Abstract
It is unclear how the immune response controls human herpesvirus 8 (HHV8; also known as Kaposi sarcoma–associated herpesvirus [KSHV]) replication and thereby prevents Kaposi sarcoma (KS). We compared CD8 T-cell responses to HHV8 latent (K12) and lytic (glycoprotein B, ORF6, ORF61, and ORF65) antigens in patients who spontaneously controlled the infection and in patients with posttransplantation, AIDS-related, or classical KS. We found that anti-HHV8 responses were frequent, diverse, and strongly differentiated toward an effector phenotype in patients who controlled the infection. Conversely, HHV8-specific CD8 cells were very rare in patients who progressed to KS, and were not recruited to the tumoral tissue, as visualized by in situ tetramer staining of KS biopsies. Last, HHV8-specific CD8 T cells were observed in a seronegative recipient of an HHV8infected graft who remained persistently aviremic and antibody negative, suggesting that specific cytotoxic T lymphocytes (CTLs) may provide protection from persistent HHV8 infection. These results support the crucial role of cellular immune responses in controlling HHV8 replication, in preventing malignancies in latently infected subjects, and in conferring genuine resistance to persistent infection. They may also have important implications for the design of prophylactic and therapeutic HHV8 vaccines, and for adoptive immunotherapy of KS.
Introduction
Human herpesvirus 8 (HHV8), a lymphotropic γ herpesvirus, is identified as the causative agent of Kaposi sarcoma1 (KS), a vascular tumor containing proliferating spindle-shaped cells of endothelial origin (for review, see Hengge et al2,3 ). KS is rare in immunocompetent individuals, but is the most common neoplasm in patients with AIDS who are not receiving highly active antiretroviral therapy (HAART). KS also occurs in immunosuppressed organ transplant recipients, and in some African populations (endemic KS) and Mediterranean populations (classical KS). The marked decline in the incidence of AIDS-related KS since the advent of HAART4 and the frequent resolution of transplantation-related KS after reduction of immunosuppression5 highlight the key role of cellular immune responses in the control of HHV8 infection.
The nature of HHV8 proteins that are expressed by infected cells and permit their recognition by the immune system is unknown. HHV8 possesses a large genome with 90 open reading frames (ORFs), of which 15 (K1 to K15) are unique to this virus. HHV8 latently infects endothelial cells and B lymphocytes, in which the viral episomes express only a limited number of genes. This implies that only a few viral peptides may be recognized by cytotoxic T lymphocytes (CTLs) on latently infected cells. When immune response declines, infected cells may proliferate unchecked and HHV8-related tumors may develop. A proportion of infected cells undergo lytic viral replication, resulting in the production of infective virions. Viral gene expression is still markedly restricted in KS lesions, and lytic cycling is restricted to a small population of endothelial spindle cells.6
HHV8-specific immune responses are poorly characterized. CTLs targeting certain latent and lytic proteins have been generated in vitro,7-9 but only a few CD8 T-cell epitopes have been identified as immunogenic in short-term in vitro culture.10-15 Ex vivo CD8 T-cell responses to HHV8 antigens have not been reported, except for glycoprotein B (gB).13
Almost half of the white population is positive for human leukocyte antigen (HLA)–A2. Studying T-cell responses in HLA-A2+ individuals is therefore not only convenient, but also highly relevant for the design of peptide vaccine trials. To further characterize cellular anti-HHV8 responses, we first identified potential HLA-A2–restricted CD8 epitopes of selected latent (K12) and lytic (gB, ORF6, ORF61, and ORF65) proteins. Using a panel of HLA-A2 tetramers specific for these epitopes, we then quantitatively and functionally assessed HHV8-specific circulating CD8 T cells in a cohort of HLA-A2+ infected patients with and without KS, and we directly visualized virus-specific CD8 T cells infiltrating KS lesions by in situ tetramer staining. We obtained new information on the specificity, magnitude, and functionality of HHV8-specific responses, which should help to identify markers of immune protection and to develop more effective vaccines.
Patients, materials, and methods
Patients
Written informed consent was obtained from each patient, and the study protocol was approved by the ethics Committee of Hôpital Saint-Louis (Paris, France). Peripheral blood mononuclear cells (PBMCs) were collected, separated by Lymphoprep gradient centrifugation (Eurobio), and cryopreserved for later analysis. We studied 48 HLA-A*0201+ patients infected with HHV8, comprising 12 HIV-seronegative kidney transplant recipients (mean age, 53 years), 24 patients with AIDS-related KS (mean age, 47 years), and 12 patients with classical KS (mean age, 61 years). The transplant recipients were all receiving conventional immunosuppression (azathioprine, methylprednisolone, and cyclosporine) and included 8 patients without KS, 2 patients with ongoing KS, and 2 patients in remission from KS after a reduction of immunosuppression. The patients with AIDS were all on HAART, and comprised 7 patients with ongoing KS and 17 patients in remission, of whom 6 had completed chemotherapy for KS at least 6 months before study. The patients with classical KS comprised 9 patients with ongoing KS and 3 patients in remission, of whom 2 had received chemotherapy. Patients with transplantation-related or classical KS had isolated cutaneous involvement, whereas 9 patients with AIDS-related KS had visceral involvement in addition to cutaneous involvement. We also studied 2 HHV8– transplant recipients whose donors were infected with HHV8. Immunofluorescence assay was used to detect latency-associated nuclear antigen (LANA) antibodies and enzyme-linked immunosorbent assay (ELISA) was used for K8.1 antibodies. HHV8 DNA load was determined by real-time polymerase chain reaction (PCR) in PBMCs, as described elsewhere.16 The positivity was defined as the linearity of 50 to 1 000 000 copies/150 000 cells for 50 μL of reaction volume (detection threshold corresponding to 10 copies/150 000 cells).
Controls consisted of PBMCs from 8 HLA-A2+ healthy donors and 18 HLA-A2– HHV8+ individuals. HLA-A2 typing was performed by flow cytometry on PBMCs using the HLA-A2–specific BB7.2 monoclonal antibody (American Tissue Cell Culture [ATCC], Rockville, MD), and was confirmed by sequence-specific primer (SSP) amplification (HLA-A2 PCR-SSP; Dynal, Oslo, Norway).
Peptides and HLA-A2 stabilization assay
Peptides were synthesized by NeoSystems (Strasbourg, France). TAP-deficient T2 cells (ATCC) were grown for 18 hours at 37°C in serum-free medium, then 106 T2 cells were pulsed with 10 μM peptide in the presence of 20 μg/mL β2 microglobulin (Sigma, St Louis, MO) and 10 μg/mL brefeldin (Sigma) for 18 hours at 37°C. After washing, cells were stained with the HLA-A2–specific BB7.2 antibody (ATCC), and then with PE-conjugated goat anti–mouse immunoglobulin (Ig). HLA-A2 expression was measured with a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA). The fluorescence index (FI) was calculated as follows: FI = (mean fluorescence intensity with peptide – mean fluorescence without peptide)/mean fluorescence without peptide.
HLA class I tetramers, antibodies, and flow cytometry
HLA-A2 tetramers were produced as described elsewhere.17 Each tetramer was titered individually, and used at the optimal concentration (5-10 μg/mL). For staining, 106 PBMCs were incubated at 37°C for 30 minutes in the dark with 0.5 to 1.0 μg of PE-labeled tetramer, then with a combination of the following antibodies: CD8-APC (Caltag, Burlingame, CA), CD3-FITC (BD/Pharmingen, San Diego, CA), CD45RA-PerCP (Beckman/Immunotech, Marseille, France), and CCR7-FITC (R&D Systems, Minneapolis, MN) or CD27-FITC (BD/Pharmingen) for 15 minutes at 4°C. Small lymphocytes were gated according to forward/side-scatter profiles, then CD3+CD8high cells were selected, and staining with 7AAD (BD PharMingen) was used to exclude dead cells. Data were collected on a FACSCalibur flow cytometer within 1 hour after staining, and were analyzed with Cell Quest software (Becton Dickinson). To determine nonspecific tetramer staining of peripheral CD8 T cells, a series of 18 HLA-A2–/HHV8+ individuals were analyzed, yielding a lower detection limit for positive staining in HLA-A2+/HHV8+ individuals (cutoff = mean frequency of tetramer+ cells in HLA-A2–/HHV8+ subjects + 2 × SD).
For intracellular perforin staining after tetramer and CD8 staining, cells were fixed for 10 minutes in 2% paraformaldehyde solution, then washed, permeabilized in 2% saponin solution, and stained with perforin-FITC antibody (BD/Pharmingen).
In situ tetramer staining
Biopsies of cutaneous lesions were available in 2 patients with classical KS. Parts of the samples were taken for diagnostic evaluation, while other parts were frozen for later in situ tetramer staining. Sections (5 μm) were cut and fixed in acetone for 10 minutes at room temperature. Nonspecific binding sites were blocked for 30 minutes with 40% bovine serum albumin (BSA)–5% fetal calf serum (FCS) in phosphate-buffered saline (PBS), followed by the Biotin-Blocking System (Dako, Glostrup, Denmark) and washing with PBS. Sections were stained overnight at 4°C with Cy3-labeled HHV8 tetramers (10 μg/mL diluted in 2% FCS in PBS). Sections were washed 3 times for 10 minutes in PBS with 0.5% Triton X-100 (Sigma), and incubated for 1 hour at room temperature with rat anti–ORF-73 of HHV8 (Caltag) diluted 1:1000. After 3 washing steps with PBS, sections were incubated for 45 minutes with FITC-conjugated goat antirat (1:100 diluted; Jackson Laboratories, West Grove, PA). After washing, sections were then incubated for 90 minutes at 4°C with mouse anti–human CD8 (1:75 diluted, clone DK25; Dako), washed 3 times, and incubated for 40 minutes with Cy5-conjugated goat antimouse (1:50 diluted; Jackson Laboratories). Slides were mounted in Vecta shield medium (Vector Laboratories, Burlingame, CA) and analyzed using a confocal laser scanning microscope with photomultipliers (TCS SPS AOBS model, Leica, Mannheim, Germany). Cells were analyzed in a minimum of 50 Z stacks per skin section by an operator who was unaware of the clinical data. Images were acquired using a 63×/1.43 numerical aperture oil objective and Leica confocal software TCS SP2 (V2.61.1537), and were analyzed using ImageJ software version 1.36b (http://rsb.info.nih.gov/ij). Each experiment included a sample taken from a KS– HLA-A2+ patient for diagnostic purpose, as negative control.
Statistical analysis
The chi-square test was used to compare the number of patients with tetramer+ response in the different groups. The nonparametric Mann-Whitney rank-sum test was used to compare frequencies between groups. When 3 or more unpaired groups were compared, the Kruskall-Wallis test was used, and the Dunn post-test was used to narrow down which group was significantly different from the others. Statistical difference was assumed when P (2-tailed) was less than .05.
Results
Identification of HLA-A*0201–binding HHV8-derived epitopes
To identify HHV8-specific CD8 T cells, we selected potential HLA-A*0201–restricted CTL epitopes from latent and lytic proteins. Among them, 3 epitopes, derived from 1 latent antigen (K1217-25) and 2 lytic antigens (gB492-500 and K8.1209-217), have previously been described as immunogenics.10,14 In addition, we attempted to identify epitopes of the LANA, and the lytic proteins ORF65, ORF6, and ORF61. LANA is responsible for persistence of the episomal genome and is detected in all HHV8-infected cells. ORF65 is a small viral capsid component that can be used to measure humoral immunity, suggesting it can also give rise to T-cell responses. ORF6 (single-stranded DNA [ssDNA]–binding protein) and ORF61 (large ribonucleotide reductase subunit) were chosen because immunodominant CTL epitopes have been described in the corresponding ORFs of the murine herpesvirus MHV-68 γ.18 To identify potential CTL epitopes within these proteins, their amino acid sequences were analyzed by computer algorithms designed to predict HLA-A*0201–binding peptides, based on the estimated half-time dissociation of the HLA-peptide complex41 or prediction of major histocompatibility complex (MHC) class I ligands.42 Selected peptides (Table 1) were synthesized, and tested for HLA-A2 binding in a conventional HLA-A2 stabilization assay with the mutant T2 cells (Table 1). The reference high-binder peptide cytomegalovirus (CMV)–pp65, which bound efficiently to T2 cells with an index of fluorescence intensity of 3.9, was used as a positive control in all experiments. Tetramers of K12, gB, ORF6, ORF61, and ORF65 peptides were generated. The K8.1 peptide did not bind to HLA-A2 and did not permit refolding of HLA-A2 monomers, and may not therefore be an HLA-A2–restricted T-cell epitope. Altogether, there was a good correlation between the capacity of a given peptide to bind HLA-A2 molecules on T2 cells, its ability to generate large amounts of stable tetramers, and the peptide-binding score determined by HLA class I ligand prediction computer programs.
Peptide characteristics and results of peptide binding assays
Peptide (location) . | Amino acid sequence . | Score,*syfpeithi/bimas . | FI† . | Tetramer . |
|---|---|---|---|---|
| K12 (17-25)‡ | LLNGWRWRL | 25/272 | 0.13 | + |
| ORF8-gB (492-500)§ | LMWYELSKI | 24/244 | 0.83 | + |
| K8.1 (204-213) | GLLLGLVLIL | 32/74 | -0.74 | - |
| K8.1 (209-217)∥ | LVLILYLCV | 20/57 | 0.04 | - |
| LANA (102-110) | ALLPPVTTS | 23/0.6 | 0.55 | - |
| LANA (191-199) | LAPSTLRSL | 22/0.2 | -0.26 | - |
| ORF6 (1050-1058) | VLGDEVLSL | 29/342 | 0.95 | + |
| ORF61 (505-513) | GLADVFAEL | 28/166 | 5.1 | + |
| ORF65 (35-43) | NMSQAEYLV | 19/50 | 0.12 | + |
| ORF65 (129-137) | TLSSGPHSL | 25/21 | 0.05 | - |
| CMV-pp65 | NLVPMVATV | 30/159 | 3.9 | + |
| HIV gag | SLYNTVATL | 31/150 | ND | + |
Peptide (location) . | Amino acid sequence . | Score,*syfpeithi/bimas . | FI† . | Tetramer . |
|---|---|---|---|---|
| K12 (17-25)‡ | LLNGWRWRL | 25/272 | 0.13 | + |
| ORF8-gB (492-500)§ | LMWYELSKI | 24/244 | 0.83 | + |
| K8.1 (204-213) | GLLLGLVLIL | 32/74 | -0.74 | - |
| K8.1 (209-217)∥ | LVLILYLCV | 20/57 | 0.04 | - |
| LANA (102-110) | ALLPPVTTS | 23/0.6 | 0.55 | - |
| LANA (191-199) | LAPSTLRSL | 22/0.2 | -0.26 | - |
| ORF6 (1050-1058) | VLGDEVLSL | 29/342 | 0.95 | + |
| ORF61 (505-513) | GLADVFAEL | 28/166 | 5.1 | + |
| ORF65 (35-43) | NMSQAEYLV | 19/50 | 0.12 | + |
| ORF65 (129-137) | TLSSGPHSL | 25/21 | 0.05 | - |
| CMV-pp65 | NLVPMVATV | 30/159 | 3.9 | + |
| HIV gag | SLYNTVATL | 31/150 | ND | + |
The HLA-A2—restricted CMV-pp65 and HIV-gag peptides were used as positive controls in CMV-infected and HIV-infected individuals, respectively.
ND indicates not determined.
Peptide-binding scores determined using algorithms based on the estimated half-time dissociation of the HLA-peptide complex (www-bimas.dcrt.nih.gov) or prediction of MHC class I ligands.
Fluorescence index (FI) = (MFI with peptide—MFI without peptide)/(MFI without peptide).
Brander et al10 first described this epitope.
Wang et al13 first described this epitope.
Bourboulia et al14 first described this epitope.
The frequency, magnitude, and diversity of HHV8-specific responses depend on the clinical setting
A series of 18 HLA-A2– individuals was analyzed for each tetramer, allowing us to define a detection limit for positive staining in HLA-A2+ individuals (cutoff = mean frequency of tetramer+ cells in HLA-A2– subjects + 2 SD; Table 2), as described in Pittet et al19 and Gannagé et al.20 Using these cutoffs, we did not detect HHV8 tetramer+ CD8 T cells in 8 HLA-A2+ healthy blood donors (data not shown). Using the CMV-pp65 tetramer as positive control, we found specific CD8 T cells in all HLA-A2+ CMV-seropositive healthy subjects (mean, 1.40%; range, 0.05%-6.3%).
Percentages of tetramer+ CD8 T cells in PBMCs of HHV8-seropositive patients
Clinical group, UPN . | Disease status* . | K12 . | gB . | ORF65 . | ORF6 . | ORF61 . | pp65 . |
|---|---|---|---|---|---|---|---|
| Cutoff value | 0.26 | 0.17 | 0.18 | 0.26 | 0.16 | 0.24 | |
| Group 1: transplant recipients | |||||||
| 1 | KS- | 0.2 | 0.07 | 0.23 | 0.3 | 0.16 | 2.96 |
| 2 | KS- | 0.5 | 0.01 | 0.3 | 0.42 | 1.1 | 0.43 |
| 3 | KS- | 0.12 | 0.09 | 0.32 | 0.41 | 0.08 | 0.12 |
| 4 | KS- | 0.19 | 0.08 | 0.25 | 0.29 | 0.06 | 0.29 |
| 5 | KS- | 0.31 | 0.34 | 0.38 | 1.06 | 0.37 | 12.9 |
| 6 | KS- | 0.27 | 0.17 | 0.38 | 1.4 | 0.38 | 0.06 |
| 7 | KS- | 0.52 | 0.1 | 0.63 | 0.11 | 0.30 | 0.53 |
| 8 | KS- | 0.26 | 0.22 | 0.17 | 0.65 | 0.2 | 0.38 |
| 9 | KSR | 0.59 | 0.3 | 0.28 | 0.03 | 0.70 | 1.72 |
| 10 | KSR | 0.31 | 0.22 | 0.30 | 0.48 | 0.12 | 0.24 |
| 11 | KS+ | 1.98 | 2.06 | 2.2 | 1.85 | 0.45 | — |
| 12 | KS+ | 0.22 | 0.09 | 0.01 | 0.23 | 0.12 | 0.15 |
| Group 2: HIV | |||||||
| 13 | KS+ | 0.18 | 0.24 | 0.23 | 0.11 | 0.05 | 0.02 |
| 14 | KS+ | 0.00 | 0.00 | 0.00 | 0.05 | 0.05 | 4.2 |
| 15 | KS+ | 0.13 | 0.18 | 0.13 | 0.24 | 0.21 | 3.2 |
| 16 | KS+ | 0.38 | 0.19 | 0.33 | 0.56 | 0.08 | — |
| 17 | KS+ | 0.04 | 0.00 | 0.02 | 0.13 | 0.04 | 2.75 |
| 18 | KS+ | 0.31 | 0.14 | 0.26 | 0.34 | 0.13 | — |
| 19 | KS+ | 0.28 | 0.2 | 0.39 | 0.46 | 0.11 | — |
| 20 | KSR | 0.08 | 0.01 | 0.01 | 0.02 | 0.07 | 5.65 |
| 21 | KSR | 0.03 | 0.00 | 0.01 | 0.11 | 0.15 | 5.33 |
| 22 | KSR | 0.04 | 0.02 | 0.02 | 0.09 | 0.15 | 3.66 |
| 23 | KSR | 0.09 | 0.03 | 0.01 | 0.06 | 0.03 | 1.00 |
| 24 | KSR | 0.04 | 0.01 | 0.02 | 0.1 | 0.03 | 0.7 |
| 25 | KSR | 0.25 | 0.06 | 0.19 | 0.05 | 0.09 | 6.98 |
| 26 | KSR | 0.01 | 0.00 | 0.01 | 0.13 | 0.24 | 0.95 |
| 27 | KSR | 0.22 | 0.04 | 0.13 | 0.04 | 0.05 | 0.16 |
| 28 | KSR | 0.1 | 0.08 | 0.07 | 0.2 | 0.1 | 4.38 |
| 29 | KSR | 0.11 | 0.03 | 0.06 | 0.06 | 0.02 | 1.3 |
| 30 | KSR | 0.2 | 0.02 | 0.1 | — | — | 1.23 |
| 31 | KSR | 0.1 | 0.1 | 0.13 | 0.04 | 0.03 | 0.09 |
| 32 | KSR | 0.25 | 0.07 | 0.1 | 0.05 | 0.12 | 0.5 |
| 33 | KSR | 0.37 | 0.07 | 0.1 | 0.03 | 0.05 | 0.02 |
| 34 | KSR | 0.25 | 0.04 | 0.07 | 0.04 | 0.04 | 2.66 |
| 35 | KSR | 0.21 | 0.1 | 0.24 | 0.33 | 0.1 | 0.54 |
| 36 | KSR | 0.2 | 0.16 | 0.2 | 0.26 | 0.17 | 6.00 |
| Group 3: classical | |||||||
| 37 | KS+ | 0.15 | 0.09 | 0.28 | 0.18 | 0.13 | 2.00 |
| 38 | KS+ | 0.17 | 0.07 | 0.15 | 0.45 | 0.11 | 0.06 |
| 39 | KS+ | 0.15 | 0.15 | 0.06 | 0.13 | 0.06 | — |
| 40 | KS+ | 0.2 | 0.14 | 0.21 | 0.28 | 0.38 | 1.29 |
| 41 | KS+ | 0.13 | 0.3 | 0.17 | 0.22 | 0.31 | 4.64 |
| 42 | KS+ | 0.52 | 0.62 | 0.57 | 0.49 | 0.39 | 0.31 |
| 43 | KS+ | 0.07 | 0.21 | 0.2 | 0.53 | 0.03 | 0.11 |
| 44 | KS+ | 0.35 | 0.2 | 0.65 | 0.35 | 0.32 | 2.85 |
| 45 | KS+ | 0.11 | 0.4 | 0.1 | 0.07 | 0.08 | 1.48 |
| 46 | KSR | 0.1 | 0.08 | 0.11 | 0.17 | 0.12 | 0.07 |
| 47 | KSR | 0.12 | 0.57 | 0.32 | — | — | — |
| 48 | KSR | 0.31 | 0.45 | 0.4 | 0.93 | 0.2 | 2.39 |
| Total patients with positive response, % | 31 | 35 | 50 | 43.5 | 32 | 74 | |
| Mean percentage of tetramer+ cells | 0.37 | 0.31 | 0.3 | 0.44 | 0.31 | 2.96 |
Clinical group, UPN . | Disease status* . | K12 . | gB . | ORF65 . | ORF6 . | ORF61 . | pp65 . |
|---|---|---|---|---|---|---|---|
| Cutoff value | 0.26 | 0.17 | 0.18 | 0.26 | 0.16 | 0.24 | |
| Group 1: transplant recipients | |||||||
| 1 | KS- | 0.2 | 0.07 | 0.23 | 0.3 | 0.16 | 2.96 |
| 2 | KS- | 0.5 | 0.01 | 0.3 | 0.42 | 1.1 | 0.43 |
| 3 | KS- | 0.12 | 0.09 | 0.32 | 0.41 | 0.08 | 0.12 |
| 4 | KS- | 0.19 | 0.08 | 0.25 | 0.29 | 0.06 | 0.29 |
| 5 | KS- | 0.31 | 0.34 | 0.38 | 1.06 | 0.37 | 12.9 |
| 6 | KS- | 0.27 | 0.17 | 0.38 | 1.4 | 0.38 | 0.06 |
| 7 | KS- | 0.52 | 0.1 | 0.63 | 0.11 | 0.30 | 0.53 |
| 8 | KS- | 0.26 | 0.22 | 0.17 | 0.65 | 0.2 | 0.38 |
| 9 | KSR | 0.59 | 0.3 | 0.28 | 0.03 | 0.70 | 1.72 |
| 10 | KSR | 0.31 | 0.22 | 0.30 | 0.48 | 0.12 | 0.24 |
| 11 | KS+ | 1.98 | 2.06 | 2.2 | 1.85 | 0.45 | — |
| 12 | KS+ | 0.22 | 0.09 | 0.01 | 0.23 | 0.12 | 0.15 |
| Group 2: HIV | |||||||
| 13 | KS+ | 0.18 | 0.24 | 0.23 | 0.11 | 0.05 | 0.02 |
| 14 | KS+ | 0.00 | 0.00 | 0.00 | 0.05 | 0.05 | 4.2 |
| 15 | KS+ | 0.13 | 0.18 | 0.13 | 0.24 | 0.21 | 3.2 |
| 16 | KS+ | 0.38 | 0.19 | 0.33 | 0.56 | 0.08 | — |
| 17 | KS+ | 0.04 | 0.00 | 0.02 | 0.13 | 0.04 | 2.75 |
| 18 | KS+ | 0.31 | 0.14 | 0.26 | 0.34 | 0.13 | — |
| 19 | KS+ | 0.28 | 0.2 | 0.39 | 0.46 | 0.11 | — |
| 20 | KSR | 0.08 | 0.01 | 0.01 | 0.02 | 0.07 | 5.65 |
| 21 | KSR | 0.03 | 0.00 | 0.01 | 0.11 | 0.15 | 5.33 |
| 22 | KSR | 0.04 | 0.02 | 0.02 | 0.09 | 0.15 | 3.66 |
| 23 | KSR | 0.09 | 0.03 | 0.01 | 0.06 | 0.03 | 1.00 |
| 24 | KSR | 0.04 | 0.01 | 0.02 | 0.1 | 0.03 | 0.7 |
| 25 | KSR | 0.25 | 0.06 | 0.19 | 0.05 | 0.09 | 6.98 |
| 26 | KSR | 0.01 | 0.00 | 0.01 | 0.13 | 0.24 | 0.95 |
| 27 | KSR | 0.22 | 0.04 | 0.13 | 0.04 | 0.05 | 0.16 |
| 28 | KSR | 0.1 | 0.08 | 0.07 | 0.2 | 0.1 | 4.38 |
| 29 | KSR | 0.11 | 0.03 | 0.06 | 0.06 | 0.02 | 1.3 |
| 30 | KSR | 0.2 | 0.02 | 0.1 | — | — | 1.23 |
| 31 | KSR | 0.1 | 0.1 | 0.13 | 0.04 | 0.03 | 0.09 |
| 32 | KSR | 0.25 | 0.07 | 0.1 | 0.05 | 0.12 | 0.5 |
| 33 | KSR | 0.37 | 0.07 | 0.1 | 0.03 | 0.05 | 0.02 |
| 34 | KSR | 0.25 | 0.04 | 0.07 | 0.04 | 0.04 | 2.66 |
| 35 | KSR | 0.21 | 0.1 | 0.24 | 0.33 | 0.1 | 0.54 |
| 36 | KSR | 0.2 | 0.16 | 0.2 | 0.26 | 0.17 | 6.00 |
| Group 3: classical | |||||||
| 37 | KS+ | 0.15 | 0.09 | 0.28 | 0.18 | 0.13 | 2.00 |
| 38 | KS+ | 0.17 | 0.07 | 0.15 | 0.45 | 0.11 | 0.06 |
| 39 | KS+ | 0.15 | 0.15 | 0.06 | 0.13 | 0.06 | — |
| 40 | KS+ | 0.2 | 0.14 | 0.21 | 0.28 | 0.38 | 1.29 |
| 41 | KS+ | 0.13 | 0.3 | 0.17 | 0.22 | 0.31 | 4.64 |
| 42 | KS+ | 0.52 | 0.62 | 0.57 | 0.49 | 0.39 | 0.31 |
| 43 | KS+ | 0.07 | 0.21 | 0.2 | 0.53 | 0.03 | 0.11 |
| 44 | KS+ | 0.35 | 0.2 | 0.65 | 0.35 | 0.32 | 2.85 |
| 45 | KS+ | 0.11 | 0.4 | 0.1 | 0.07 | 0.08 | 1.48 |
| 46 | KSR | 0.1 | 0.08 | 0.11 | 0.17 | 0.12 | 0.07 |
| 47 | KSR | 0.12 | 0.57 | 0.32 | — | — | — |
| 48 | KSR | 0.31 | 0.45 | 0.4 | 0.93 | 0.2 | 2.39 |
| Total patients with positive response, % | 31 | 35 | 50 | 43.5 | 32 | 74 | |
| Mean percentage of tetramer+ cells | 0.37 | 0.31 | 0.3 | 0.44 | 0.31 | 2.96 |
Cutoff value was defined for each tetramer as the mean frequency + 2 SD of tetramer+ cells in 18 HLA-A2- HHV8+ individuals. Responses in patients were considered positive if the percentage of tetramer+ cells was above the cutoff value. Positive values are shown in italics.
UPN indicates unique patient number; and —, not done.
Disease status at time of analysis, defined as patients with ongoing KS (KS+), patients with KS in remission (KSR), and patients without KS lesions (KS-).
The frequency and diversity of HHV8-specific CD8 T-cell responses in 48 patients infected with HHV8 is shown in Table 2. Positive responses to at least 1 latent or lytic HHV8 epitope were detected in 67% of patients, with tetramer+ cell frequencies ranging from 0.16% to 2.2% of CD8 T cells. A mean of 2 HHV8 peptides were recognized per patient. These results indicate that anti-HHV8 CD8 T-cell responses are frequent in infected individuals, and show their diversity and lack of immunodominance. Responses to the CMV-pp65 peptide were detected in 76% of patients (0.24%-16.3% of CD8 T cells).
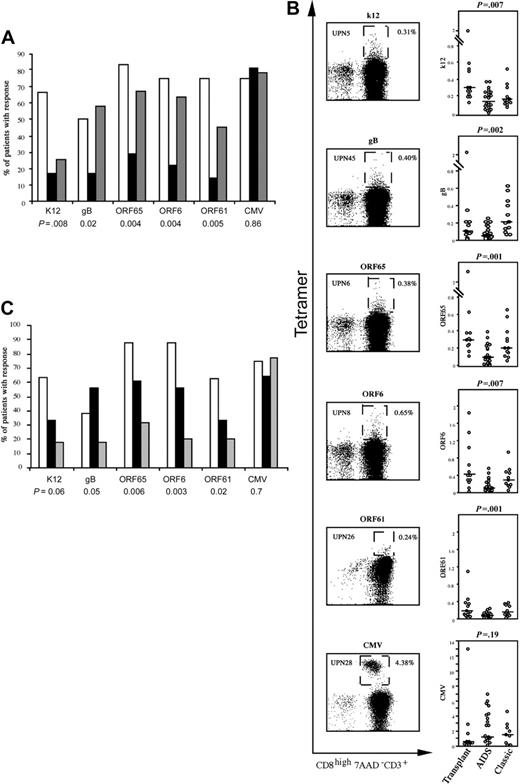
Because the context in which HHV8 infection occurred was not the same among the different patients, we examined whether HHV8-specific CD8 responses varied by comparing transplant recipients, patients with AIDS-related KS, and patients with classical KS. HHV8-specific T-cell responses were less frequent in AIDS-related KS than in transplant recipients or patients with classical KS (Figure 1A). Moreover, responses in AIDS-related KS were of lower magnitude (Figure 1B) and diversity: a mean of 0.96 HHV8 peptides were recognized per HIV-infected patient, compared with 2.5 peptides in patients with classical KS and 3.4 peptides in transplant recipients (P < .001). To ensure that the weak responses in HIV-infected patients were not related to a general weakness of antiviral T-cell immunity, we measured HIV- and CMV-specific responses by using relevant tetramers. Responses to the HLA-A*0201–restricted HIV gag epitope were low, as expected in patients on HAART. However, responses to the immunodominant pp65 peptide of CMV were strong in more than 80% of patients (0.5%-6.3% of CD8 T cells).21 Likewise, the weak HHV8-specific response was not associated with HIV-related CD4 T-cell depletion, as 55% of patients with CD4 cell counts of more than 200/mm3 had no responses, compared with 33% of patients with counts below this value (P = .61; Fisher exact test). The HIV-infected patients were younger than the transplant recipients and the patients with classical KS (P = .005), indicating that weak anti-HHV8 responses were not related to an age-associated decline in virus-specific T-cell responses. Together, these results show that patients with AIDS-related KS have a lower capacity to mount anti-HHV8 responses than transplant recipients and patients with classical KS.
HHV8-specific responses are impaired in patients who progress to KS
As the course of HHV8 infection was not the same among the different groups of patients, we examined whether HHV8-specific responses could influence the progression of viral infection by grouping the patients according to the absence or presence of KS. The first group was composed of 8 transplant recipients who were HHV8 seropositive at the time of grafting but remained consistently PCR– and did not develop KS after a mean follow-up of 24 months. They were compared with 40 patients who progressed to KS (4 kidney transplant recipients, 12 patients with classic KS, and 24 patients with AIDS-related KS). The patients who did not develop KS had more frequent anti-HHV8 T-cell responses than the patients who developed KS, while the frequency of CMV-specific responses was similar in the 2 groups (Figure 1C), suggesting that these differences did not reflect global CD8 T-cell dysfunction in the patients with KS. In addition, the patients who did not develop KS had a broader repertoire of HHV8-specific responses than the patients who progressed to KS: a mean of 3.6 and 1.6 HHV8 peptides, respectively, were recognized per patient (P = .007, nonparametric analysis of variance [ANOVA]). Although age is an established risk factor for KS, the patients with KS were not older than the patients without KS (52 and 54 years, respectively). These data suggest that multiepitopic HHV8-specific CD8 T cells contribute to HHV8 control and protection from KS. However, because patients who did not develop KS were all transplant recipients, it cannot be excluded that their higher frequency of HHV8-specific response is due to the clinical context in which HHV8 infection occurred. Indeed, our preliminary results in a series of HIV+ HHV8+ KS– patients showed weak HHV8-specific CD8 T-cell responses, of the same order of magnitude that responses observed in HIV+ KS+ patients (Table S1, available at the Blood website; see the Supplemental Table link at the top of the online article), suggesting that HIV infection by itself may be deleterious for HHV8 response.
The frequencies of HHV8-specific responses depend on the context and course of infection. (A) The number of patients with HHV8-specific CD8 T-cell responses were compared in 12 transplant recipients (□), 24 patients with AIDS-related KS (▪) and 12 patients with classical KS (▦) by chi-square test. The histograms show the percentages of patients with positive responses to the indicated peptide. (B) For each peptide, a representative example of the staining of the tetramer+ population is shown in the left panels, and grouped data for the frequency of the corresponding tetramer+ CD8 T cells are compared in the right panels. Boxes in the left panels delineate the tetramer+ CD8 T-cell population. The cut-off value for positive staining was defined by the mean + 2 SD frequency of tetramer+ cells in HLA-A2–/HHV8+ patients. Horizontal bars represent the median percentages of tetramer+ cells. P values are shown for the comparison between the 3 patient groups (nonparametric Kruskall-Wallis test). (C) Comparison of anti-HHV8 responses in patients without KS (n = 8; □), patients with KS with persistent lesions (n = 18; ▪) and patients with KS in remission (n = 22; ▦). The results are the percentages of patients with positive responses to the indicated peptide. P values are shown for the comparison between the 3 groups.
The frequencies of HHV8-specific responses depend on the context and course of infection. (A) The number of patients with HHV8-specific CD8 T-cell responses were compared in 12 transplant recipients (□), 24 patients with AIDS-related KS (▪) and 12 patients with classical KS (▦) by chi-square test. The histograms show the percentages of patients with positive responses to the indicated peptide. (B) For each peptide, a representative example of the staining of the tetramer+ population is shown in the left panels, and grouped data for the frequency of the corresponding tetramer+ CD8 T cells are compared in the right panels. Boxes in the left panels delineate the tetramer+ CD8 T-cell population. The cut-off value for positive staining was defined by the mean + 2 SD frequency of tetramer+ cells in HLA-A2–/HHV8+ patients. Horizontal bars represent the median percentages of tetramer+ cells. P values are shown for the comparison between the 3 patient groups (nonparametric Kruskall-Wallis test). (C) Comparison of anti-HHV8 responses in patients without KS (n = 8; □), patients with KS with persistent lesions (n = 18; ▪) and patients with KS in remission (n = 22; ▦). The results are the percentages of patients with positive responses to the indicated peptide. P values are shown for the comparison between the 3 groups.
Patients with KS were further subdivided into those (n = 22) who were in remission from KS and who had undetectable HHV8 viral load for at least 6 months at the time of the study (mean length of remission, 57 months), and those (n = 18) with persistent KS lesions. Anti-HHV8 CD8 T-cell responses were more frequent in patients with persistent lesions than in patients in remission. There was no correlation between the frequency of HHV8-specific responses and the duration of KS remission.
HHV8-specific CD8 T cells are functionally relevant
To determine whether the observed differences in the magnitude and diversity of HHV8-specific responses were functionally relevant, we characterized the phenotype of CD8 T cells directed against the predominant epitope(s) (ie, the epitope(s) associated with the highest percentage of tetramer+ cells in each patient). Significant differences in the extent of CD45RA, CD27, and CCR7 positivity among HHV8-tetramer+ cells were found between transplant recipients without KS, patients in remission from KS, and patients with ongoing KS (Figure 2A). We observed strong polarization toward a late CD45RA+/CCR7– or CD45RA+/CD27– phenotype in transplant recipients who spontaneously controlled HHV8 infection compared with patients with ongoing KS and patients in remission. Conversely, transplant recipients who spontaneously controlled HHV8 infection had lower percentages of early/intermediate differentiated CD8 T cells expressing a CD45RA–/CCR7– phenotype (Figure 2B). There were no differences according to whether cells responded to latent or lytic HHV8 epitopes. CMV tetramer+ cells were predominantly late differentiated effector cells (not shown). Of note, there was no correlation between HHV8 viral load and the number or differentiation phenotype of tetramer+ cells.
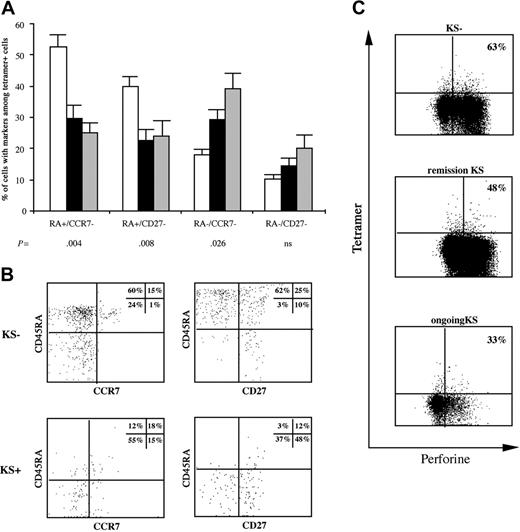
Phenotype of HHV8-specific cells according to the outcome of infection. (A) Tetramer+ cells were compared for the presence of CD45RA, CCR7, and CD27 markers in 14 patients from whom enough material was available (□ indicates 6 patients without KS; ▪, 7 patients with ongoing KS; and ▦, 5 patients in KS remission). The histograms show the percentages of tetramer+ cells (grouped data) with the markers indicated (mean ± SEM). (B) Flow cytometric analysis of CD45RA and CCR7 or CD27 coexpression gated on CD8+ tetramer+ cells. Each display represents results from a representative patient in each group. (C) Intracellular perforin content was determined in tetramer+ cells. Dot plots are gated on live CD8 T cells and show tetramer and perforin staining. The percentages indicated in the quadrants are representative of grouped data for patients without KS (median, 63%; range, 12%-99%), patients in KS remission (median, 48%; range, 25%-88%, nonsignificant), and patients with ongoing KS (median, 33%; range, 13%-66%, P = .04 for comparison with patients without KS).
Phenotype of HHV8-specific cells according to the outcome of infection. (A) Tetramer+ cells were compared for the presence of CD45RA, CCR7, and CD27 markers in 14 patients from whom enough material was available (□ indicates 6 patients without KS; ▪, 7 patients with ongoing KS; and ▦, 5 patients in KS remission). The histograms show the percentages of tetramer+ cells (grouped data) with the markers indicated (mean ± SEM). (B) Flow cytometric analysis of CD45RA and CCR7 or CD27 coexpression gated on CD8+ tetramer+ cells. Each display represents results from a representative patient in each group. (C) Intracellular perforin content was determined in tetramer+ cells. Dot plots are gated on live CD8 T cells and show tetramer and perforin staining. The percentages indicated in the quadrants are representative of grouped data for patients without KS (median, 63%; range, 12%-99%), patients in KS remission (median, 48%; range, 25%-88%, nonsignificant), and patients with ongoing KS (median, 33%; range, 13%-66%, P = .04 for comparison with patients without KS).
We then studied the relationship between the memory cell phenotype and perforin expression, which reflects immediate effector function (Figure 2C). Perforin levels were variable in terms of both the number of perforin-expressing tetramer+ cells and the staining intensity (low to high perforin content). However, we observed more perforin-expressing tetramer+ cells in patients without KS than in patients with KS. Thus, although their cytotoxic efficiency may vary, HHV8-specific CD8 T cells exhibit effector functions and may thus control progression to KS.
In situ tetramer staining of HHV8-specific CD8 T cells in KS biopsies. CD8 cells appear in red; HHV8-specific tetramer+ cells appear in green; and LANA-specific cells appear in blue. Large 3-channel merged images are shown. Compared with large tetramer– CD8 T-cell infiltrates in the vicinity of LANA+ tumoral cells (left panel), double-stained HHV8-specific CD8 T cells appear as isolated yellow cells (arrow) scarcely dispersed within the LANA– tissue (middle panel). Detection of LANA+ cells by immunohistochemistry is shown as control (right panel). Magnification, × 63.
In situ tetramer staining of HHV8-specific CD8 T cells in KS biopsies. CD8 cells appear in red; HHV8-specific tetramer+ cells appear in green; and LANA-specific cells appear in blue. Large 3-channel merged images are shown. Compared with large tetramer– CD8 T-cell infiltrates in the vicinity of LANA+ tumoral cells (left panel), double-stained HHV8-specific CD8 T cells appear as isolated yellow cells (arrow) scarcely dispersed within the LANA– tissue (middle panel). Detection of LANA+ cells by immunohistochemistry is shown as control (right panel). Magnification, × 63.
In situ visualization of HHV8-specific CD8 T cells
To determine whether low frequency of HHV8-specific CD8 T cells in the peripheral blood of patients with KS is secondary to their specific migration within the tumor, we used in situ tetramer staining and confocal laser scanning microscopy to analyze KS biopsy specimens taken from 2 patients who had detectable circulating HHV8-specific responses at the same time. Large tetramer– CD8 T-cell infiltrates were observed in the vicinity of LANA+ infected cells (Figure 3). However, only very few CD8 T cells exhibited costaining with HHV8 tetramers, and these tetramer+ cells did not colocalize with tumor cells, but were scattered among LANA– areas. A control biopsy performed in a KS– HLA-A2+ patient for diagnostic purposes showed no staining with the tetramers (not shown). This indicates that HHV8 epitope–specific CD8 T cells are not recruited within inflamed tumoral tissue, which may at least in part explain the lack of T-cell–mediated tumor immunity in patients with KS.
CD8 T-cell phenotype shifts following changes in viral load and clinical events
To determine the functional relevance of HHV8-specific T cells in vivo, we studied their changes over time, with respect to clinical events and viral load changes, in a transplant recipient who presented with recurrent remission/relapse episodes of KS lesions (UPN11; Figure 4). Expansions (3- to 6-fold) of both lytic and latent HHV8-tetramer+ cells were observed over periods of about 3 months, and these changes coincided with HHV8 viral load negativization and complete regression of KS lesions. More than 90% of these expanded HHV8-specific T cells had a late effector cell phenotype and abundantly expressed perforin. After a few months of clinical remission, viral load rebounded (except at month 8, when it was still undetectable), tetramer+ cell numbers fell, and KS lesions relapsed. Although restricted to 1 patient, these data indicate that rapid variations in HHV8-specific CD8 T-cell numbers may result from antigen-specific cell proliferation due to viral reactivation. This also indicates that HHV8-specific CD8 T cells participate in the control of KS lesions.
Time course of HHV8-specific CD8 T cells in a transplant recipient with recurring KS. The figure illustrates the dynamics of HHV8 tetramer+ cells tested at approximately 3-month intervals over 2 years (beginning 4 years after grafting, indicated as M0), and shows major clinical events and viral load variations during the same period. × indicates gB; ▴, ORF65; ○, k12; ⋄, ORF6; and ▪, ORF61. • indicates the time course of HHV8 DNA load (copies/mL). Clinical relapses are indicated by arrows. The quadrant shows the phenotype profile and perforin expression of ORF6 tetramer+ cells at the indicated time.
Time course of HHV8-specific CD8 T cells in a transplant recipient with recurring KS. The figure illustrates the dynamics of HHV8 tetramer+ cells tested at approximately 3-month intervals over 2 years (beginning 4 years after grafting, indicated as M0), and shows major clinical events and viral load variations during the same period. × indicates gB; ▴, ORF65; ○, k12; ⋄, ORF6; and ▪, ORF61. • indicates the time course of HHV8 DNA load (copies/mL). Clinical relapses are indicated by arrows. The quadrant shows the phenotype profile and perforin expression of ORF6 tetramer+ cells at the indicated time.
HHV8-exposed but uninfected individuals can mount specific CD8 T-cell responses
Because primary HHV8 infection is usually asymptomatic, it is difficult to follow initial immune responses to the virus. We were able to examine 2 HLA-A2+ HHV8-seronegative patients who received kidney grafts from HHV8-infected donors. Both recipients remained consistently aviremic and antibody negative for 24 and 11 months after grafting, respectively. Interestingly, HLA-A2–restricted HHV8-specific tetramer+ cells were found in the recipient who received a graft from an HLA-A2+ donor (Figure 5A), while no response was detected in the other recipient, whose donor was HLA-A2–. The presence of tetramer+ cells specific for all lytic epitopes in the first recipient suggests that viral replication at very low levels, below those detected by current assays, primed the immune response. As most tetramer+ cells were phenotypically characterized as late effector cells expressing perforin (Figure 5B-C), it is likely that the generation of HHV8-specific CTLs can lead to abortive infection. Furthermore, the absence of detectable responses in the recipient with an HLA-A2– donor suggests that HHV8-specific CD8 T cells are restricted by donor HLA alleles. Therefore, the presence of specific CTLs may provide protection from persistent HHV8 infection.
Discussion
Several viruses, such as Epstein-Barr virus (EBV), CMV, and hepatitis C virus (HCV) can establish persistent infection, their replication being tightly controlled by specific CTLs.22-24 Immune responses to HHV8, the most recently identified human herpesvirus, are far less well documented. Our results indicate that HLA-A2–restricted HHV8-specific CTLs can both abort the primary infection and control the latent infection, thereby protecting the host from progression to KS. As almost half of the white population is positive for HLA-A2, these observations have strong implications for the design of prophylactic and therapeutic vaccine strategies.
We found that HHV8-infected subjects generated polyclonal CD8 T cells targeting a variety of viral proteins. Lytic (gB, ORF6, ORF61, and ORF65) or latent (K12) antigen-specific cells were detected in more than two-thirds of patients, and together could account for up to 8% of the circulating CD8 pool. The magnitude, diversity, and functional capacity of these cells varied, however, according to the clinical context (circumstances and outcome of primary infection). HHV8-specific CD8 T cells were more frequent and more diverse in patients who controlled HHV8 infection than in those who progressed to KS. Moreover, we observed a marked enrichment of late effector cells with cytotoxic potential in HHV8+ transplant recipients who did not progress to KS, whereas patients with AIDS-related or classical KS showed a substantial enrichment in cells with an earlier phenotype and weak cytotoxic potential. As the HHV8+ transplant recipients remained free of KS for at least 2 years after grafting, it is likely that they will never develop KS.25 If resistance to KS is indeed mediated by HHV8-specific CTLs, then the protection they confer seems to last long enough to avoid tumor development during the first months after grafting, when immunosuppression is maximal. A kinetic analysis of a transplant recipient with recurring KS further confirmed the functionality of antiviral CTLs in vivo. Indeed, a fall in the frequency of HHV8-specific CD8 T cells coincided with viremic rebound and with clinical relapse.
HHV8-specific CD8 T cells in a seronegative transplant recipient exposed to the virus through the graft. The figure shows the staining of the tetramer+ population for the indicated peptides (A), their perforin expression (B), and their phenotype profile (C), performed 12 months after transplantation with a kidney graft from an HHV8-infected donor. (A) Boxes delineate the tetramer+ CD8 T-cell population.
HHV8-specific CD8 T cells in a seronegative transplant recipient exposed to the virus through the graft. The figure shows the staining of the tetramer+ population for the indicated peptides (A), their perforin expression (B), and their phenotype profile (C), performed 12 months after transplantation with a kidney graft from an HHV8-infected donor. (A) Boxes delineate the tetramer+ CD8 T-cell population.
Why were HHV8-specific responses impaired in some patients? Because HAART also controls HHV8 replication,26 anti-HHV8 responses might wane over time in patients on HAART who enter remission from KS, in keeping with the dependency of specific CTLs on continued viral replication.27 In a recent study, responses to ORF73 and ORF65 were observed in HIV-coinfected patients with active KS and elevated HHV8 load, but whether the detected responses were CD4 or CD8 mediated was not known.28 We found here that HHV8-specific responses could persist in transplant recipients at relatively high levels in the absence of detectable circulating virus. Therefore, it is likely that HIV infection itself impairs cellular responses to HHV8. Indeed, our preliminary results in HIV+ HHV8+ KS– patients, which showed weak HHV8-specific CD8 T-cell responses, support this hypothesis. The reason for the weak responses observed in patients with classical KS is more puzzling, but may relate to differences in the route of antigen exposure.29 Saliva is the source of most infections in endemic areas. KS in transplant recipients is primarily due to virus reactivation, but de novo infection by the allograft has also been described. As HHV8 load (DNA copies) is higher in saliva than in blood or semen, the distinct patterns of CD8 T-cell differentiation observed here might reflect route- and inoculum-dependent differences in the nature or efficiency of antigen processing, or in the repertoire of CD8 T cells that interact with antigen-presenting cells at the site of priming. In addition, HHV8-specific T cells might be prevented from exerting their antitumoral effector function by the multiple immunoregulatory strategies that KS cells use to avoid immune recognition, such as down-regulation of MHC class I and costimulatory molecules.30-35 Such mechanisms could also explain the defect of migration and recruitment of virus-specific CTLs to the tumoral tissue.
Of particular interest is the case of the transplant recipient who was exposed to HHV8 through the graft, and who had persistent HHV8-specific responses several months after grafting without developing detectable HHV8 infection. This patient may have had a very low level viral replication that primed the immune response, as observed recently in seronegative subjects who are at risk for HHV8 infection28 and in persons who are repeatedly exposed to HIV or HCV but who do not seroconvert,36-38 This case also raises a conceptual issue—how the HHV8-specific repertoire is shaped by donor MHC alleles. Recognition of the infected allograft may involve recipient-derived T cells either via HLA molecules shared with the donor, or through expansion of T cells that are solely restricted by the donor's HLA molecules. We found that this patient's HHV8 tetramer+ cells were apparently restricted by donor HLA molecules, as described in liver transplant recipients whose allograft is infected by HCV.39 These HHV8 tetramer+ cells were not simply alloreactive cells, as they did not bind an irrelevant HLA-A2 tetramer containing HIV gag peptide (not shown). They were late effector cells, capable of invading peripheral tissues, including the infected graft. These findings support the hypothesis that an HHV8-infected graft is capable of stimulating and expanding naive recipient CD8 T cells, either directly via presentation of HHV8 antigens by infected B cells or endothelial cells, or indirectly via cross-presentation of HHV8 peptides by donor-derived dendritic cells. Whatever the mechanism, de novo acquisition of HHV8-specific CD8 T cells that mediate protection from persistent HHV8 infection in the recipient has major implications for HHV8 vaccine development and immunotherapy. It was recently shown that KS progenitor cells, which are thought to be infected by the virus, can be present in solid grafts and undergo neoplastic transformation in the recipient,40 suggesting that KS tumor cells form continually in infected individuals but are suppressed or killed through normal immune surveillance. Our data strongly corroborate this idea.
In conclusion, although a larger selection of patients with varied HLA types needs to be evaluated, our study highlights hitherto undescribed differences in CD8 T-cell responses between patients who resolve or control HHV8 infection and patients who progress to KS. We confirm the crucial role of HHV8-specific CTLs as effectors of long-term immune surveillance against a persistent virus, not only for controlling viral replication and preventing malignancies associated with latent infection, but also for generating genuine resistance to primary infection. This suggests that interventions aimed at controlling HHV8 infection should be started early. There is currently no way of predicting the individual risk of progression to KS in HHV8-infected individuals, and it will be interesting to see if anti-HHV8 responses could serve this purpose. Finally, our data support the use of vaccines or adoptive strategies to boost HHV8-specific CTL responses in patients with KS.
Authorship
The authors declare no competing financial interests.
M.L. and M.G. contributed equally to the study.
Prepublished online as Blood First Edition Paper, August 22, 2006; DOI 10.1182/blood-2006-03-014225.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by a grant from Sidaction (Paris, France). M.L. was supported by the Association pour la Recherche contre le Cancer (ARC); M.G. and M.A. were supported by the Institut Nationale de la Santé et de la Recherche Médicale (INSERM).




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal