The α globin gene cluster, which contains embryonic (ζ) and fetal/adult (α) genes, has been highly conserved throughout evolution.1 Four putative regulatory elements (called MCS-R1 to MCS-R4; Figure 1) corresponding to erythroid-specific DNase1 hypersensitive sites (HSs)2 are found upstream (30-160 kb) of the α genes in most mammalian species, but the relative importance of each of these elements in each species has not been fully elucidated. In humans, characterization of natural deletions,3-5 analysis of interspecific hybrids,3,6 stable transfectants,6 and studies of transgenic mice7 all indicate that the most highly conserved element (MCS-2, which corresponds to HS-40 in humans) is the major regulatory element; on their own, the other elements (MCS-1, -3, and -4) cannot drive substantial levels of α globin expression. Targeted deletion of HS-40 in a hybrid containing human chromosome 16 in a mouse erythroleukemia (MEL) background reduces human α globin expression to less than 5% of normal.6 It was therefore surprising to find that deletion of the highly conserved MCS-R2 element (mHS-26) in mice, using homologous recombination, only reduced α globin expression to approximately 50% of normal,8 suggesting that the 4 elements may play different roles in different species and casting some doubt on the value of the mouse as a faithful model of human α globin gene expression.8 At present, the only way to determine the role of individual regulatory elements in situ, in the normal human erythroid environment, is to identify natural mutants in human populations.
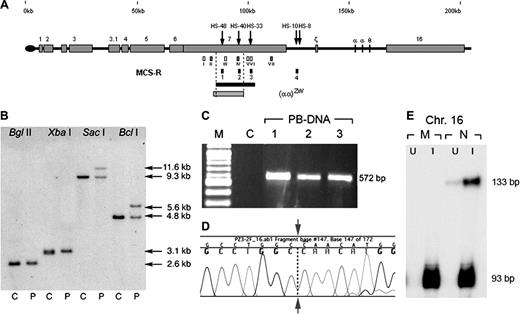
Summary of the molecular analysis of a novel interstitial upstream deletion. (A) The (αα)ZW deletion and its relative position within chromosome 16p13.3. Coordinates (in kb) according to GenBank no. NT_037 887.4 are depicted above the line. Erythroid DHS are shown as black arrows. Numbered gray boxes represent known genes9 located in this subtelomeric region in addition to the α globin genes (labeled with Greek symbols). The small gray bar below the line represents (αα)ZW deletion and the black bar represents the shortest region of overlap (20.4 kb) derived from previously characterized upstream deletions.5 The (αα)ZW deletion provides a new 3′ limit for minimal upstream sequences required for full α globin expression (14.7 kb, dashed line). The small white and gray boxes below the line represent probes used for Southern blotting (white) and MLPA (gray):10 (I) 757 × 758, (87 755-88 337); (II) 4a, (90 548-90 603); (III) 755 × 756, (96 671-97 389); (IV) 5a (103 695-103 774); (V) 767 × 768, (107 503-108 177); (VI) 769 × 770 (108 517-109 159); and (VII) 6a (120 541-120 590). Four small black boxes show the multispecies conserved sequences-regulatory elements (MCS-R1 to -R4). (B) Characterization of (αα)ZW breakpoint. The 3′ breakpoint of the (αα)ZW chromosome was characterized after BglII, XbaI, SacI, and BclI digestion, hybridized with combined probes V and VI. Breakpoint fragments were identified with SacI and BclI demonstrating that the 3′ breakpoint lies between coordinates 105 036-107 043. No breakpoint fragment was detected using probe III, showing that this region was deleted from the (αα)ZW chromosome. The 5′ breakpoint (coordinate 90 512-91 701) was identified using NcoI, PstI, HpaI, and HindII using probe I (not shown). C indicates control; P, patient. (C) Gap-polymerase chain reaction (PCR) amplified a 572-bp fragment specific to the (αα)ZW chromosome. Using forward (ZW-2F; 5′-GCTTAGGGGAAACTGCAGGTG-3′) and reverse (ZW-2R; 5′-AGGCAGACTGCACTTCATTGTTTA-3′) primers, the PCR product was amplified in peripheral blood DNA (PB-DNA) in triplicate. The PCR did not amplify a normal chromosome. M indicates 100-bp marker (New England Biolabs, Hitchin, United Kingdom). (D) Direct sequencing of the breakpoint fragment demonstrates the sequence adjoining between coordinates 90 777 and 106 774 (leftward and rightward to the arrow respectively). (E) Marked decrease in mRNA levels from the (αα)ZW chromosome. An RNase protection assay was used to analyze total RNA extracted from chromosome 16 x mouse erythroleukemia, interspecific hybrids. This showed a severely reduced level of human α globin expression (133-bp protected band) in both uninduced (U) and hemin-induced (I) cells containing the mutant (M) but not the normal (N) chromosome 16. The 93-bp protected fragment represents the mouse α globin mRNA as an internal control.
Summary of the molecular analysis of a novel interstitial upstream deletion. (A) The (αα)ZW deletion and its relative position within chromosome 16p13.3. Coordinates (in kb) according to GenBank no. NT_037 887.4 are depicted above the line. Erythroid DHS are shown as black arrows. Numbered gray boxes represent known genes9 located in this subtelomeric region in addition to the α globin genes (labeled with Greek symbols). The small gray bar below the line represents (αα)ZW deletion and the black bar represents the shortest region of overlap (20.4 kb) derived from previously characterized upstream deletions.5 The (αα)ZW deletion provides a new 3′ limit for minimal upstream sequences required for full α globin expression (14.7 kb, dashed line). The small white and gray boxes below the line represent probes used for Southern blotting (white) and MLPA (gray):10 (I) 757 × 758, (87 755-88 337); (II) 4a, (90 548-90 603); (III) 755 × 756, (96 671-97 389); (IV) 5a (103 695-103 774); (V) 767 × 768, (107 503-108 177); (VI) 769 × 770 (108 517-109 159); and (VII) 6a (120 541-120 590). Four small black boxes show the multispecies conserved sequences-regulatory elements (MCS-R1 to -R4). (B) Characterization of (αα)ZW breakpoint. The 3′ breakpoint of the (αα)ZW chromosome was characterized after BglII, XbaI, SacI, and BclI digestion, hybridized with combined probes V and VI. Breakpoint fragments were identified with SacI and BclI demonstrating that the 3′ breakpoint lies between coordinates 105 036-107 043. No breakpoint fragment was detected using probe III, showing that this region was deleted from the (αα)ZW chromosome. The 5′ breakpoint (coordinate 90 512-91 701) was identified using NcoI, PstI, HpaI, and HindII using probe I (not shown). C indicates control; P, patient. (C) Gap-polymerase chain reaction (PCR) amplified a 572-bp fragment specific to the (αα)ZW chromosome. Using forward (ZW-2F; 5′-GCTTAGGGGAAACTGCAGGTG-3′) and reverse (ZW-2R; 5′-AGGCAGACTGCACTTCATTGTTTA-3′) primers, the PCR product was amplified in peripheral blood DNA (PB-DNA) in triplicate. The PCR did not amplify a normal chromosome. M indicates 100-bp marker (New England Biolabs, Hitchin, United Kingdom). (D) Direct sequencing of the breakpoint fragment demonstrates the sequence adjoining between coordinates 90 777 and 106 774 (leftward and rightward to the arrow respectively). (E) Marked decrease in mRNA levels from the (αα)ZW chromosome. An RNase protection assay was used to analyze total RNA extracted from chromosome 16 x mouse erythroleukemia, interspecific hybrids. This showed a severely reduced level of human α globin expression (133-bp protected band) in both uninduced (U) and hemin-induced (I) cells containing the mutant (M) but not the normal (N) chromosome 16. The 93-bp protected fragment represents the mouse α globin mRNA as an internal control.
We have previously characterized 9 deletions that remove various combinations of the upstream regulatory elements and cause α-thalassemia.5 In all cases, MCS-R1, -R2, and -R3 are removed and in 4 cases, MCS-4 is also deleted. Here we describe a small, approximately 16-kb interstitial deletion that removes only MCS-R1 (HS-48) and MCS-R2 (HS-40) and that caused α-thalassemia in a Filipino girl. The combination of severely reduced red cell indices (hemoglobin [Hb], 113 g/L [11.3 g/dL]; mean corpuscular volume [MCV], 65 fL; and mean corpuscular hemoglobin [MCH], 19.48 pg), normal Hb analysis (HbF < 1% and HbA2 = 2.2%), demonstrable HbH inclusion bodies, and a significantly reduced α/β globin chain synthesis ratio (0.71) suggested that she had α-thalassemia with inactivation of at least 2 of the normal 4 α globin genes. Analysis of interspecific hybrids confirmed that expression from the affected chromosome is reduced to less than 1% of normal (Figure 1). Although we have confirmed that the α genes are structurally normal (data not shown), analysis of the upstream region using multiplex ligation-dependent probe amplification (MPLA) analysis identified a deletion removing MCS-R2 (HS-40).10 DNA sequence across the breakpoint shows that this deletion (αα)ZW extends for 15 997 bp (coordinates, 90 778-106 773 bp) and that it arose via recombination between 2 Alu repetitive elements (Figure 1), as observed for many other deletions in this region of the genome.
This patient provides the first example of an upstream deletion in which MCS-R3 (HS-33) remains intact (Figure 1A), demonstrating that this element alone or in combination with MCS-R4 has no significant positive effect on α globin expression. To fully demonstrate that, in humans, in contrast to mice, HS-40 is the dominant α globin regulatory element, it now only remains to be seen whether MCS-R1 (HS-48) can act alone or in combination with MCS-R2 (HS-40) in human erythroid cells. Future identification of an upstream deletion, including MCS-R2, in which MCS-R1 remains intact, would therefore complete the search for such deletions andhelp clarify the apparent differences between the regulation of α globin expression in mice and humans.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal