) confirmed lineage-specific maturation with the emergence of erythroid, granulocytic, or macrophage features, respectively (Figure 5C-E). In vitro differentiation of CD34+ cells revealed down-regulation of MN1 transcripts by day 4 in cultures supplemented with lineage-specific cytokines to levels found in CD34– cells, whereas CD34 transcripts were down-regulated to levels found in CD34– cells by day 8 (Figure 5A-B). Maintenance culture using lineage-unspecific cytokines did not delay down-regulation of MN1 transcripts, whereas CD34 expression was maintained at elevated levels compared with CD34– cells through day 16 (Figure 5A-B). Collectively, these data show that MN1 is highly expressed in cell populations enriched for primitive hematopoietic cells, is down-regulated on differentiation, and can be found at elevated levels in acute leukemias of both myeloid and lymphoid lineages.
Mean MN1 expression (▪) and range in hematopoietic progenitor and differentiated cells determined by real-time RT-PCR. Sorted cells from G-CSF–mobilized peripheral blood of healthy donors were analyzed for expression levels of MN1 (□) and CD34 ( ), and mean expression was compared by Student t test (CD34– and CD34+ cell population, n = 4, respectively; CD34+/CD38– cell population, n = 2). Boxes represent interquartile range; error bars, outliers.
), and mean expression was compared by Student t test (CD34– and CD34+ cell population, n = 4, respectively; CD34+/CD38– cell population, n = 2). Boxes represent interquartile range; error bars, outliers.
Mean MN1 expression (▪) and range in hematopoietic progenitor and differentiated cells determined by real-time RT-PCR. Sorted cells from G-CSF–mobilized peripheral blood of healthy donors were analyzed for expression levels of MN1 (□) and CD34 ( ), and mean expression was compared by Student t test (CD34– and CD34+ cell population, n = 4, respectively; CD34+/CD38– cell population, n = 2). Boxes represent interquartile range; error bars, outliers.
), and mean expression was compared by Student t test (CD34– and CD34+ cell population, n = 4, respectively; CD34+/CD38– cell population, n = 2). Boxes represent interquartile range; error bars, outliers.
MN1 and CD34 expression during in vitro differentiation of CD34+ progenitor cells. Real-time RT-PCR was performed on days 0, 4, 8, 12, and 16 of in vitro differentiation using differentiation-inducing growth conditions (G-CSF, M-CSF, or EPO) or maintenance culture conditions with lineage-unspecific cytokines (SCF, TPO, FL ± IL-6). Mean ± SD is shown for 3 independent experiments. Expression of MN1 and CD34 in CD34– cell populations was analyzed at day 0. (A) MN1 expression levels during in vitro differentiation. (B) CD34 expression levels during in vitro differentiation. Giemsa staining of cytospin preparations shows typical morphologic features of granulocytic (G-CSF; C), macrophage (M-CSF; D), and erythroid (EPO; E) maturation.
MN1 and CD34 expression during in vitro differentiation of CD34+ progenitor cells. Real-time RT-PCR was performed on days 0, 4, 8, 12, and 16 of in vitro differentiation using differentiation-inducing growth conditions (G-CSF, M-CSF, or EPO) or maintenance culture conditions with lineage-unspecific cytokines (SCF, TPO, FL ± IL-6). Mean ± SD is shown for 3 independent experiments. Expression of MN1 and CD34 in CD34– cell populations was analyzed at day 0. (A) MN1 expression levels during in vitro differentiation. (B) CD34 expression levels during in vitro differentiation. Giemsa staining of cytospin preparations shows typical morphologic features of granulocytic (G-CSF; C), macrophage (M-CSF; D), and erythroid (EPO; E) maturation.
Discussion
We evaluated the prognostic significance of MN1 mRNA expression levels in 142 adult patients with acute myeloid leukemia with normal cytogenetics and established, for the first time, MN1 as an independent prognostic marker in this patient group. The most important finding is that MN1 overexpression is associated with treatment failure, specifically a significantly worse day 15 response rate, higher relapse rate, and shorter relapse-free and overall survivals. In addition, we found MN1 highly expressed in cell populations enriched for primitive hematopoietic cells and rapidly down-regulated on differentiation of CD34+ cells.
MN1 expression has not been specifically studied in AML, but we have found MN1 overexpression as part of a gene profile characterizing patients with poor response compared with patients with good response with AML,10 and the translocation involving MN1 and TEL has been associated with treatment failure.24 With the number of prognostic markers growing, the relative contribution of each becomes important. Between high and low MN1 expression groups dichotomized at the median expression level, no significant correlations with disease characteristics were found, including blast percentage in PB or BM at diagnosis. No correlation was found between MN1 expression and the prognostically important mutations FLT3 ITD and MLL.16,17,25-27 However, low MN1 expression was highly associated with mutated NPM1 and the mutation status NPM1mutated/FLT3ITDnegative. Recent reports show that mutated NPM1 is associated with a high CR rate and, at least in the absence of FLT3 ITD, with improved event-free survival, RFS, or OS.11,23,28-31 In the subgroup analysis in which patients with the favorable mutation status NPM1mutated/FLT3ITDnegative were excluded, patients with high MN1 expression showed a trend toward shorter RFS compared with patients with low MN1 expression. Thus, MN1 expression levels may add prognostic information to the patient group with unmutated NPM1 or mutated NPM1/positive FLT3ITD, although the analysis is limited by a low number of patients. To estimate the contribution of prognostic factors significant in univariate analysis (MN1 expression level, age, ECOG performance status, and FLT3 ITD), multivariate analysis was performed. It revealed that MN1 is an independent prognostic parameter for overall survival in addition to age and ECOG performance status. CD34 expression has been suggested as a prognostic marker in AML.32 Because MN1 and CD34 expressions were highly correlated in normal hematopoietic cells, the prognostic significance of MN1 and CD34 was compared in patients with AML. As expected the expression levels of these 2 genes were significantly correlated. Whereas patients with low CD34 expression had a significantly shorter RFS compared with patients with high CD34 expression, bivariate analysis of MN1 and CD34 revealed that MN1 is an independent prognostic factor. Moreover, CD34 expression was not a significant prognostic marker for OS. Another important prognostic marker in AML is the mutation status of CEBPA.33-35 We have not included this marker in our analysis, because we could analyze fewer than 40% of patients for CEBPA mutations because of limited patient material. Similarly to MN1 overexpression, up-regulation of BAALC,36-38 WT1,39,40 and EVI141 have been reported as poor prognostic markers in AML. Future studies need to address whether these markers have independent prognostic significance or whether they are jointly up-regulated as a consequence of a transforming event.
Relapse-free survival for patients with good response was significantly shorter in patients with high MN1 expression compared with low MN1 expression. In the future, this may allow stratification of treatment for patients with good response according to their relapse risk at this early time point and to apply either the same treatment strategies for patients with good response and high MN1 expression because they are used already for patients with poor response, or to allocate these patients to experimental strategies.
To address the question of whether allogeneic transplantation would benefit patients with high MN1 expression, donor versus no donor analysis was performed. There was no benefit of allogeneic transplantation in patients with high MN1 expression. The role of allogeneic transplantation in AML with intermediate risk cytogenetics in first CR has been challenged.42,43 Data suggested that only patients younger than age 35 years benefit from allogeneic transplantation.43 In patients with high MN1 expression and age younger than 40 years, no advantage of allogeneic transplantation could be detected, although this analysis is restricted by a low patient number.
The mechanism through which MN1 overexpression contributes to treatment failure or leukemogenesis remains unknown. We found MN1 expressed at higher levels in samples not only of AML but also of acute lymphoblastic leukemia and normal committed and uncommitted hematopoietic progenitor cells (HPCs) compared with chronic leukemias or normal differentiated cells. Similarly, other prognostic markers in AML such as EVI1 or BAALC are overexpressed in normal HPCs.44,45 Up-regulation of these genes may reflect activation of a stem cell–specific pathway leading to an early differentiation block in leukemic cells, which has been associated with a poor prognosis in AML.46,47 Identification of genes involved in such a pathway, such as MN1, will allow molecular dissection, functional characterization, and potential targeting of this pathway. In contrast to the CD34 transcript expression, MN1 expression cannot be sustained on culturing CD34+ progenitor cells in lineage-unspecific cytokines supporting proliferation of early-stage progenitors. MN1 function might therefore be most important at the uncommitted progenitor cell stage. Data to support a functional role of MN1 in HPCs were recently published by Kawagoe and Grosveld.4 The fusion protein MN1-TEL promoted the repopulating ability of myeloid progenitors in vitro in a conditional MN1-TEL transgenic mouse model and partially inhibited granulocytic differentiation.4 Evidence for a more direct role of MN1 in leukemogenesis was provided by cytogenetic analyses, whereby the translocation t(12;22) involving MN1 and TEL was found in myeloid leukemia and myelodysplastic syndrome.2 This has been substantiated by the recent finding that the fusion gene MN1-TEL, in cooperation with HOXA9, rapidly induces myeloid leukemia.5 Although the contribution of MN1 in this context is not known, these data support the hypothesis that MN1 plays a role in leukemogenesis either by blocking differentiation or by conferring a proliferation/self-renewal advantage to leukemic cells. As shown for other genes such as HOXA948 or CDX2,49 increasing the transcript dosage by mere overexpression may be sufficient as a first leukemogenic event.
In conclusion, our data suggest that MN1 overexpression is a new prognostic marker in AML with normal cytogenetics, which is associated with poor induction response, shorter relapse-free survival, and shorter overall survival. Confirmation of these results in a prospective trial may ultimately lead to improved risk stratification of this heterogeneous group of patients with AML.
Authorship
M.H., J.K., N.v.N., B.S., and A.G. designed the research; M.H., G.B., J.K., and K.D. performed the research; M.H., N.v.N, and A.G. analyzed the data; and M.H. and A.G. wrote the paper. All authors checked the final version of the manuscript.
The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, August 15, 2006; DOI 10.1182/blood-2006-04-014845.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank the patients participating in this study and the members of the AML-SHG Study Group for providing leukemia specimens. We thank Elvira Lux and Irina Schäfer for their support in sample and data acquisition, Matthias Ballmaier for sorting hematopoietic cell populations (Cell Sorting Core Facility, Hannover Medical School, Germany), and Gerd Wegener and Ludwig Hoy for very valuable statistical advice (Hematology, Hemostasis, and Oncology and Institute of Biometry, Hannover Medical School, Germany, respectively).
This work was supported by the Gottfried Arndt-Stiftung Hannover, Germany, and the Bundesministerium für Bildung und Forschung (Kompetenznetz “Akute und chronische Leukämie”), Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal