Abstract
Human cytomegalovirus (HCMV) establishes and maintains a latent infection in myeloid cells and can reactivate to cause serious disease in allograft recipients. To better understand the molecular events associated with the establishment of latency, we tracked the virus following infection of primary human myeloid progenitor cells at days 1, 2, 3, 5, and 11. At all time points, the viral genome was maintained in most cells at approximately 10 copies. Infectious virus was not detected, but virus could be reactivated by extended fibroblast coculture. In contrast to wild-type HCMV, the viral genome was rapidly lost from myeloid progenitors infected with ultraviolet (UV)–inactivated virus, suggesting viral gene expression was required for efficient establishment of latency. To identify viral genes associated with the establishment phase, RNA from each time point was interrogated using custom-made HCMV gene microarrays. Using this approach, we detected expression of viral RNAs at all time points. The pattern of expression differed from that which occurs during productive infection, and decreased over time. This study provides evidence that a molecular pathway into latency is associated with expression of a unique subset of viral transcripts. Viral genes expressed during the establishment phase may serve as targets for therapies to interrupt this process.
Introduction
Human cytomegalovirus (HCMV) is a species-specific betaherpesvirus carried by a majority of the population. Following primary infection, HCMV has the ability to establish a lifelong latent infection, from which it periodically reactivates to form new infectious virus that can be transmitted to other susceptible individuals.1 Although primary and reactivated infections are usually mild or asymptomatic in healthy persons, primary infection is a leading cause of congenital infection leading to neurologic damage in children, and reactivated infection is a major cause of serious disease in immunocompromised individuals, such as allogeneic transplant recipients.2 Reactivation from latency during immunosuppression is often followed by disseminated HCMV, which can lead to severe gastrointestinal tract infections, hepatitis, pneumonia, accelerated development of coronary artery atherosclerosis, chronic rejection in solid organ transplant recipients, and chronic graft-versus-host disease in bone marrow allograft recipients.2 The ability of HCMV to establish, maintain, and reactivate from a latent state contributes significantly to its success as a human pathogen, yet these phases of infection remain poorly understood.
HCMV latent infection can be separated into establishment and maintenance phases, where the establishment phase refers to the early events that lead to the maintenance of lifelong latency. The majority of studies on latency have sought to assess various aspects relating to the maintenance phase. Studies have demonstrated that viral DNA can be detected in myeloid cells from peripheral blood of healthy seropositive individuals in the absence of detectable infectious virus.3-5 In addition, the virus has been reactivated from terminally differentiated monocyte-derived macrophages6 or differentiated dendritic cell precursors,7 implicating myeloid lineage cells as an important site of latent infection. The HCMV genome is maintained during latency in mononuclear cells as a circular episome8 and it has been estimated that the viral genome is carried in between 0.004% and 0.01% of naturally infected mononuclear cells, at an average of approximately 2 to 13 genome copies per latently infected cell.9 Due to this low level of natural HCMV latent infection and its strict species specificity, experimental models have been developed that have proved to be valuable in providing new insights into HCMV latency.10-12 In an effort to further characterize the interaction of HCMV with myeloid cells, viral gene expression during latent infection has been examined using experimentally infected granulocyte macrophage progenitor cells (GM-Ps). These studies identified novel HCMV latency-associated transcripts (CLTs) encoded within the major immediate early (MIE) region of the genome.9,10,13-15 These MIE region CLTs have also been detected in HCMV seropositive donors, and sera collected from these donors have antibodies to predicted latency-associated open reading frames (ORFs).14,16 The functions encoded by MIE CLTs and their respective ORFs have not yet been defined.17 We recently identified a novel transcript derived from the UL111A region of the viral genome that was expressed during both experimental latent infection of GM-Ps and in naturally infected mobilized peripheral blood and bone marrow samples.18 This UL111A region transcript encodes a protein with homology to the potent immunosuppressor human interleukin-10 (IL-10). Transcriptional activity from the US28 region has also been reported following nonproductive infection of undifferentiated THP-1 monocytic cells.19 In a broader assessment of viral gene expression during experimental latent infection of hematopoietic progenitor cells, more extensive expression of viral transcripts has been reported.11,20 These studies demonstrate that the virus remains transcriptionally active during the latent phase of infection.
In this study we sought to better understand the molecular events that accompany the establishment of latency in primary human myeloid progenitor cells. We applied a combination of cell-dilution PCR and quantitative-competitive–polymerase chain reaction (QC-PCR) to track the distribution and quantity of viral genomes in myeloid progenitor cells over a time course, starting at early times after infection, and contrasted this with productive infection of human fibroblasts (HFs). We also compared viable and UV-inactivated viruses for their ability to successfully establish latency, and showed that viral gene expression appears to be a prerequisite for this process to occur. Finally, to identify viral genes involved in this process, we surveyed RNA extracted over a time course of infection of myeloid progenitor cells against custommade HCMV gene microarrays to both identify and map the kinetics of expression of a subset of viral genes that accompany the establishment of a reactivatable latent infection.
Materials and methods
Cells and virus culture
Human granulocyte macrophage progenitor cells (GM-Ps) were freshly isolated from human fetal liver samples as previously described10 and further purified for early CD34+ progenitor cells by magnetic bead separation twice in LS+ columns according to the manufacturer's instructions (Direct CD34 Progenitor Cell Isolation Kit; Miltenyi Biotec, Auburn, CA). Purity of isolated cells was assessed by flow cytometry using a CD34 monoclonal antibody conjugated to PE (Miltenyi Biotec) and IgG2a-PE antibodies (Becton Dickinson, Mountain View, CA) as an isotype control. On day 3 of culture in GM-P media,10 cells were either mock infected or infected with cell-free stocks of HCMV strain TownevarRIT321,22 at a multiplicity of infection (MOI) of 3 for 3 hours. Cells were washed 3 times in Hanks balanced salt solution (HBSS) and maintained in GM-P media. HFs were used for the generation of positive control productively infected cells, virus propagation, and plaque assay. To generate UV-inactivated virus stocks, cell-free HCMV was exposed to 0.72 J of UV light using a UV Stratalinker 1800 (Stratagene, La Jolla, CA). Complete loss of virus viability was confirmed by a lack of any detectable cytopathic effect (CPE) following inoculation of HF monolayers.
Flow cytometry
Cells (1.0 × 105) were first washed and then suspended in 100 μL fluorescence-activated cell sorting (FACS) staining buffer (phosphate-buffered saline [PBS] with 1% fetal bovine serum, 0.2% sodium azide). Anti–CD34-PE (Miltenyi Biotec) or anti–CD33-PE antibody (Becton Dickinson) was added to a final 1:20 dilution and incubated for 30 minutes at 4°C. In all experiments, mock- and HCMV-infected cells were incubated with isotype control antibodies to control for nonspecific antibody binding. Cells were washed with FACS staining buffer and suspended in FACS staining buffer with 1% paraformaldehyde for evaluation with a FACSCalibur flow cytometer (Becton Dickinson).
Quantitative-competitive–polymerase chain reaction (QC-PCR) and cell-dilution PCR
Cells were suspended in lysis buffer10 at a concentration of 1.0 × 104 cells per 10 μL, incubated for 16 hours at 65°C, followed by 10 minutes at 98°C to inactivate Proteinase K. For QC-PCR, each 2 μL aliquot of cell lysate was analyzed in the presence of between 1.0 and 1.0 × 106 copies of a denatured HCMV ie1/ie2 cDNA competitor pON2347.10 PCR amplification for 40 cycles (94°C for 1 minute, 62°C for 1 minute, and 72°C for 2 minutes) was performed with primers IEP3C and IEP4BII.9,10 After amplification, 20% of each reaction was analyzed by electrophoresis in 2.5% agarose gels. The relative quantities of PCR products derived from genomic and competitor templates were estimated after staining with ethidium bromide. For cell-dilution PCR, serial dilutions of cell lysates were amplified for 40 cycles with primers IEP3C and IEP4BII using the conditions described for QC-PCR.
In some experiments, acid stripping of cell-surface–bound extraneous membranes was performed in a stripping medium consisting of cold Dulbecco modified Eagle medium (DMEM), supplemented with 1% bovine serum albumin (BSA) and adjusted to pH 2.8 with HCl. Cells were washed 3 times in cold PBS, centrifuged at 270g for 7 minutes, and then resuspended in stripping medium for 3 minutes at 4°C. Stripping was stopped by the addition of neutralizing wash medium DMEM supplemented with 100 mM HEPES, pH 7.2, followed by 3 consecutive washes in this medium. Cells were resuspended in GM-P media. This procedure removes any extraneous cellular membranes bound to cells without significantly altering cell viability or the membrane expression of cell-surface molecules.23,24
Generation of HCMV gene microarrays
HCMV gene-specific primers were used to amplify fragments representing the full complement of HCMV genes based on the original annotation of the HCMV strain AD169 genome,25 together with additional genes identified in HCMV strain TownevarRIT3 and HCMV strain Toledo.26 PCR products were carefully checked for successful and specific amplification by 3% agarose gel electrophoresis before being purified using Multiscreen96 PCR plates (Millipore). Purified PCR products were spotted in duplicate onto poly-l-lysine glass slides using a high-precision microarrayer robot (ESI, Toronto, ON, Canada). In total, 194 of 202 HCMV strain TownevarRIT3-common genes were successfully amplified and spotted down, together with 13 of 19 additional genes from HCMV strain Toledo and 94 assorted human cellular cDNAs (Table S1, available at the Blood website, includes a listing of HCMV primer sequences; see the Supplemental Table link at the top of the online article).
RNA extraction, labeling, and microarray hybridization
Total RNA was extracted from mock and infected myeloid progenitor cells using RNAqueous (Ambion, Austin, TX) and the mRNA amplified using MessageAmp aRNA kit (Ambion), labeled by reverse transcription with either Cya3-dUTP (mock) or Cya5-dUTP (infected) and hybridized to microarrays for 16 hours at 65°C as previously described.27 Following hybridization, microarrays were washed sequentially for 2 minutes in 2 × SSC plus 0.1% SDS, 1 × SSC and 0.2 × SSC, scanned using a GenePix 4000B-series dual laser scanner (Axon Instruments, Union City, CA), and data were acquired using the integrated software package GenePix Pro. The median normalized pixel intensity for both Cya3 and Cya5 channels was determined and processed using Bioconductor R (Fred Hutchinson Cancer Research Center, Seattle, WA; http://www.r-project.org). Presentation of microarray-related information was based on the Minimal Information About a Microarray Experiment (MIAME) standard.28 In some experiments, RNA was reverse transcribed with random primers in the absence of Cya-dyes to generate cDNA for confirmation of microarray data by RT-PCR.
Statistical analysis of microarray data
For each day after infection, a penalized t statistic was calculated for each gene using the method of Efron et al29 and Smyth et al.30 The penalized t statistic is given by
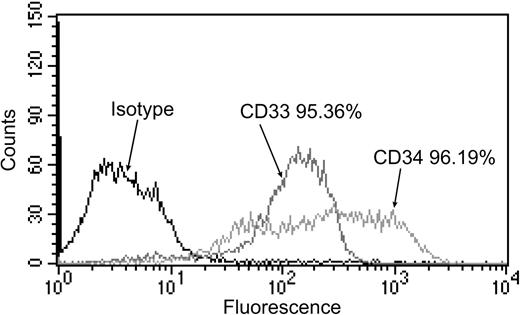
Flow cytometry analysis of cell-surface CD33 and CD34 expression on myeloid progenitor cells. On the day of infection, magnetic bead–enriched, nonadherent progenitor cells were stained with either isotype control antibody, anti-CD33 antibody, or anti-CD34 antibody. Expression is shown as a histogram, with the percentage of cells expressing cell-surface CD33 or CD34 above isotype control staining indicated.
Flow cytometry analysis of cell-surface CD33 and CD34 expression on myeloid progenitor cells. On the day of infection, magnetic bead–enriched, nonadherent progenitor cells were stained with either isotype control antibody, anti-CD33 antibody, or anti-CD34 antibody. Expression is shown as a histogram, with the percentage of cells expressing cell-surface CD33 or CD34 above isotype control staining indicated.
Results
Quantitation of viral genomes in HCMV-infected myeloid progenitors over a time course of infection
Human fetal liver–derived granulocyte macrophage progenitor cells (GM-Ps) have been used in a number of studies as the basis for a model of HCMV latent infection.9,10,13-15 At some point after exposure to virus, latency is established and cells from 14 days after infection had been shown to harbor latent infection. In both experimentally infected GM-Ps and in naturally infected mononuclear cells the viral genome is maintained at approximately 10 copies per infected cell in the absence of detectable, productive infection.9 In this study, GM-Ps were further purified for CD34+ progenitor cells by magnetic bead separation. Prior to infection, suspension cells were immunostained for CD34 and for the myeloid lineage-committed cell-surface marker CD33, and analyzed by flow cytometry. By comparison to the isotype antibody control, more than 95% of cells expressed both CD33 and CD34, demonstrating that these cells were myeloid progenitors (Figure 1).
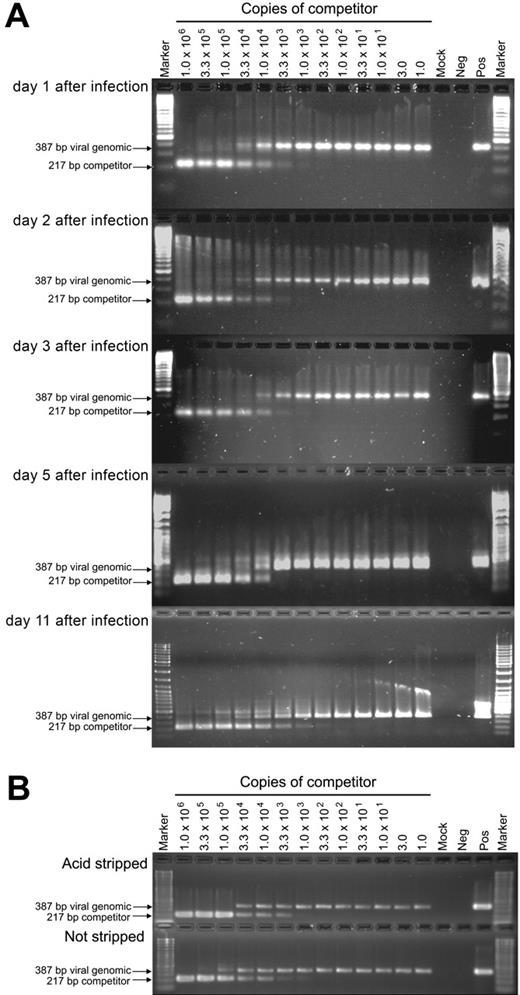
To better understand the events that accompany the establishment of latency in myeloid progenitors, and to contrast these with productive infection of a permissive cell type (HFs), we assessed the distribution and quantity of viral genomes over a time course of infection using a combination of cell-dilution PCR and quantitative-competitive PCR. Myeloid progenitor cells or HFs were infected with HCMV strain TownevarRIT3 at a MOI of 3 and DNA extracted on days 1, 2, 3, 5, and 11 after infection. Infected myeloid progenitor cell lysates were diluted such that each reaction contained either 500, 80, 16, 8, 4, 2, 1, 0.1, or 0.01 cell equivalents. Uninfected cell DNA was added to each dilution such that the total amount of DNA per reaction remained constant. PCR amplification was performed with primers IEP3C and IEP4BII. In myeloid progenitor cells, a 387-bp product corresponding to amplified viral genomic DNA was detected at all time points after infection, down to a dilution of 1 cell equivalent, but became undetectable at 0.1 and 0.01 cell equivalents (Figure 2A). Similarly, viral DNA was detected down to 1 cell equivalent in dilutions of productively infected HFs on all days tested (Figure 2B). Amplification was not detected in mock-infected myeloid progenitor cells or HFs. These data indicate that following infection at an MOI of 3, viral DNA was maintained in all myeloid progenitor cells throughout the time course.
Cell-dilution PCR across a time course of infection of myeloid progenitor cells or HFs. Lysates of myeloid progenitor cells or HFs harvested on days 1, 2, 3, 5, and 11 after infection were diluted, and were analyzed by PCR with primers IEP3C and IEP4BII. The position of the 387-bp product derived from HCMV genomic DNA is indicated by arrows. Samples from either mock-infected myeloid progenitor cell cultures or HFs (Mock), productively infected HFs (Pos), or samples without any added DNA template (Neg) were included as controls. Marker was GeneRuler DNA ladder mix (Fermentas, Burlington, ON, Canada).
Cell-dilution PCR across a time course of infection of myeloid progenitor cells or HFs. Lysates of myeloid progenitor cells or HFs harvested on days 1, 2, 3, 5, and 11 after infection were diluted, and were analyzed by PCR with primers IEP3C and IEP4BII. The position of the 387-bp product derived from HCMV genomic DNA is indicated by arrows. Samples from either mock-infected myeloid progenitor cell cultures or HFs (Mock), productively infected HFs (Pos), or samples without any added DNA template (Neg) were included as controls. Marker was GeneRuler DNA ladder mix (Fermentas, Burlington, ON, Canada).
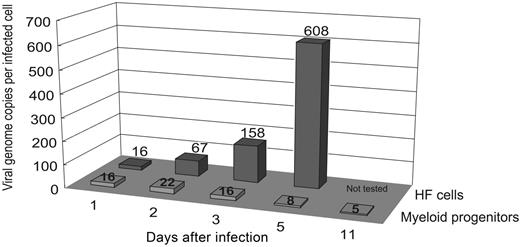
We next used QC-PCR to determine the number of viral genome copies per cell across the same time course. At each time point, cell lysates of 1000 myeloid progenitor cells or HFs were mixed with between 1.0 and 1.0 × 106 copies of a competitive template, the ie1/ie2 cDNA plasmid clone pON2347, and subjected to 40 rounds of PCR amplification with primers IEP3C and IEP4BII. Cells (1.0 × 103) from mock-infected cultures and a sample lacking cell or competitor DNAs were included as controls. After amplification, both the 387-bp genomic and 217-bp competitor PCR products were readily detectable (Figure 3A). Based on the relative amounts of the genomic and competitor PCR products, and the amount of competitor template added to the reaction, the number of viral genomes per cell could be calculated. As all cells were infected, this number also corresponded to the number of viral genomes per infected cell. The number of genome copies per cell from 2 independent replicate time course experiments was averaged (Figure 4). As expected, this number increased rapidly over time in productively infected HFs, with an increase from 16 to 608 copies per cell between day 1 and day 5 after infection. In contrast, the number of viral genome copies per myeloid progenitor cell remained low and relatively constant throughout the time course, with little change from the initial 16 copies per cell detected on day 1 after infection. To confirm that the PCR data were not affected by virus particles stuck to the outer cell-surface membranes of myeloid progenitors, QC-PCR was repeated using a well-characterized acid stripping protocol to remove surface-bound material from infected myeloid progenitors (day 3 after infection, MOI = 3). As a control, myeloid progenitors were not subjected to acid stripping, but were washed in culture media and analyzed in parallel with acid-stripped cells. There was no significant difference in the number of viral genomes per cell in either acid-stripped or unstripped HCMV-infected myeloid progenitors, with both cell populations yielding approximately 10 viral genome equivalents per cell (Figure 3B).
HCMV DNA copy number determination by QC-PCR across a time course of infection of myeloid progenitor cells. (A) Lysates of 1.0 × 103 cells from HCMV-infected myeloid progenitor cell cultures harvested on days 1, 2, 3, 5, and 11 after infection were each analyzed by PCR in the presence of between 1.0 and 1.0 × 106 copies of competitor template, an HCMV ie1/ie2 cDNA plasmid pON2347. The number of copies of competitor template added to each reaction is indicated at the top of lanes. The position of the 387-bp product derived from HCMV genomic DNA and the 217-bp product derived from the cDNA competitor template are indicated by arrows. Samples from either mock-infected myeloid progenitor cell cultures (Mock), productively infected HFs (Pos), or samples without any added DNA template (Neg) were included as controls. Marker was GeneRuler DNA ladder mix (Fermentas). (B) To determine whether the PCR data were affected by virus particles stuck to the outer cell-surface membranes of myeloid progenitors, QC-PCR was repeated using an acid-stripping protocol to remove surface-bound material from infected myeloid progenitors (day 3 after infection, MOI = 3). Non–acid-stripped cells were examined in parallel.
HCMV DNA copy number determination by QC-PCR across a time course of infection of myeloid progenitor cells. (A) Lysates of 1.0 × 103 cells from HCMV-infected myeloid progenitor cell cultures harvested on days 1, 2, 3, 5, and 11 after infection were each analyzed by PCR in the presence of between 1.0 and 1.0 × 106 copies of competitor template, an HCMV ie1/ie2 cDNA plasmid pON2347. The number of copies of competitor template added to each reaction is indicated at the top of lanes. The position of the 387-bp product derived from HCMV genomic DNA and the 217-bp product derived from the cDNA competitor template are indicated by arrows. Samples from either mock-infected myeloid progenitor cell cultures (Mock), productively infected HFs (Pos), or samples without any added DNA template (Neg) were included as controls. Marker was GeneRuler DNA ladder mix (Fermentas). (B) To determine whether the PCR data were affected by virus particles stuck to the outer cell-surface membranes of myeloid progenitors, QC-PCR was repeated using an acid-stripping protocol to remove surface-bound material from infected myeloid progenitors (day 3 after infection, MOI = 3). Non–acid-stripped cells were examined in parallel.
In a parallel series of experiments, lysates of 1.0 × 104 myeloid progenitor cells from each time point were tested for the presence of infectious virus by plaque assay on permissive HFs. No evidence of plaque formation was detected at any time point from these myeloid progenitor cell lysates (data not shown). To determine whether the sonication method used to prepare cell lysates disrupted infectivity, diluted virus inoculum was either sonicated or left nonsonicated prior to plating onto HF monolayers. On day 7, the resulting plaques were counted. In 2 independent replicate experiments, we detected 42 and 34 plaques, respectively (mean 38 plaques) in sonicated samples and 36 and 32 plaques (mean 34 plaques) in nonsonicated samples, demonstrating that the method used to prepare cell lysates did not disrupt infectivity. Taken together, these results are consistent with the establishment of a latent infection in myeloid progenitor cells in the absence of detectable productive infection and with little change in input viral genome copy number.
Summary of number of viral genomes per infected cell across a time course of infection of myeloid progenitor cells and HFs. At the indicated time points after infection, cells from HCMV-infected myeloid progenitor cell or HF cultures (MOI = 3) were analyzed by cell-dilution PCR and QC-PCR to determine number of viral genome copies per infected cell. The average from 2 independent replicate experiments is shown.
Summary of number of viral genomes per infected cell across a time course of infection of myeloid progenitor cells and HFs. At the indicated time points after infection, cells from HCMV-infected myeloid progenitor cell or HF cultures (MOI = 3) were analyzed by cell-dilution PCR and QC-PCR to determine number of viral genome copies per infected cell. The average from 2 independent replicate experiments is shown.
UV-inactivated HCMV is unable to successfully establish a latent infection in myeloid progenitor cells
To determine whether HCMV latency could be established in the absence of viral gene expression, myeloid progenitor cells were infected with either HCMV strain TownevarRIT3 at a MOI of 3, or with the same virus inactivated by UV-irradiation. After incubation for 3 hours with each inoculum, cells were thoroughly washed and placed into culture. At 3, 6, 12, 24, 72, and 120 hours after infection, cells were harvested for PCR analysis. The number of infected cells (ie, HCMV DNA–positive cells) was estimated by cell-dilution PCR. At the earliest time sampled after infection (3 hours after infection) the number of infected cells was the same between viable virus and UV-inactivated virus infections, with both yielding viral DNA detection down to a dilution of 1 cell equivalent (Figure 5). These data indicate comparable levels of infection with both viable and UV-inactivated viruses. However, from 6 hours after infection and onward, viral DNA became less abundant in cells infected with UV-inactivated virus. By 72 hours after infection this difference was considerable, with UV-inactivated virus DNA being 80-fold less abundant than viable virus DNA, as the ability to amplify HCMV DNA was lost once the reaction sample contained fewer than 80 cell equivalents, whereas HCMV DNA continued to be successfully amplified in viable virus-infected samples down to 1 cell equivalent. The magnitude of the drop in genome number following infection with UV-inactivated virus was much greater than the less than 3-fold increase in cell number observed during the culture period, demonstrating that the inability to maintain viral genomes following UV-inactivated virus infection was not due to a failure to partition with dividing cells. Rather, the rapid loss of viral genomes from UV-inactivated myeloid progenitor cells in comparison to their viable virus-infected counterparts indicated that viral gene expression was required for the successful establishment of latency within these cells.
Maintenance of the viral genome in myeloid progenitor cells infected with either viable or UV-inactivated virus. Lysates of myeloid progenitor cells harvested at 3, 6, 12, 24, 72, and 120 hours after infection with either HCMV strain TownevarRIT3 (MOI = 3) or its UV-inactivated equivalent were serially diluted such that each reaction contained between 1 and 500 cell equivalents, and were analyzed by PCR for 40 cycles with primers IEP3C and IEP4BII. The dilution containing the lowest number of cell equivalents at which HCMV DNA was detected following infection with viable (▪) or UV-inactivated virus (□) was plotted against time after infection.
Maintenance of the viral genome in myeloid progenitor cells infected with either viable or UV-inactivated virus. Lysates of myeloid progenitor cells harvested at 3, 6, 12, 24, 72, and 120 hours after infection with either HCMV strain TownevarRIT3 (MOI = 3) or its UV-inactivated equivalent were serially diluted such that each reaction contained between 1 and 500 cell equivalents, and were analyzed by PCR for 40 cycles with primers IEP3C and IEP4BII. The dilution containing the lowest number of cell equivalents at which HCMV DNA was detected following infection with viable (▪) or UV-inactivated virus (□) was plotted against time after infection.
Detection of viral gene expression during the establishment of latency in myeloid progenitor cells
The inability of UV-inactivated virus to successfully establish latency indicated a requirement for viral gene expression in this process. We generated HCMV gene microarrays bearing almost all known HCMV genes as a means to assess the viral genome for expression during the establishment of latency. The ability of these microarrays to successfully detect viral gene expression was first tested using RNA extracted from productively infected HFs. As early as 2 hours after the addition of virus, expression from the major immediate early genes ie1/ie2 (UL123 and UL122) became detectable, and over a 7-day time course, expression from 95% of genes common to HCMV strain Towne was detected. No hybridization to HCMV strain Toledo-specific genes was detected (data not shown). These results demonstrate that the viral gene microarrays used in this study provided a sensitive and specific means to detect global HCMV gene expression.
To determine whether viral genes were expressed during the establishment of latency, and to identify those involved in this process, we next assessed RNA extracted from myeloid progenitor cells over a time course of infection. Cells were either mock infected or infected with HCMV strain TownevarRIT3 at a MOI of 3. On days 1, 2, 3, 5, and 11 after infection, total RNA was extracted and mRNA linearly amplified using a poly dT-T7 RNA polymerase-based in vitro transcription technique. The success of amplification was confirmed by agarose gel electrophoresis of amplified products as previously described27 (data not shown). Amplified RNA (aRNA) was labeled with either Cya3 (mock) or Cya5 (infected) and hybridized to viral gene microarrays before being washed and scanned with a dual laser scanner. To identify viral RNAs that were reproducibly expressed at each time point after infection, we performed several replicates at each time point (day 1: 12 replicates; day 2: 4 replicates; day 3: 5 replicates; day 5: 2 replicates; day 11: 2 replicates) using freshly prepared myeloid progenitor cultures divided into 2 aliquots for mock or HCMV infection. For each day after infection, a penalized t statistic was calculated for each gene using the method of Efron et al29 and Smyth et al.30
These analyses revealed the expression of subsets of viral RNAs throughout the time course of infection (Table 1). Relative to the total number of viral genes encoded by the HCMV genome, the number of viral RNAs that were expressed during infection of myeloid progenitor cells was small, with the highest number detected on day 1 after infection. At this early time after infection, viral RNAs representative of 25 different genes were detected. Although many of these remained detectable at multiple time points, with the exception of the time point 3 days after infection, the number of viral RNAs detected decreased over the time course of infection. By the final time point on day 11 after infection, expression was only detected from 7 viral RNAs.
Microarray-based determination of HCMV gene RNAs expressed in myeloid progenitor cells over a time course of infection
. | Time after infection . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| HCMV gene . | Day 1 . | Day 2 . | Day 3 . | Day 5 . | Day 11 . | ||||
| RL2 | 2.25 | 3.92 | 3.53 | 3.48 | — | ||||
| RL3 | 3.58 | 2.33 | 4.89 | 4.29 | — | ||||
| RL4 | 16.56 | 4.23 | 7.36 | 5.39 | — | ||||
| RL5 | 3.03 | — | — | — | — | ||||
| RL6 | 2.66 | 2.60 | 7.67 | — | — | ||||
| RL7 | 9.71 | 2.91 | 6.77 | 4.00 | 2.00 | ||||
| RL11 | — | — | 2.50 | — | — | ||||
| UL5 | 2.08 | — | — | — | — | ||||
| UL23 | 2.89 | — | — | — | — | ||||
| UL30 | 3.53 | — | — | 2.00 | — | ||||
| UL39 | 4.20 | — | — | — | — | ||||
| UL53 | — | — | 2.43 | — | — | ||||
| UL56 | — | — | 2.00 | — | — | ||||
| UL62 | 2.28 | — | — | — | — | ||||
| UL64 | 2.93 | — | — | — | — | ||||
| UL66 | — | — | 3.46 | — | — | ||||
| UL67 | 2.87 | — | 2.99 | — | 2.79 | ||||
| UL68 | 6.45 | 2.22 | 7.62 | 7.52 | 4.06 | ||||
| UL71 | — | 2.00 | — | — | — | ||||
| UL81 | 2.95 | — | — | — | — | ||||
| UL89 | 2.10 | — | — | — | — | ||||
| UL90 | — | 2.00 | — | — | — | ||||
| UL93 | — | 2.00 | — | — | — | ||||
| UL99 | 2.50 | — | 2.00 | — | — | ||||
| UL108 | 4.50 | 2.20 | 3.25 | — | 2.41 | ||||
| UL110 | 8.17 | — | 6.87 | 6.59 | 2.55 | ||||
| UL111 | 2.23 | — | 2.22 | 2.03 | — | ||||
| UL111A | — | — | — | — | 2.07 | ||||
| UL115 | — | — | — | — | 2.04 | ||||
| UL123 | — | — | 2.10 | — | — | ||||
| UL132 | 2.07 | — | — | — | — | ||||
| UL147 | 2.01 | — | — | — | — | ||||
| UL153 | 9.45 | 5.03 | 7.52 | 4.79 | — | ||||
| UL154 | 5.82 | 2.11 | 2.51 | 2.25 | — | ||||
| US12 | 2.38 | — | — | — | — | ||||
| US28 | — | — | 2.22 | — | — | ||||
| US32 | — | — | — | 2.00 | — | ||||
. | Time after infection . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| HCMV gene . | Day 1 . | Day 2 . | Day 3 . | Day 5 . | Day 11 . | ||||
| RL2 | 2.25 | 3.92 | 3.53 | 3.48 | — | ||||
| RL3 | 3.58 | 2.33 | 4.89 | 4.29 | — | ||||
| RL4 | 16.56 | 4.23 | 7.36 | 5.39 | — | ||||
| RL5 | 3.03 | — | — | — | — | ||||
| RL6 | 2.66 | 2.60 | 7.67 | — | — | ||||
| RL7 | 9.71 | 2.91 | 6.77 | 4.00 | 2.00 | ||||
| RL11 | — | — | 2.50 | — | — | ||||
| UL5 | 2.08 | — | — | — | — | ||||
| UL23 | 2.89 | — | — | — | — | ||||
| UL30 | 3.53 | — | — | 2.00 | — | ||||
| UL39 | 4.20 | — | — | — | — | ||||
| UL53 | — | — | 2.43 | — | — | ||||
| UL56 | — | — | 2.00 | — | — | ||||
| UL62 | 2.28 | — | — | — | — | ||||
| UL64 | 2.93 | — | — | — | — | ||||
| UL66 | — | — | 3.46 | — | — | ||||
| UL67 | 2.87 | — | 2.99 | — | 2.79 | ||||
| UL68 | 6.45 | 2.22 | 7.62 | 7.52 | 4.06 | ||||
| UL71 | — | 2.00 | — | — | — | ||||
| UL81 | 2.95 | — | — | — | — | ||||
| UL89 | 2.10 | — | — | — | — | ||||
| UL90 | — | 2.00 | — | — | — | ||||
| UL93 | — | 2.00 | — | — | — | ||||
| UL99 | 2.50 | — | 2.00 | — | — | ||||
| UL108 | 4.50 | 2.20 | 3.25 | — | 2.41 | ||||
| UL110 | 8.17 | — | 6.87 | 6.59 | 2.55 | ||||
| UL111 | 2.23 | — | 2.22 | 2.03 | — | ||||
| UL111A | — | — | — | — | 2.07 | ||||
| UL115 | — | — | — | — | 2.04 | ||||
| UL123 | — | — | 2.10 | — | — | ||||
| UL132 | 2.07 | — | — | — | — | ||||
| UL147 | 2.01 | — | — | — | — | ||||
| UL153 | 9.45 | 5.03 | 7.52 | 4.79 | — | ||||
| UL154 | 5.82 | 2.11 | 2.51 | 2.25 | — | ||||
| US12 | 2.38 | — | — | — | — | ||||
| US28 | — | — | 2.22 | — | — | ||||
| US32 | — | — | — | 2.00 | — | ||||
Normalized intensity ratio values are shown for viral RNAs defined as being expresed.
— indicated not expressed.
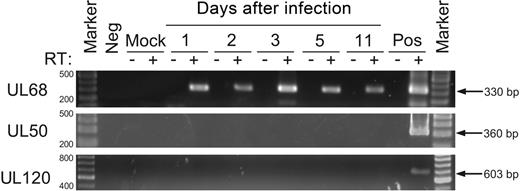
We confirmed the presence or absence of expression of a subset of HCMV RNAs using RT-PCR (Figure 6). UL50 and UL120 were selected, as both were defined as not being expressed in our microarray experiments, and UL68 was selected on the basis that it was defined by microarray as one of the most abundantly expressed transcripts. RNA extracted on days 1, 2, 3, 5, and 11 after infection was reverse transcribed with random primers before being subjected to PCR amplification with HCMV gene-specific primers. Controls included RNA extracted from mock-infected myeloid progenitors, the omission of RNA template, the omission of reverse transcriptase, and the inclusion of RNA from productively infected HFs. Expression of UL68 was detected by both microarray and RT-PCR whereas expression of UL50 and UL120 was not detected by either method. These results provide validation of microarray-based findings.
Confirmation of the presence or absence of HCMV gene expression in myeloid progenitors by RT-PCR. RNA extracted from myeloid progenitors on days 1, 2, 3, 5, and 11 after infection was examined by RT-PCR for the expression of UL68, UL50, and UL120 transcripts for 40 cycles of amplication. Controls included the omission of reverse transcriptase (RT), samples from either mock-infected myeloid progenitor cells or HFs (Mock), productively infected HFs (Pos), or samples without any added template (Neg). Marker was GeneRuler DNA ladder mix (Fermentas).
Confirmation of the presence or absence of HCMV gene expression in myeloid progenitors by RT-PCR. RNA extracted from myeloid progenitors on days 1, 2, 3, 5, and 11 after infection was examined by RT-PCR for the expression of UL68, UL50, and UL120 transcripts for 40 cycles of amplication. Controls included the omission of reverse transcriptase (RT), samples from either mock-infected myeloid progenitor cells or HFs (Mock), productively infected HFs (Pos), or samples without any added template (Neg). Marker was GeneRuler DNA ladder mix (Fermentas).
Reactivation of HCMV from latently infected myeloid progenitor cells
Exposure of myeloid progenitor cells to HCMV resulted in the establishment of a nonproductive infection. To determine whether virus could be reactivated from these cells, we employed a coculture assay with permissive HFs, which has been shown previously to induce virus reactivation from experimentally infected myeloid progenitor cells.10,11,15,17 Aliquots of 5 × 104 infected myeloid progenitor cells harvested on days 1, 2, 3, 5, and 11 after infection were cocultured with uninfected HF monolayers which were monitored for the appearance of cytopathic effect (CPE) over a period of 20 days. At the time coculture was initiated, infected myeloid progenitor cells from all time points after infection did not show any signs of productive infection, as determined by a lack of detectable infectivity (ie, lack of CPE) in HFs incubated with lysates of 5 × 104 infected myeloid progenitor cells (Table 2). However, intact myeloid progenitor cells from cultures harvested on one of days 1, 2, 3, 5, or 11 after infection yielded reactivated virus between day 10 and day 14 after coculture with HFs as indicated by the detection of the earliest signs of CPE in HFs during this time period. In contrast, positive controls consisting of productively infected HFs harvested on day 1, 2, 3, or 5 after infection rapidly induced CPE within 1 to 3 days after coculture with uninfected HFs. The frequency of infected myeloid progenitor cells, which reactivated and produced infectious virus, was determined by quantitation of infectious centers that were formed within 4 days of the first signs of CPE at each time point after infection (Table 2). The ability to induce reactivation from myeloid progenitor cells nonproductively infected with HCMV following extended coculture with HFs demonstrates that these cells had supported a latent rather than abortive infection.
Reactivation of latent HCMV from myeloid progenitor cells by coculture with permissive HFs
. | Day after coculture at which cytopathic effect (CPE) is first detected . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Days after infection . | HCMV-infected myeloid progenitor cells (frequency of reactivation*) . | HCMV-infected myeloid progenitor cell lysate . | Mock-infected myeloid progenitor cells . | Mock-infected myeloid progenitor cell lysate . | HCMV-infected HF cells, productive infection . | Mock-infected HF cells . | |||||
| 1 | 14 (1.4 × 10-3) | No CPE | No CPE | No CPE | 3 | No CPE | |||||
| 2 | 13 (1.3 × 10-3) | No CPE | No CPE | No CPE | 2 | No CPE | |||||
| 3 | 13 (5.8 × 10-4) | No CPE | No CPE | No CPE | 1 | No CPE | |||||
| 5 | 11 (2.8 × 10-4) | No CPE | No CPE | No CPE | 1 | No CPE | |||||
| 11 | 10 (1.8 × 10-4) | No CPE | No CPE | No CPE | Not done | No CPE | |||||
. | Day after coculture at which cytopathic effect (CPE) is first detected . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Days after infection . | HCMV-infected myeloid progenitor cells (frequency of reactivation*) . | HCMV-infected myeloid progenitor cell lysate . | Mock-infected myeloid progenitor cells . | Mock-infected myeloid progenitor cell lysate . | HCMV-infected HF cells, productive infection . | Mock-infected HF cells . | |||||
| 1 | 14 (1.4 × 10-3) | No CPE | No CPE | No CPE | 3 | No CPE | |||||
| 2 | 13 (1.3 × 10-3) | No CPE | No CPE | No CPE | 2 | No CPE | |||||
| 3 | 13 (5.8 × 10-4) | No CPE | No CPE | No CPE | 1 | No CPE | |||||
| 5 | 11 (2.8 × 10-4) | No CPE | No CPE | No CPE | 1 | No CPE | |||||
| 11 | 10 (1.8 × 10-4) | No CPE | No CPE | No CPE | Not done | No CPE | |||||
No CPE indicates no cytopathic effect observed after 20 days coculture with permissive HFs.
Frequency of myeloid progenitors that reactivate and produce infectious virus, determined by infectious center assay.
Discussion
In this study we tracked the virus over time following infection of primary human myeloid progenitor cells to better define the molecular events that occur during the establishment of latency within these cells. There are 3 possible pathways leading to the establishment of latency. First, after attachment and entry, the virus may enter directly into a latent state without de novo viral gene expression. Second, after entry the virus may initiate a productive infection that is prematurely interrupted at some point, leading to the establishment of latency. The full replicative cycle is characterized by immediate early, early, and late class genes being expressed sequentially in an ordered cascade, and so latency resulting from an interruption of this process would share features of such a cascade. A third alternative is that following entry the virus expresses a subset of viral genes that are not associated with productive infection but are required for the successful establishment of latency. The results of this study support this third pathway.
There was little change observed from initial levels of 16 copies per cell throughout the time course of infection of myeloid progenitors, a level consistent with that found in naturally infected mononuclear cells from healthy donors.9 In contrast, infection of HFs, a cell type that supports the full replicative cycle, resulted in a rapid increase in viral genomes, from an initial 16 copies per cell on day 1 to an average of 608 copies per cell by day 5 after infection. We were not able to detect infectious virus in myeloid progenitors at any time point after infection but were able to recover virus after extended coculture of infected myeloid progenitor cells with uninfected HFs. These data are consistent with the virus establishing a reactivatable latent infection within myeloid progenitor cells without increasing the number of viral genomes per infected cell. Although the number of HCMV genome copies per infected cell did not increase over time, preliminary analysis of a mutant virus carrying a large deletion that removes the ie1/ie2 modulator region has raised the possibility that this region may act as a latent origin of replication for viral genome maintenance in dividing progenitor cells.32
Importantly, when myeloid progenitor cells exposed to UV-inactivated virus were examined, the proportion of cells in which the viral genome could be detected dropped rapidly over time. From an equal initial level of infection in both UV-inactivated and viable virus-infected cells, by day 3 after infection only about 1% of cells exposed to UV-inactivated virus remained genome positive compared with viable virus-infected cells in which viral DNA remained present in all cells. Thus, infected myeloid progenitors retained viral DNA in a manner indicating that the successful establishment of latency is dependent on viral gene expression. Similarly, latency-associated gene products encoded by other human herpes viruses, including herpes simplex type 1 (HSV-1) latency-associated transcripts (LATs) and the varicella zoster virus (VZV) ORF63 and ORF4 proteins, have been reported to play significant roles in the establishment of latency.33-37
Using a viral gene microarray-based approach to analyze RNA extracted from myeloid progenitors over a time course of infection, 37 viral RNAs were detected at some point during an 11-day time course, with 25 of these being expressed on day 1 and only 7 regions (RL7, UL67, UL68, UL108, UL110, UL111A, UL115) detectable on day 11 after infection. One of these, UL111A, corresponds to a region shown to express a latency-associated homologue of IL-1018 (LAcmvIL-10). The functions of the remaining 6 RNAs and indeed most of the RNAs expressed at the other time points have not yet been elucidated in any cell type. The point at which the virus transitions from the establishment phase to the maintenance phase of latency is unclear, but these data show that the majority of detected viral RNAs are expressed early after infection and that this number decreases over time, consistent with a model of the establishment of latency in which upon entry the virus expresses a subset of viral genes that help to facilitate this process. These results also indicate that the virus does not initiate a productive infection within cells prior to the establishment of latency. Defining the functions of viral genes expressed in myeloid progenitor cells during the establishment of latency will be an important component of future work, as these may play essential roles in this process.
Viral RNAs at each time point were expressed in the absence of detectable, productive infection and the pattern of viral gene expression differed significantly from that observed during productive phase infection in so far as the number of viral RNAs expressed in myeloid progenitors decreased over time and did not follow the pattern of coordinate expression of viral genes assigned to different kinetic classes which is characteristic of productive replication in permissive HFs.1 Furthermore, this pattern was inconsistent with the interruption of a productive infection gene expression cascade, suggesting that the establishment of latency is accompanied by the expression of a unique subset of viral RNAs.
The construction of the viral gene microarrays used in this study was based on the original annotation of the HCMV strain AD169 genome with the potential to encode 208 ORFs,25 together with additional ORFs identified in HCMV strains Towne and Toledo.26 In recent years, the protein-coding potential of HCMV across a range of virus strains and isolates has been reassessed, with current estimates ranging from 165 to 252 protein-encoding ORFs, depending on the criteria used for sequence analysis and virus strains and isolates examined.38-40 Based on these re-evaluations, almost half of the RNAs we detected in myeloid progenitors were expressed from regions encoding ORFs predicted (in at least one study) to lack protein-coding potential. This raises the intriguing possibility that HCMV may express nontranslated LATs that themselves may function during the establishment and/or maintenance of a latent infection. In this respect, HSV-1 LATs are expressed abundantly in neurons during latent infection yet do not encode a detectable protein product, but have been reported to play roles in the establishment and maintenance of latency,33-35 spontaneous reactivation in latently infected rabbits,41-43 the inhibition of apoptosis,44-46 and to enhance the frequency of encephalitis in mice.47
Whilst the viral RNAs detected in myeloid progenitor cells may represent the ORFs currently identified as corresponding to the regions from which expression was detected, it remains possible that transcripts encode as-yet-unidentified latency-specific ORFs, as a result of different sites of transcriptional initiation or termination, or alternate transcript splicing. In this regard, HCMV latency associated transcripts (CLTs) identified from both the MIE region and the UL111A region have both been shown to encode unique ORFs that differ from those originally identified during productive infection due to alternate splicing patterns and/or different start sites of transcription.14,18
In addition, as the viral gene microarrays used to identify virus transcription were composed of denatured, double-stranded PCR products, expression of transcripts antisense to known ORFs cannot be excluded. Perhaps the best examples of antisense transcripts expressed during latency which overlap ORFs expressed during productive infection are the HSV-1 LATs, which are expressed antisense to ICP0,48 the antisense HCMV MIE region CLTs,14 and most recently UL81-82ast, a UL81-82 antisense transcript isolated from a cDNA library of monocytes from a healthy HCMV seropositive donor.49 Interestingly, in the current study, expression was detected from the UL81 region. Cloning and mapping of HCMV RNAs from infected myeloid progenitor cells will better define the nature of viral transcripts expressed during the establishment and maintenance phases of HCMV latency.
We have previously shown that there is a dissociation between latency-associated transcription and the presence of the viral genome. MIE region CLTs are expressed in approximately 2% of latently infected GM-Ps9 and between 1% and 12% of GM-Ps from latently infected cultures expressed detectable UL111A region CLTs.18 The distribution of viral transcripts identified in the current study remains to be determined, and it is possible that differential expression occurs in different subpopulations of latently infected myeloid progenitor cells with potentially different biologic functions. In this respect, Goodrum and colleagues examined experimental infection of primitive CD34+ hematopoietic progenitor cells11 and more recently, subpopulations of these CD34+ cells, and found that the outcome of infection was dependent on the nature of the CD34+ cell populations infected, with CD34+/CD38– cells supporting infection with the hallmarks of latency.20 Our study used a different model of latent HCMV infection that focused on the kinetics of infection of myeloid lineage-committed CD34+/CD33+ progenitor cells, which made up more than 95% of cells, although we also determined the percentage of cells that were CD34+/CD38– and CD34+/c-kit+, as these also mark primitive progenitors. We found 86.6% ± 1.8% of cells were CD34+/CD38– and 38.4% ± 12.1% were CD34+/c-kit+ at the time of infection (data not shown). Although these proportions indicate that the composition of our progenitor cell cultures differ from those of Goodrum and colleagues,20 a number of the viral RNAs we detected were common to both culture systems. This likely indicates robust expression of some viral RNAs during infection of cells at different stages of differentiation, from early pluripotent CD34+ hematopoietic cells to lineage-committed myeloid progenitor cells, and also suggests that the expression of other viral RNAs may be myeloid progenitor cell–specific.
Although it has been reported that the number of viral genomes per cell increases at 10 days after infection of CD34+/CD38– cells, indicating that limited DNA replication may have occurred,20 we did not observe any evidence of viral DNA amplification over the 11-day culture period. Certainly there are both similarities and differences in the types of cells infected in each experimental system that may influence outcome, and it is possible that different progenitor populations support latent infections that differ with respect to initial viral genome amplification. Similarly, while viral gene expression in purified CD34+/CD38– cells has been reported to be detectable between 1 and 8 days after infection but becomes undetectable by day 10 after infection,20 we continued to detect expression of a subset of viral RNAs on day 11 (the latest time point assessed) following infection of CD33+ myeloid progenitor cells. Together with the cell type examined and the state of cellular differentiation, the relative sensitivities of the assays used to detect viral gene expression may account for some of these differences. The viral gene microarray assay we employed was sensitive enough to reproducibly detect major immediate early region ie1/ie2 (UL123/UL122) RNA expression as early as 2 hours after addition of virus to permissive HFs (data not shown). However, we were able to detect MIE region CLTs in RNA extracts from latently infected myeloid progenitors by RT-PCR (data not shown), even though MIE region transcription was almost never detected by microarray in these cells. This is consistent with a very low level of expression of MIE CLTs, as previously reported in latently infected GM-Ps, which requires PCR-based detection methods.9,10,14,15,17 It also suggests that there may be additional, as-yet-unidentified CMV LATs that remain below the limits of detection of microarray-based techniques. Ultimately, the full complement of viral RNAs expressed by HCMV during latent infection will require a detailed analysis of ex vivo cells from healthy, seropositive donors, an approach severely complicated by the very low levels of latent infection and the requirement for highly sensitive detection techniques.6,7,10,14,18,49 Consequently, experimental models of latent infection of primary hematopoietic cell types will continue to play a major role in guiding studies of HCMV latency by providing important new insights into the interaction of this virus with cells of the myeloid lineage.10-12
The ability of HCMV to establish and maintain a latent infection enables this virus to successfully persist in cells of the myeloid lineage, from which it can reactivate years later to cause disease. The identification of viral RNAs expressed early after infection of myeloid progenitor cells implicates viral gene expression in the establishment of latency and provides potential targets for the development of antiviral therapies to prevent or inhibit this process and the serious consequences that arise from reactivation from latency.
Authorship
The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, August 24, 2006; DOI 10.1182/blood-2005-12-026682.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We would like to thank Bronwyn Robertson and staff from the Clive and Vera Ramaciotti Centre for Gene Function Analysis for assistance in the generation of viral gene microarrays, and Karen Byth and Chris Bye for assistance with microarray data analysis.
This work was supported by Australian National Health and Medical Research Council grants 301943 and 358399. A.K.L.C. was the holder of an Australian Postgraduate Award and a Westmead Millennium Foundation Stipend Enhancement Award.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal