To identify the molecular basis underlying the functions of tumor-associated macrophages (TAMs), we characterized the gene expression profile of TAMs isolated from a murine fibrosarcoma in comparison with peritoneal macrophages (PECs) and myeloid suppressor cells (MSCs), using a cDNA microarray technology. Among the differentially expressed genes, 15 genes relevant to inflammation and immunity were validated by real-time polymerase chain reaction (PCR) and protein production. Resting TAMs showed a characteristic gene expression pattern with higher expression of genes coding for the immunosuppressive cytokine IL-10, phagocytosis-related receptors/molecules (Msr2 and C1q), and inflammatory chemokines (CCL2 and CCL5) as expected, as well as, unexpectedly, IFN-inducible chemokines (CXCL9, CXCL10, CXCL16). Immunohistology confirmed and extended the in vitro analysis by showing that TAMs express M2-associated molecules (eg, IL-10 and MGL1), as well as CCL2, CCL5, CXCL9, CXCL10, and CXCL16, but no appreciable NOS2. Lipopolysaccharide (LPS)-mediated activation of TAMs resulted in defective expression of several proinflammatory cytokines (eg, IL-1β, IL-6, TNF-α) and chemokines (eg, CCL3), as opposed to a strong up-regulation of immunosuppressive cytokines (IL-10, TGFβ) and IFN-inducible chemokines (CCL5, CXCL9, CXCL10, CXCL16). Thus, profiling of TAMs from a murine sarcoma revealed unexpected expression of IFN-inducible chemokines, associated with an M2 phenotype (IL-10high, IL-12low), and divergent regulation of the NF-κB versus the IRF-3/STAT1 pathway.
Introduction
Macrophages are versatile, plastic cells that respond to environmental signals with diverse functional programs. In addition to classical or M1 macrophage activation in response to microbial products and interferon-γ, it was recently observed that anti-inflammatory molecules, such as glucocorticoid hormones, IL-4, IL-13, and IL-10, are more than simple inhibitors of macrophage activation in that they induce distinct M2 activation programs.1-5 M1 macrophages are involved in type 1 reactions and are classically activated by microbial products, killing microorganisms and producing reactive oxygen and nitrogen intermediates. In contrast, M2 cells, involved in type 2 reactions,4,6 tune inflammation and adaptive immunity; promote cell proliferation by producing growth factors and products of the arginase pathway (ornithine and polyamines); scavenge debris by expressing scavenger receptors; and promote angiogenesis, tissue remodeling, and repair. M1 and M2 cells are extremes in a continuum of functional states. For instance, different forms of M2 cells have been described sharing an IL-12low/IL-10high phenotype with variable capacity to produce TNF, IL-1, and IL-6.6 Tumors are diverse and so are their associated inflammatory reactions. When associated with established neoplasia, inflammation is usually polarized in a type 2 direction.
Tumors represent a unique example of the plasticity of macrophages, reflected by the ambivalent relationship between tumor-associated macrophages (TAMs) and cancer cells, originally expressed in the “macrophage balance” hypothesis.4,5 TAMs originate from circulating monocytes and their recruitment into tumors is driven by tumor-derived chemotactic factors.4,7,8 Among these, CCL2 has been identified as a major chemokine inducing the recruitment of macrophages in a variety of murine and human tumors.3,7,8 In addition, evidence suggests that the tumor microenvironment may also divert macrophage functions toward a protumoral phenotype.4 These tumor-diverted macrophages play a key role in subversion of adaptive immunity and in inflammatory circuits that promote tumor growth and progression.4,6,9,10
Despite the fact that TAMs originate from prototypical inflammatory cells, a number of reports providing a partial characterization of their phenotype have suggested that these cells are strongly impaired in various functions related to inflammation, such as production of various inflammatory mediators.4,11,12 Available information is consistent with a protumoral role for these cells, mainly based on their functional similarities with the immunosuppressive, M2-polarized macrophages.6,10 It has been hypothesized that TAMs, besides contributing actively to tumor-induced immunosuppression through subversion of innate and adaptive immunity, are also capable of affecting diverse aspects of neoplastic development, including vascularization, growth rate and metastasis, stroma formation, and dissolution.4 A protumoral role of TAMs is consistent with studies from humans, wherein a high density/number of TAMs is associated with poor prognosis in different cancers (cervix, prostate, breast, bladder).13,14
The present study was designed to characterize the transcriptional profile of TAMs obtained from a murine sarcoma11 where these cells exert a protumor function.15
The transcriptional program of TAMs was compared with those expressed by peritoneal macrophages (PECs) and myeloid suppressor cells (MSCs). PECs and TAMs are mature macrophage populations originating from common precursors, the circulating monocytes, whereas MSCs represent a prototypical M2 myeloid population.16 Lipopolysaccharide (LPS) was used in the present study as a classic model of activation stimulus of macrophages interacting with TLR4,17 a receptor that may also be triggered by components present in the tumor microenvironment (eg, hsp and derivatives of fibrinogen).18-21
Profiling of TAMs from a murine sarcoma revealed unexpected expression of IFN-inducible chemokines associated with an M2 phenotype (IL-10high, IL-12low) and divergent regulation of the NF-κB versus the IRF-3/STAT1 pathway.
Materials and methods
Cell culture
PECs, TAMs, and MSC-2 cells were cultured in RPMI 1640 medium containing 10% FCS, 2 mM glutamine, and 100 U/mL penicillin-streptomycin. The concentrations for the different agents used to activate the macrophage cultures were as follows: IFN-γ (500 U/mL; Hoffmann-LaRoche, Basilea, Switzerland); LPS (100 ng/mL; lipopolysaccharide from Escherichia coli strain B05.55; Sigma, St Louis, MO). The MSC-2 line originated from Gr-1+ splenocytes from mice immunized 6 days earlier with a recombinant vaccinia virus encoding mouse IL-2, as previously described.22
Preparation of PECs and TAMs
The study was reviewed and approved by the Istituto di Ricerche Farmacologiche Mario Negri (IRFMN) Animal Care and Use Committee (IACUC), which includes members ad hoc for ethical issues, in compliance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and European Union (EU) directives and guidelines.
Male 8-week-old C57Bl/6 mice were obtained from Charles River Breeding Laboratories (Calco, Italy). PECs and TAMs were isolated on the same day under the same culture conditions. Mice were inoculated intramuscularly in the left hind limb with a total of 105 MN/MCA1 fibrosarcoma11 cells per mouse. Tumor take was monitored and diameter of growing tumors was measured in centimeters twice a week by caliper. TAMs were isolated 3 weeks after tumor implantation, as described previously.11 Briefly, solid tumors were disaggregated by stirring with 0.125% (wt/vol) Trypsin (Sigma) for 40 minutes at 37°C, followed by washing twice in incomplete RPMI 1640 medium. Cells (70 × 106) were seeded in 140-mm Petri dishes (Cel Cult; Sterilin, Feltham, United Kingdom) in a final volume of 20 mL incomplete medium and, after 1 hour of incubation, nonadherent cells were vigorously washed off. The adherent cells were greater than 95% macrophages as assessed by morphologic and functional criteria. Contaminating cells were tumor cells, unidentified small mononuclear cells, and polymorphonuclear leukocytes. Immunofluorescence staining for macrophage markers (F4/80 and CD68) also confirmed the identity of the adherent populations. All culture reagents contained less than 0.125 endotoxin unit/mL as checked by Limulus amebocyte lysate assay (BioWhittaker, Walkersville, MD).
Peritoneal exudate cells were harvested from mice that had received injections of 500 μL 3% (wt/vol) thioglycollate medium (Difco, Detroit, MI) 4 days prior to isolation, as described previously.11 A total of 20 × 106 macrophages were seeded in 140-mm Petri dishes (Cel Cult; Sterilin) in a final volume of 20 mL incomplete medium and, after 1 hour of incubation, nonadherent cells were thoroughly washed off with jets of medium. Monolayers were greater than 95% macrophages, assessed by morphologic criteria and by a monoclonal antibody to mouse CD68 (macrophage marker, clone FA-11, dilution 1:50; Hycult Biotechnology, Uden, The Netherlands) or an anti-mouse F4/80 antigen monoclonal antibody (clone C1:A3-1, dilution 1:50; Serotec, Oxford, United Kingdom). All cultures were maintained in complete RPMI 1640 medium. After adherence, cells were rested for 1 hour in standard culture conditions and subsequently used in our experiments.
Possible effects of trypsin (used to disaggregate the tumors) on the gene expression profile were ruled out based on the results of gene expression experiments performed on normal macrophages in the presence or absence of trypsin, under exactly the same conditions as used for tumor disaggregation. Microarray as well as real-time polymerase chain reaction (PCR) for some representative TAM genes showed no significant change in response to trypsin treatment (data not shown).
RNA isolation and cDNA microarray
Microarray analysis of PECs and TAMs was done using the 10K murine cDNA array slides provided by the Molecular Technology Center (National Cancer Institute, Frederick, MD). Briefly, RNA from PECs or TAMs was extracted by the TRIzol method, quantified by optical density (OD) measurement, and checked for degradation. Before using for microarray, reverse transcriptase-PCR (RT-PCR) was run on IFN-γ/LPS-treated PEC and TAM RNA to check the IL-12p40+/IL-10- phenotype for TAMs. Total RNA from each sample (20 μg) was labeled and purified using the LabelStar Array kit (Qiagen gmbH, Hilden, Germany) as per manufacturer's protocol. Hybridization of the denatured labeled cDNA probes onto the 10K array slides was carried out by incubation at 65°C overnight in a hybridization chamber, followed by serial stringency washing (1 minute in 1 × SSC, 1 minute in 0.2 × SSC, 10 seconds in 0.05 × SSC) to remove excess dye. Thereafter, the dried slides were scanned with an Axon 4000 scanner (Axon Instruments, Foster City, CA) at a resolution of 10 μm. The reference RNA was labeled by using cyanine 3-dUTP (Cy3), whereas the experimental RNA was labeled with cyanine 5-dUTP (Cy5), except for the reverse-labeled experiments, which were performed to remove dye bias from the analysis. In most cases, PEC samples were labeled for Cy3 dye, TAM samples were labeled for Cy5 dye (for PEC versus TAM experiments), untreated samples were labeled for Cy3 dye, and LPS-treated samples were labeled for Cy5 dye (for untreated versus LPS-treated experiments in PECs and TAMs).
Data mining and statistical analysis
Image analysis and the calculation of average foreground signal adjusted for local channel-specific background was performed with GenePix Pro 4.0 (Axon Instruments) software. Global normalization of each array was separately done to make the median value of log 2 ratios equal to zero. Finally, the image and signal intensity data were uploaded onto the online MADB facility23 for data analysis. Cy5/Cy3 intensity ratios from each gene were calculated and subsequently normalized to ratios of overall signal intensity from the corresponding channel in each hybridization. The Cy5/Cy3 ratio represents the relative abundance of the genes in each experimental sample compared with the reference sample and hence provides quantitative measurements of the relative gene expression levels across all experimental samples. For all data filtering processes, genes qualifying for Cy5/Cy3 ratios greater than or equal to 2.5, or less than or equal to 0.4 were used as indicators of significantly different gene expression levels between 2 samples hybridized to the same array spot. All statistical analysis for the arrays was carried out using the MADB online facility.23 Each experiment was performed twice with reciprocal labeling and 3 such independent experiments were performed to meet the statistical requirements. Minimum information about a Microarray Experiment (MIAME) and the entire data set for all microarray experiments are available on the MADB website23 and NCBI-GEO24 (accession no. GSE2098).
Real-time PCR
RT reaction from 1 μg RNA template was performed using TaqMan reverse transcription reagents (Applied Biosystems, Piscataway, NJ) as per manufacturer's instructions. Real-time PCR was done using SyBr Green PCR Master Mix (Applied Biosystem, Piscataway, NJ) and detected by ABI-Prism 5700 Sequence Detector (Applied Biosystems, Foster City, CA). Data were processed using the GeneAmp software (Applied Biosystems, Foster City, CA) and normalized by actin gene expression levels. All real-time results were expressed as fold changes in mRNA expression with respect to the control cells. All results were normalized to the expression of the housekeeping gene β-actin in the PCR reactions. Data represented are from 3 independent experiments done in triplicate.
Immunoblotting
After the indicated treatments, PECs or TAMs were washed with ice-cold phosphate-buffered saline (PBS) containing 1 mM Na3VO4, then lysed in 50 μL of lysis buffer (20 mM Tris-HCl, pH 8; 137 mM NaCl; 10% glycerol [vol/vol]; 1% Triton X-100 [vol/vol]; 1 mM Na3VO4; 2 mM EDTA; 1 mM PMSF; 20 μM leupeptin; and 0.15 U/mL aprotinin) for 20 minutes at 4°C. The lysates were centrifuged at 13 000g at 4°C for 15 minutes, and the supernatants (containing Triton X-100-soluble proteins) were run on a 10% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 50 μg protein/lane). Separated proteins were transferred onto a nitrocellulose membrane (1 h at 125 mA) and immunoblotted for specific antibodies as per manufacturer's instructions. Blocking was done with 5% (wt/vol) bovine serum albumin (BSA) in TBS-0.1% Tween (TBST) for 1 hour at room temperature. All antibody dilutions were prepared in 5% (wt/vol) BSA-TBST. Primary antibodies were used at 1:1000 dilution for overnight at 4°C. HRP-conjugated antirabbit secondary antibody (Amersham, Arlington Heights, IL) was used at 1:10 000 dilution for 1 hour at room temperature. Blots were visualized using an enhanced chemiluminescence (ECL) kit (Amersham, Arlington Heights, IL). The antibodies used were anti-phospho-STAT1 (Cell Signaling Technologies, Beverly, MA), anti-STAT1, anti-p65 NF-κB, and antiactin (Santa Cruz Biotechnologies, Santa Cruz, CA). For the nuclear translocation of p65 NF-κB proteins, extraction of nuclear proteins was done as described earlier.25
ELISA
Cell-free supernatants from untreated or LPS-treated (100 ng/mL) PECs and TAMs were harvested after 24-hour incubation and tested in sandwich enzyme-linked immunosorbent assay (ELISA) for the indicated cytokines/chemokines. IL-6, TNF-α, IL-10, CCL2, CCL5, CXCL9, CXCL10, and CXCL16 were quantified by ELISA kits from R&D Systems (Minneapolis, MN), as per manufacturer's instructions. Results were normalized between PECs and TAMs and expressed as ng/mL/2 × 106 cells. Data are representative of 3 independent experiments done in triplicate.
Laser confocal microscopy
After indicated treatments, macrophage monolayers were cultured on rounded pyrogen-free glass coverslips, washed with permeabilized PBS, fixed with 4% (wt/vol) paraformaldehyde (SIGMA-Aldrich, St Louis, MO) for 20 minutes at room temperature, permeabilized, and blocked with 0.03% Triton-X100, normal goat serum (SIGMA-Aldrich), and 2% BSA (Amersham, Little Chalfont, United Kingdom) for 1 hour at room temperature. Thereafter, sections were washed with 0.2% BSA, 0.05% Tween 20 (Merck, West Point, PA) PBS and incubated with a rabbit polyclonal antibody (Ab) anti-IRF-3 (gift from Dr Takashi Fujita, Tokyo) at a dilution of 1:50 for 60 minutes or with the indicated antibody. Sections were extensively washed and subsequently incubated with an anti-rabbit IgG cross-adsorbed Alexa Fluor 647 conjugated antibody (Molecular Probes, Eugene, OR) diluted 1:1000 for 45 minutes at room temperature. For DNA detection, green SYTO 13 (Molecular Probes) plus RNAsi (SIGMA-Aldrich) was used. Sections were extensively washed with 0.2% BSA, 0.05% Tween 20 PBS; mounted with the FluorSave reagent (Calbiochem, San Diego, CA); and analyzed using an Olympus Fluoview 500 laser scanning confocal microscope (Olympus, Hamburg, Germany) and a UPLANSApo 60 ×/1.35 numeric aperture oil objective (Olympus). Images were captured and processed using Fluoview TV10-ASW application software for spectral analysis.
Immunohistochemistry and tissue laser confocal microscopy
For immunohistochemistry and tissue laser confocal microscopy, samples from tumor tissues were collected and frozen in liquid nitrogen. Nine-micrometer sections were cut and mounted on poly-l-lysine-coated slides. After fixation with acetone/chloroform for 3 minutes, sections were incubated for 2 hours with the following antibodies: monoclonal antibody to mouse CD68 (macrophage marker, clone FA-11, dilution 1:50; Hycult Biotechnology), goat biotinylated anti-mouse CRG-2/IP10 antibody (dilution 1:25; R&D Systems), monoclonal antibody to mouse Dectin-1 (clone 2A11, dilution 1:50; Hycult Biotechnology), rat monoclonal antibody to mouse IL-10 (clone JES5-2A5, dilution 1:200; Serotec), rabbit polyclonal to mouse NOS 2 (N-20; dilution 1:10; Santa Cruz Biotechnology), monoclonal antibody to mouse MGL1 (dilution 1:10; a kind gift of Dr Pieter Leenen, Erasmus MC, Rotterdam; and Dr Geert Raes, Vrije Universiteit Brussel),26 goat polyclonal biotinylated anti-mouse CXCL9/MIG antibody (dilution 1:10; R&D Systems), goat polyclonal biotinylated anti-mouse CXCL16 antibody (dilution 1:5; R&D Systems), goat polyclonal biotinylated anti-mouse CCL2/JE/MCP-1 antibody (dilution 1:5; R&D Systems), goat polyclonal anti-mouse CCL5/RANTES antibody (dilution 1:20; R&D Systems). For immunohistochemistry, the staining was revealed by using specific secondary antibodies with 3-3′ diamino-benzidine as chromogen. For laser confocal analysis, the specific secondary antibodies were used: Alexa Fluor 488 goat antimouse, antirabbit, and anti-rabbit streptavidin conjugated, and Alexa 647 antimouse, antirabbit, and anti-rabbit streptavidin conjugated. Negative controls were obtained by using isotype-matched primary antibodies.
Results
TAM transcriptome shows a characteristic gene expression profile
RNA from TAMs and thioglycollate-elicited PECs (used as control population) was subjected to microarray analysis using a murine 10 000-gene National Cancer Institute (NCI) slide as described in “RNA isolation and cDNA microarray.” The comparison between PECs and TAMs revealed qualitative and quantitative differences in their transcriptomes and only the significantly modulated genes of immunologic relevance were further investigated. As summarized in Table 1, the resting TAM transcriptome was characterized by the high expression of genes coding for chemokines (Ccl2, Ccl5, and Cxcl10); scavenging receptors and phagocytosis-associated molecules (macrophage scavenger receptor 2 or scavenger receptor type II [Msr2] and complement 1q, polypeptide alpha and gamma [C1qa, g]); surface molecules/markers (lymphocyte antigen 6 complex locus A, E [Ly6a, e], CD81 [Cd81], major histocompatibility complex [MHC] II [H2DMa, H2Eb1]); miscellaneous genes for growth regulators (migration inhibitory factor [Mif], allograft inflammatory factor 1 [Aif1], transforming growth factor beta-induced [Tgfbi]); and transcription factors (interferon responsive element 7 [Irf7], T-box 6 [Tbx6], and inhibitor of DNA binding 3 [Idb3]). In contrast, though PECs expressed very low but detectable levels of proinflammatory chemokines/cytokine genes such as Ccl3, Ccl4, Ccl6, Ccl9, Il1b, and tumor necrosis factor alpha (Tnfa), the basal expression of these genes was significantly down-regulated in TAMs. Other notably down-regulated genes were those coding for proteases and their inhibitors (cathepsins B, L [Ctsl, Ctsb], tissue inhibitor of metalloprotease 1 [Timp1], secretory leukocyte protease inhibitor [Slpi]; Table 1). As shown in Figure 1, hierarchical clustering confirmed that the TAM profile was consistently observed in 4 different experiments.
Selected gene list defining the TAM transcriptome
Feature ID . | Gene . | Description . | Average log ratio . |
|---|---|---|---|
| Up-regulated genes | |||
| Receptors/surface molecules | |||
| IMAGE:534886 | Ly6a* | Lymphocyte antigen 6 complex, locus A | 8.70 |
| IMAGE:582679 | Ly6e | Lymphocyte antigen 6 complex, locus E | 7.05 |
| IMAGE:775893 | Cd81* | CD81 antigen | 5.05 |
| IMAGE:1477073 | Msr2* | Macrophage scavenger receptor 2 | 5.50 |
| IMAGE:1367026 | Sema4d | Sema domain, (semaphorin) 4D | 3.85 |
| IMAGE:597157 | C1qa* | Complement component 1, q subcomponent, alpha polypeptide | 4.69 |
| IMAGE:426010 | C1qg | Complement component 1, q subcomponent, gamma polypeptide | 2.95 |
| IMAGE:524391 | Axl | AXL receptor tyrosine kinase | 3.60 |
| IMAGE:1382670 | H2-DMa | Histocompatibility 2, class II, locus DMa | 2.95 |
| IMAGE:1092081 | H2-Eb1* | Histocompatibility 2, class II, antigen E beta 1 | 2.70 |
| IMAGE:406272 | Crabp1 | Cellular retinoic acid binding protein I | 3.10 |
| IMAGE:576062 | Fegr1 | Fe receptor, IgG, high affinity I | 4.30 |
| Chemokines | |||
| IMAGE:1446589 | Cxcl10* | Chemokine (C-X-C motif) ligand 10 | 4.10 |
| IMAGE:832342 | Ccl5* | Chemokine (C-C motif) ligand 5 | 2.75 |
| IMAGE:573898 | Ccl2* | Chemokine (C-C motif) ligand 2 | 2.51 |
| Transcriptional factors | |||
| IMAGE:1430219 | Irf7 | Interferon regulatory factor 7 | 5.40 |
| IMAGE:521100 | Idb3 | Inhibitor of DNA binding 3 | 3.10 |
| IMAGE:1446422 | Tbx6 | T-box 6 | 2.90 |
| IMAGE:1222882 | Hdac6 | Histone deacetylase 6 | 3.20 |
| IMAGE:934509 | Hist2h2aa1 | Histone 2, H2aa1 | 3.10 |
| Growth factors/miscellaneous | |||
| IMAGE:734101 | Tgfbi* | Transforming growth factor, beta induced | 4.70 |
| IMAGE:315447 | Mif* | Macrophage migration inhibitory factor | 6.00 |
| IMAGE:1382096 | Aif1 | Allograft inflammatory factor 1 | 3.50 |
| IMAGE:819851 | Gstp2 | Glutathioņe S-transferase, pi 2 | 3.10 |
| IMAGE:570994 | Igfbp4 | Insulin-like growth factor binding protein 4 | 2.70 |
| IMAGE:1228288 | Ucp2 | Uncoupling protein 2, mitochondrial | 2.60 |
| IMAGE:537489 | Ccnd1 | Cyclin D1 | 2.93 |
| IMAGE:440572 | Col13a1 | Procollagen, type XIII, alpha 1 | 4.00 |
| IMAGE:864318 | Lst1 | Leukocyte-specific transcript 1 | 2.80 |
| IMAGE:581981 | Unknown | 3.20 | |
| IMAGE:1247540 | Dapk3 | Death associated protein kinase 3 | 5.80 |
| IMAGE:1210036 | Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | 2.50 |
| Proteases and their inhibitors | |||
| IMAGE:1227398 | Spint1 | Serine protease inhibitor, Kunitz type 1 | 2.80 |
| IMAGE:1077734 | Serpinh1 | Serine (or cysteine) proteinase inhibitor, clade H, member 1 | 2.50 |
| Down-regulated genes | |||
| Receptors/surface molecules | |||
| IMAGE:764624 | Cd5l | CD5 antigen-like | 0.18 |
| IMAGE:440614 | Gp49b | Glycoprotein 49 B | 0.29 |
| IMAGE:404131 | Plaur | Urokinase plasminogen activator receptor | 0.25 |
| IMAGE:518979 | Hbb-y | Hemoglobin Y, beta-like embryonic chain | 0.28 |
| IMAGE:1617481 | Cd83 | CD83 antigen | 0.30 |
| IMAGE:437512 | Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b | 0.50 |
| IMAGE:1068952 | Folr2 | Folate receptor 2 (fetal) | 0.21 |
| IMAGE:366999 | Il11ra1 | Interleukin 11 receptor, alpha chain 1 | 0.49 |
| IMAGE:1349690 | Il11ra2 | Interleukin 11 receptor, alpha chain 2 | 0.43 |
| IMAGE:405832 | P2ry2 | Purinergic receptor P2Y, G-protein coupled 2 | 0.39 |
| IMAGE:1227418 | C4 | Complement component 4 (within H-2S) | 0.35 |
| IMAGE:1224549 | Igf2r | Insulin-like growth factor 2 receptor | 0.43 |
| IMAGE:533799 | Lbp | Lipopolysaccharide binding protein | 0.36 |
| Chemokines | |||
| IMAGE:751833 | Ccl3* | Chemokine (C-C motif) ligand 3 | 0.44 |
| IMAGE:1078018 | Ccl4 | Chemokine (C-C motif) ligand 4 | 0.47 |
| IMAGE:1077529 | Ccl6 | Chemokine (C-C motif) ligand 6 | 0.14 |
| IMAGE:1148463 | Ccl9 | Chemokine (C-C motif) ligand 9 | 0.38 |
| Transcriptional factors | |||
| IMAGE:403789 | Idb2 | Inhibitor of DNA binding 2 | 0.26 |
| Growth factors/miscellaneous | |||
| IMAGE:1139544 | Il1b* | Interleukin 1 beta | 0.43 |
| IMAGE:1327679 | Tnf* | Tumor necrosis factor | 0.32 |
| IMAGE:1001011 | Fcna | Ficolin A | 0.07 |
| IMAGE:1195776 | Retnla | Resistin-like alpha (Fizzl) | 0.06 |
| IMAGE:1264951 | Saa3 | Serum amyloid A 3 | 0.04 |
| IMAGE:990152 | Fabp5 | Fatty acid binding protein 5, epidermal | 0.21 |
| IMAGE:523460 | Fabp4 | Fatty acid binding protein 4, adipocyte | 0.22 |
| IMAGE:368778 | Pmp22 | Peripheral myelin protein | 0.21 |
| IMAGE:368524 | Mfge8 | Milk fat globule-EGF factor 8 protein | 0.19 |
| IMAGE:1001588 | Pltp | Phospholipid transfer protein | 0.19 |
| IMAGE:1227378 | Lpl | Lipoprotein lipase | 0.05 |
| IMAGE:819960 | Arhb | ras homolog gene family, member B | 0.22 |
| IMAGE:2101183 | Arhc | ras homolog gene family, member C | 0.26 |
| Proteases and their inhibitors | |||
| IMAGE:976659 | Ctsl | Cathepsin L | 0.34 |
| IMAGE:437755 | Ctsb | Cathepsin B | 0.33 |
| IMAGE:1077399 | Sdc1 | Syndecan 1 | 0.16 |
| IMAGE:1367299 | Slpi | Secretory leukocyte protease inhibitor | 0.04 |
| IMAGE:622732 | Timp1 | Tissue inhibitor of metalloproteinase 1 | 0.47 |
| IMAGE:1037661 | Mmp12 | Matrix metalloproteinase 12 | 0.25 |
Feature ID . | Gene . | Description . | Average log ratio . |
|---|---|---|---|
| Up-regulated genes | |||
| Receptors/surface molecules | |||
| IMAGE:534886 | Ly6a* | Lymphocyte antigen 6 complex, locus A | 8.70 |
| IMAGE:582679 | Ly6e | Lymphocyte antigen 6 complex, locus E | 7.05 |
| IMAGE:775893 | Cd81* | CD81 antigen | 5.05 |
| IMAGE:1477073 | Msr2* | Macrophage scavenger receptor 2 | 5.50 |
| IMAGE:1367026 | Sema4d | Sema domain, (semaphorin) 4D | 3.85 |
| IMAGE:597157 | C1qa* | Complement component 1, q subcomponent, alpha polypeptide | 4.69 |
| IMAGE:426010 | C1qg | Complement component 1, q subcomponent, gamma polypeptide | 2.95 |
| IMAGE:524391 | Axl | AXL receptor tyrosine kinase | 3.60 |
| IMAGE:1382670 | H2-DMa | Histocompatibility 2, class II, locus DMa | 2.95 |
| IMAGE:1092081 | H2-Eb1* | Histocompatibility 2, class II, antigen E beta 1 | 2.70 |
| IMAGE:406272 | Crabp1 | Cellular retinoic acid binding protein I | 3.10 |
| IMAGE:576062 | Fegr1 | Fe receptor, IgG, high affinity I | 4.30 |
| Chemokines | |||
| IMAGE:1446589 | Cxcl10* | Chemokine (C-X-C motif) ligand 10 | 4.10 |
| IMAGE:832342 | Ccl5* | Chemokine (C-C motif) ligand 5 | 2.75 |
| IMAGE:573898 | Ccl2* | Chemokine (C-C motif) ligand 2 | 2.51 |
| Transcriptional factors | |||
| IMAGE:1430219 | Irf7 | Interferon regulatory factor 7 | 5.40 |
| IMAGE:521100 | Idb3 | Inhibitor of DNA binding 3 | 3.10 |
| IMAGE:1446422 | Tbx6 | T-box 6 | 2.90 |
| IMAGE:1222882 | Hdac6 | Histone deacetylase 6 | 3.20 |
| IMAGE:934509 | Hist2h2aa1 | Histone 2, H2aa1 | 3.10 |
| Growth factors/miscellaneous | |||
| IMAGE:734101 | Tgfbi* | Transforming growth factor, beta induced | 4.70 |
| IMAGE:315447 | Mif* | Macrophage migration inhibitory factor | 6.00 |
| IMAGE:1382096 | Aif1 | Allograft inflammatory factor 1 | 3.50 |
| IMAGE:819851 | Gstp2 | Glutathioņe S-transferase, pi 2 | 3.10 |
| IMAGE:570994 | Igfbp4 | Insulin-like growth factor binding protein 4 | 2.70 |
| IMAGE:1228288 | Ucp2 | Uncoupling protein 2, mitochondrial | 2.60 |
| IMAGE:537489 | Ccnd1 | Cyclin D1 | 2.93 |
| IMAGE:440572 | Col13a1 | Procollagen, type XIII, alpha 1 | 4.00 |
| IMAGE:864318 | Lst1 | Leukocyte-specific transcript 1 | 2.80 |
| IMAGE:581981 | Unknown | 3.20 | |
| IMAGE:1247540 | Dapk3 | Death associated protein kinase 3 | 5.80 |
| IMAGE:1210036 | Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | 2.50 |
| Proteases and their inhibitors | |||
| IMAGE:1227398 | Spint1 | Serine protease inhibitor, Kunitz type 1 | 2.80 |
| IMAGE:1077734 | Serpinh1 | Serine (or cysteine) proteinase inhibitor, clade H, member 1 | 2.50 |
| Down-regulated genes | |||
| Receptors/surface molecules | |||
| IMAGE:764624 | Cd5l | CD5 antigen-like | 0.18 |
| IMAGE:440614 | Gp49b | Glycoprotein 49 B | 0.29 |
| IMAGE:404131 | Plaur | Urokinase plasminogen activator receptor | 0.25 |
| IMAGE:518979 | Hbb-y | Hemoglobin Y, beta-like embryonic chain | 0.28 |
| IMAGE:1617481 | Cd83 | CD83 antigen | 0.30 |
| IMAGE:437512 | Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b | 0.50 |
| IMAGE:1068952 | Folr2 | Folate receptor 2 (fetal) | 0.21 |
| IMAGE:366999 | Il11ra1 | Interleukin 11 receptor, alpha chain 1 | 0.49 |
| IMAGE:1349690 | Il11ra2 | Interleukin 11 receptor, alpha chain 2 | 0.43 |
| IMAGE:405832 | P2ry2 | Purinergic receptor P2Y, G-protein coupled 2 | 0.39 |
| IMAGE:1227418 | C4 | Complement component 4 (within H-2S) | 0.35 |
| IMAGE:1224549 | Igf2r | Insulin-like growth factor 2 receptor | 0.43 |
| IMAGE:533799 | Lbp | Lipopolysaccharide binding protein | 0.36 |
| Chemokines | |||
| IMAGE:751833 | Ccl3* | Chemokine (C-C motif) ligand 3 | 0.44 |
| IMAGE:1078018 | Ccl4 | Chemokine (C-C motif) ligand 4 | 0.47 |
| IMAGE:1077529 | Ccl6 | Chemokine (C-C motif) ligand 6 | 0.14 |
| IMAGE:1148463 | Ccl9 | Chemokine (C-C motif) ligand 9 | 0.38 |
| Transcriptional factors | |||
| IMAGE:403789 | Idb2 | Inhibitor of DNA binding 2 | 0.26 |
| Growth factors/miscellaneous | |||
| IMAGE:1139544 | Il1b* | Interleukin 1 beta | 0.43 |
| IMAGE:1327679 | Tnf* | Tumor necrosis factor | 0.32 |
| IMAGE:1001011 | Fcna | Ficolin A | 0.07 |
| IMAGE:1195776 | Retnla | Resistin-like alpha (Fizzl) | 0.06 |
| IMAGE:1264951 | Saa3 | Serum amyloid A 3 | 0.04 |
| IMAGE:990152 | Fabp5 | Fatty acid binding protein 5, epidermal | 0.21 |
| IMAGE:523460 | Fabp4 | Fatty acid binding protein 4, adipocyte | 0.22 |
| IMAGE:368778 | Pmp22 | Peripheral myelin protein | 0.21 |
| IMAGE:368524 | Mfge8 | Milk fat globule-EGF factor 8 protein | 0.19 |
| IMAGE:1001588 | Pltp | Phospholipid transfer protein | 0.19 |
| IMAGE:1227378 | Lpl | Lipoprotein lipase | 0.05 |
| IMAGE:819960 | Arhb | ras homolog gene family, member B | 0.22 |
| IMAGE:2101183 | Arhc | ras homolog gene family, member C | 0.26 |
| Proteases and their inhibitors | |||
| IMAGE:976659 | Ctsl | Cathepsin L | 0.34 |
| IMAGE:437755 | Ctsb | Cathepsin B | 0.33 |
| IMAGE:1077399 | Sdc1 | Syndecan 1 | 0.16 |
| IMAGE:1367299 | Slpi | Secretory leukocyte protease inhibitor | 0.04 |
| IMAGE:622732 | Timp1 | Tissue inhibitor of metalloproteinase 1 | 0.47 |
| IMAGE:1037661 | Mmp12 | Matrix metalloproteinase 12 | 0.25 |
The values corresponding to each gene represent its expression levels in terms of the averaged log 2 ratio of Cy5/Cy3 intensities across all the microarray experiments. Stringency limits for significant gene modulation were as follows: mean ratio of Cy5/Cy3 intensity ≥2.5 for up-regulated genes and ≤0.4 for down-regulated genes.
Genes were validated by RT-PCR. Data are representative of 4 independent experiments
Based on the TAM microarray profile, further investigation on expression of the genes coding for chemokines/cytokines/growth factors (Ccl2, Ccl5, Cxcl10, Il-10, Mif) and surface molecules (Cd81, Msr2, C1qa, Ly6e and H2Eb1 [MHC-II]) was performed by real-time PCR. Figure 2A shows the relative mRNA expression levels of these genes in TAMs and PECs. The results demonstrated consistently higher mRNA expression for all the above genes in resting TAMs compared with PECs. Similar results were obtained when resident peritoneal macrophages were used (not shown). In addition, the up-regulation of 2 other interferon-inducible chemokine genes, Cxcl9 and Cxcl16, was also detected in TAMs by RT-PCR. The gene expression profile for TAMs was also reflected in terms of protein secretion. ELISA for the chemokine/cytokine-associated genes clearly confirmed significantly higher levels of CCL2, CCL5, CXCL9, CXCL10, CXCL16, and IL-10 proteins in the supernatants from TAMs compared with PECs (Figure 2B).
Characterization of TAMs in vivo
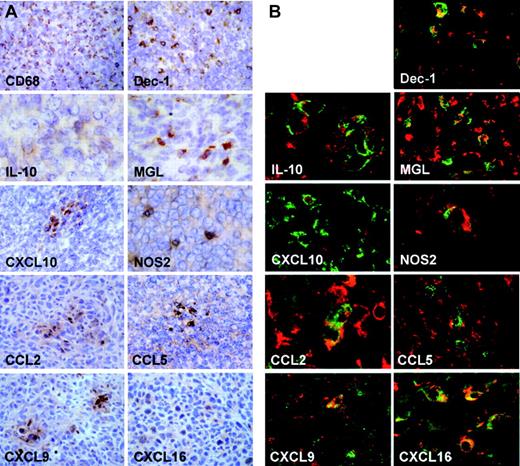
In an effort to assess the in vivo relevance of the present, as well as previous, in vitro characterization of TAMs, we performed immunohistochemical evaluation of M1 and M2 markers on frozen tumor sections. Dectin-1, MGL1, and IL-10 were chosen as M2 markers.4,26,27 We also investigated NOS2, CCL2, CCL5, CXCL9, CXCL10, and CXCL16, which are generally associated with an M1 phenotype although they are also expressed in TAMs. Dectin-1 is the major macrophage receptor for beta-glucans and was found to be highly up-regulated by GM-CSF and by the cytokines that induce alternative macrophage activation, IL-4 and IL-13.27 MGL1 is a member of the mouse macrophage galactose-type C-type lectin gene family and is induced in diverse M2 macrophage population during infection with the protozoan Trypanosoma brucei or the Helminth Taenia crassiceps or exposure to IL-4 and IL-13.26 As shown in Figure 3A, the MN/MCA1 fibrosarcoma was highly infiltrated by CD68+ cells, corresponding to TAMs. In addition, we observed a high number of Dectin-1+ cells as well as a significant positivity for MGL1 and IL-10. As confirmed by confocal microscopy (Figure 3B), CD68 expression colocalized with Dectin-1, MGL, and IL-10 on a significant number of cells. As shown, we also observed islets of either CCL2, CCL5, CXCL9, CXCL10, and CXCL16, or NOS2-positive cells (Figure 3A). To note, while CCL2, CCL5, CXCL9, CXCL10, and CXCL16 colocalized with CD68 on a significant number of cells, only sporadically could we observe colocalization of small spots of NOS2 on CD68+ cells (Figure 3B). Thus, analysis of TAMs in vivo is consistent with in vitro profiling and suggests that TAMs express a mixed phenotype, with key properties of M2 cells (eg, MGL-1high, scavenger receptorhigh IL-10high, IL-12low), coexpressed with IFN-inducible chemokines.
LPS responsiveness of TAMs
We have previously shown that TAMs express defective IL-12 production and NF-κB activation.11 Based on this, it was important to characterize the transcriptome profile of TAMs in response to proinflammatory signals, such as LPS. Three types of array experiments were performed: untreated PECs versus LPS-treated PECs, untreated TAMs versus LPS-treated TAMs, and LPS-treated PECs versus LPS-treated TAMs. Results from these experiments showed the induction of a common set of LPS-inducible genes in both PECs and TAMs. However, quantitative estimate of the total LPS-inducible genes in both of the transcriptomes showed a significantly lower number of induced genes in TAMs (263) compared with PECs (692; Figure 4A). More importantly, among the common set of LPS-inducible genes, the TAM profile showed a significantly lower level of expression of several proinflammatory genes such as Tnfa; Il1b; Ccl3, 6, and 9; Slpi; and ROI-related genes like Sod2 and Mt2 compared with PECs (Table 2). Real-time PCR analysis for several proinflammatory genes validated the above gene expression profile. Figure 4B demonstrates that LPS-treated TAMs not only expressed lower mRNA levels for Tnfa, Il1b, Il6, and Ccl3 genes but high mRNA expression of anti-inflammatory cytokines IL-10 and TGFβ1. For the IL-12p40 gene, mRNA expression was studied in response to IFN-γ plus LPS treatment, as stimulation with LPS alone yielded poor mRNA expression. In accordance to an earlier report,11 TAMs showed decreased IL-12p40 mRNA expression compared with the PECs. High mRNA expression of Ccl2, Ccl5, Cxcl9, Cxcl10, and Cxcl16 in the LPS-treated TAMs, as opposed to their PEC counterpart, was also detectable by RT-PCR. Finally, confirmation of the above results by ELISA demonstrated impaired expression of proinflammatory molecules IL-12p70, TNF-α, IL-6, and CCL3 but high expression of anti-inflammatory cytokine IL-10 as well as CCL2 and the IFN-inducible chemokines CCL5, CXCL9, CXCL10, and CXCL16 in the LPS-stimulated TAM supernatants (Figure 4C).
Profiling of the response of TAMs and PECs to LPS
. | . | . | Average log 2 ratio . | . | . | ||
|---|---|---|---|---|---|---|---|
| Genes . | Description . | Feature ID . | PECs, Unt vs LPS . | TAMs, Unt vs LPS . | TAM-LPS vs PEC-LPS . | ||
| Cytokines/chemokines/growth factors | |||||||
| Tnf | Tumor necrosis factor | IMAGE:1327679 | 3.57 | 1.62 | 0.20 | ||
| Il1b | Interleukin 1 beta | IMAGE:1139544 | 4.94 | 2.18 | 0.14 | ||
| Saa3 | Serum amyloid A 3 | IMAGE:1264951 | 2.85 | 1.78 | 0.21 | ||
| Ccl3 | Chemokine (C-C motif) ligand 3 | IMAGE:751833 | 4.94 | 2.53 | 0.08 | ||
| Ccl5 | Chemokine (C-C motif) ligand 5 | IMAGE:832342 | 4.35 | 4.54 | 2.71 | ||
| Ccl6 | Chemokine (C-C motif) ligand 6 | IMAGE:1077529 | 4.04 | 0.30 | 0.14 | ||
| Ccl9 | Chemokine (C-C motif) ligand 9 | IMAGE:1148463 | 2.61 | 0.30 | 0.14 | ||
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | IMAGE:1446589 | 3.99 | 3.48 | 2.55 | ||
| Tgfbi | Transforming growth factor, beta induced | IMAGE:734101 | 3.20 | 1.80 | 9.45 | ||
| Il12b | Interleukin 12 beta* | RT-PCR | 11059.75 | 151.70 | 0.01 | ||
| Il10 | Interleukin 10* | RT-PCR | 4.31 | 12.37 | 2.87 | ||
| ROI/RNI metabolism | |||||||
| Sod2 | Superoxide dismutase 2, mitochondrial | IMAGE:791140 | 2.26 | 1.10 | 0.34 | ||
| Nos2 | Nitric oxide synthase 2, inducible, macrophage | IMAGE:922250 | 3.57 | 4.11 | 1.16 | ||
| Atox1 | ATX1 (antioxidant protein 1) homolog 1 (yeast) | IMAGE:479066 | 1.74 | 0.69 | 0.83 | ||
| Mt1 | Metallothionein 1 | IMAGE:1037652 | 3.43 | 0.48 | 1.05 | ||
| Mt2 | Metallothionein 2 | IMAGE:334351 | 3.58 | 1.30 | 0.45 | ||
| Lyzs | Lysozyme | IMAGE:1382758 | 1.52 | 0.32 | 0.22 | ||
| Surface molecules/receptors | |||||||
| Il17r | Interleukin 17 receptor | IMAGE:1139646 | 2.31 | 0.23 | 0.18 | ||
| Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b | IMAGE:437512 | 4.54 | 3.22 | 0.30 | ||
| Tnfrsf5 | Tumor necrosis factor receptor superfamily, member 5 | IMAGE:477641 | 2.77 | 2.13 | 0.41 | ||
| Ccrl2 | Chemokine (C-C motif) receptor-like 2 | IMAGE:442765 | 2.40 | 2.22 | 0.38 | ||
| Icam1 | Intercellular adhesion molecule | IMAGE:1045389 | 2.13 | 1.13 | 1.18 | ||
| Signaling molecules/transcription factors | |||||||
| Nfkbib | Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, beta | IMAGE:946105 | 2.47 | 1.55 | 0.75 | ||
| Socs3 | Suppressor of cytokine signaling 3 | IMAGE:988726 | 3.71 | 2.14 | 0.30 | ||
| Stat3 | Signal transducer and activator of transcription 3 | IMAGE:479013 | 1.43 | 2.05 | 0.68 | ||
| Tbx6 | T-box 6 | IMAGE:1446422 | 3.27 | 3.32 | 0.46 | ||
| Irf1 | Interferon regulatory factor 1 | IMAGE:1344724 | 1.50 | 0.2 | 0.45 | ||
| Irf7 | Interferon regulatory factor 7 | IMAGE:1430219 | 2.54 | 1.61 | 0.62 | ||
| Proteases and their inhibitors | |||||||
| Slpi | Secretory leukocyte protease inhibitor | IMAGE:1367299 | 2.46 | 0.12 | 0.12 | ||
| Timp1 | Tissue inhibitor of metalloproteinase 1 | IMAGE:622732 | 3.73 | 0.31 | 0.08 | ||
| Mmp14 | Matrix metalloproteinase 14 (membrane-inserted) | IMAGE:1209994 | 2.38 | 1.15 | 0.90 | ||
. | . | . | Average log 2 ratio . | . | . | ||
|---|---|---|---|---|---|---|---|
| Genes . | Description . | Feature ID . | PECs, Unt vs LPS . | TAMs, Unt vs LPS . | TAM-LPS vs PEC-LPS . | ||
| Cytokines/chemokines/growth factors | |||||||
| Tnf | Tumor necrosis factor | IMAGE:1327679 | 3.57 | 1.62 | 0.20 | ||
| Il1b | Interleukin 1 beta | IMAGE:1139544 | 4.94 | 2.18 | 0.14 | ||
| Saa3 | Serum amyloid A 3 | IMAGE:1264951 | 2.85 | 1.78 | 0.21 | ||
| Ccl3 | Chemokine (C-C motif) ligand 3 | IMAGE:751833 | 4.94 | 2.53 | 0.08 | ||
| Ccl5 | Chemokine (C-C motif) ligand 5 | IMAGE:832342 | 4.35 | 4.54 | 2.71 | ||
| Ccl6 | Chemokine (C-C motif) ligand 6 | IMAGE:1077529 | 4.04 | 0.30 | 0.14 | ||
| Ccl9 | Chemokine (C-C motif) ligand 9 | IMAGE:1148463 | 2.61 | 0.30 | 0.14 | ||
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | IMAGE:1446589 | 3.99 | 3.48 | 2.55 | ||
| Tgfbi | Transforming growth factor, beta induced | IMAGE:734101 | 3.20 | 1.80 | 9.45 | ||
| Il12b | Interleukin 12 beta* | RT-PCR | 11059.75 | 151.70 | 0.01 | ||
| Il10 | Interleukin 10* | RT-PCR | 4.31 | 12.37 | 2.87 | ||
| ROI/RNI metabolism | |||||||
| Sod2 | Superoxide dismutase 2, mitochondrial | IMAGE:791140 | 2.26 | 1.10 | 0.34 | ||
| Nos2 | Nitric oxide synthase 2, inducible, macrophage | IMAGE:922250 | 3.57 | 4.11 | 1.16 | ||
| Atox1 | ATX1 (antioxidant protein 1) homolog 1 (yeast) | IMAGE:479066 | 1.74 | 0.69 | 0.83 | ||
| Mt1 | Metallothionein 1 | IMAGE:1037652 | 3.43 | 0.48 | 1.05 | ||
| Mt2 | Metallothionein 2 | IMAGE:334351 | 3.58 | 1.30 | 0.45 | ||
| Lyzs | Lysozyme | IMAGE:1382758 | 1.52 | 0.32 | 0.22 | ||
| Surface molecules/receptors | |||||||
| Il17r | Interleukin 17 receptor | IMAGE:1139646 | 2.31 | 0.23 | 0.18 | ||
| Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b | IMAGE:437512 | 4.54 | 3.22 | 0.30 | ||
| Tnfrsf5 | Tumor necrosis factor receptor superfamily, member 5 | IMAGE:477641 | 2.77 | 2.13 | 0.41 | ||
| Ccrl2 | Chemokine (C-C motif) receptor-like 2 | IMAGE:442765 | 2.40 | 2.22 | 0.38 | ||
| Icam1 | Intercellular adhesion molecule | IMAGE:1045389 | 2.13 | 1.13 | 1.18 | ||
| Signaling molecules/transcription factors | |||||||
| Nfkbib | Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, beta | IMAGE:946105 | 2.47 | 1.55 | 0.75 | ||
| Socs3 | Suppressor of cytokine signaling 3 | IMAGE:988726 | 3.71 | 2.14 | 0.30 | ||
| Stat3 | Signal transducer and activator of transcription 3 | IMAGE:479013 | 1.43 | 2.05 | 0.68 | ||
| Tbx6 | T-box 6 | IMAGE:1446422 | 3.27 | 3.32 | 0.46 | ||
| Irf1 | Interferon regulatory factor 1 | IMAGE:1344724 | 1.50 | 0.2 | 0.45 | ||
| Irf7 | Interferon regulatory factor 7 | IMAGE:1430219 | 2.54 | 1.61 | 0.62 | ||
| Proteases and their inhibitors | |||||||
| Slpi | Secretory leukocyte protease inhibitor | IMAGE:1367299 | 2.46 | 0.12 | 0.12 | ||
| Timp1 | Tissue inhibitor of metalloproteinase 1 | IMAGE:622732 | 3.73 | 0.31 | 0.08 | ||
| Mmp14 | Matrix metalloproteinase 14 (membrane-inserted) | IMAGE:1209994 | 2.38 | 1.15 | 0.90 | ||
The values corresponding to each gene represents gene modulation expressed in terms of the averaged log 2 ratio of Cy5/Cy3 intensities across all the microarray experiments. Stringency limits for significant gene modulation were as follows: mean ratio of Cy5/Cy3 intensities ≥ 2.5 for up-regulated genes and ≤ 0.4 for down-regulated genes. Data shown are representative of 6 independent experiments.
Expression values shown for the gene checked by real-time PCR
Hierarchical clustering of the TAM transcriptome in different experiments. Significantly modulated genes from the TAM transcriptome were clustered into a hierarchical representation, generated by the online NCI-MADB facility.23 Each column represents a particular experiment, whereas each row corresponds to the expression of a particular gene across different experiments. Gene names are indicated on the right. The color bar at the bottom left corner of the diagram correlates the degree of gene expression with the color scheme: up-regulated genes are represented in red, down-regulated genes in green, and unmodulated genes in black.
Hierarchical clustering of the TAM transcriptome in different experiments. Significantly modulated genes from the TAM transcriptome were clustered into a hierarchical representation, generated by the online NCI-MADB facility.23 Each column represents a particular experiment, whereas each row corresponds to the expression of a particular gene across different experiments. Gene names are indicated on the right. The color bar at the bottom left corner of the diagram correlates the degree of gene expression with the color scheme: up-regulated genes are represented in red, down-regulated genes in green, and unmodulated genes in black.
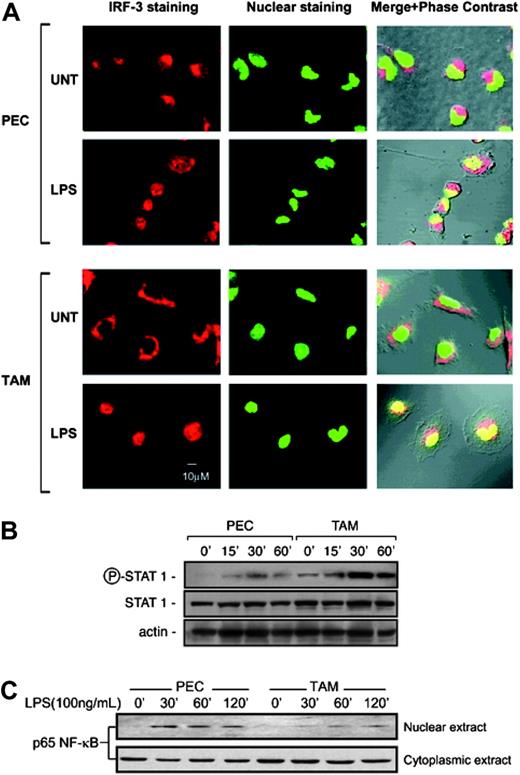
TAMs express defective NF-κB and enhanced IRF-3/STAT1 activation
To investigate the mechanistic basis of the TAM phenotype, we carried out a biochemical dissection of the signal transduction process in both PECs and TAMs upon activation with LPS. LPS signaling through the TLR4 receptor leads to the MyD88-dependent activation of NF-κB, a key transcription factor for the expression of most proinflammatory genes.28 In parallel, TLR4 engagement also promotes the MyD88-independent transcription of interferon-inducible chemokines (eg, CCL2, CCL5, CXCL10) through the activation of IRF-3 and STAT1.17,29,30 The activation of IRF-3 was studied by visualizing its nuclear translocation in TAMs by laser confocal microscopy. Untreated and LPS-treated PECs were used as controls. Figure 5A (bottom panel) shows massive nuclear translocation of IRF-3 at 2 hours following stimulation of TAMs with 100 ng/mL LPS, indicating its activation. Expression of interferon-inducible chemokines requires activation of STAT1 transcription factor.31 We investigated the activation of STAT1 and its expression in untreated or LPS-treated PECs and TAMs. As shown in Figure 5B, time kinetics for STAT1 activation using an anti-phospho-STAT1 antibody revealed a significantly higher STAT1 phosphorylation in LPS-treated TAMs (30-60 minutes of stimulation) than in treated PECs. Furthermore, constitutive phospho-STAT1 expression was detectable in unstimulated TAMs as opposed to PECs. Reblotting for STAT1 and actin expression confirmed equal loading.
Validation of selected components of the TAM transcriptome by RT-PCR and ELISA. (A) Real-time PCR of few highly expressed genes from the TAM transcriptome. The same pool of PEC and TAM RNA used in the array analysis was subjected to real-time PCR for the indicated genes. Representative results are given as fold increases in mRNA expression with respect to (wrt) the PECs. Data were normalized to actin gene expression as mentioned in “Materials and methods.” (B) ELISA detection for CCL2, CCL5, CXCL9, CXCL10, CXCL16, and IL-10 proteins in the culture supernatants of untreated PECs and TAMs cultured overnight under standard conditions. ND indicates not determined. Data are mean ± standard deviation (SD), representative of 3 independent experiments done in triplicate (PECs vs TAMs, P < .05).
Validation of selected components of the TAM transcriptome by RT-PCR and ELISA. (A) Real-time PCR of few highly expressed genes from the TAM transcriptome. The same pool of PEC and TAM RNA used in the array analysis was subjected to real-time PCR for the indicated genes. Representative results are given as fold increases in mRNA expression with respect to (wrt) the PECs. Data were normalized to actin gene expression as mentioned in “Materials and methods.” (B) ELISA detection for CCL2, CCL5, CXCL9, CXCL10, CXCL16, and IL-10 proteins in the culture supernatants of untreated PECs and TAMs cultured overnight under standard conditions. ND indicates not determined. Data are mean ± standard deviation (SD), representative of 3 independent experiments done in triplicate (PECs vs TAMs, P < .05).
Immunohistochemical and microscopy evaluation of frozen tumor sections. (A) Immunohistochemical evaluation of frozen tumor sections. The micrograph shows the following: high number of macrophages (brown cytoplasm staining to monoclonal CD68 antibody) infiltrating neoplastic tissue; monoclonal antibody to mouse Dectin-1 identifies a high number of small immunopositive cells (cells with brown cytoplasm) intermingled with neoplastic elements; immunohistochemistry with monoclonal antibodies to mouse interleukin-10, mMGL1, CXCL16, and NOS2 identifies isolated positive elements (brown cells); immunohistochemistry with biotinylated anti-mouse CRG-2/IP10 antibody (CXCL10) shows a cluster of positive small cells, whereas isolated elements are not observed; immunohistochemistry to CCL2, CCL5, and CXCL9 shows similar patterns with clusters of positive brown cells and few isolated positive cells. Original magnifications: × 20 (CD68, Dectin-1, CXCL10), × 40 (IL-10, mMGL1, NOS2, CCL2, CCL5, CXCL9, CXCL16). (B) Laser scanning confocal microscopy evaluation of frozen tumor sections: double immunostaining with monoclonal antibodies to mouse CD68 (red) and Dectin-1, IL-10, CRG2/IP10/CXCL10, mMGL1, NOS2, CCL2, CCL5, CXCL9, CXCL16 (all green staining). As shown in the micrographs, Dectin-1, IL-10, CRG-2/IP10, mMGL1, CCL2, CCL5, CXCL9, CXCL16 are expressed in macrophages but not in the totality of the CD68+ elements (red staining); NOS2-CD68 colocalization is observed in very rare cells.
Immunohistochemical and microscopy evaluation of frozen tumor sections. (A) Immunohistochemical evaluation of frozen tumor sections. The micrograph shows the following: high number of macrophages (brown cytoplasm staining to monoclonal CD68 antibody) infiltrating neoplastic tissue; monoclonal antibody to mouse Dectin-1 identifies a high number of small immunopositive cells (cells with brown cytoplasm) intermingled with neoplastic elements; immunohistochemistry with monoclonal antibodies to mouse interleukin-10, mMGL1, CXCL16, and NOS2 identifies isolated positive elements (brown cells); immunohistochemistry with biotinylated anti-mouse CRG-2/IP10 antibody (CXCL10) shows a cluster of positive small cells, whereas isolated elements are not observed; immunohistochemistry to CCL2, CCL5, and CXCL9 shows similar patterns with clusters of positive brown cells and few isolated positive cells. Original magnifications: × 20 (CD68, Dectin-1, CXCL10), × 40 (IL-10, mMGL1, NOS2, CCL2, CCL5, CXCL9, CXCL16). (B) Laser scanning confocal microscopy evaluation of frozen tumor sections: double immunostaining with monoclonal antibodies to mouse CD68 (red) and Dectin-1, IL-10, CRG2/IP10/CXCL10, mMGL1, NOS2, CCL2, CCL5, CXCL9, CXCL16 (all green staining). As shown in the micrographs, Dectin-1, IL-10, CRG-2/IP10, mMGL1, CCL2, CCL5, CXCL9, CXCL16 are expressed in macrophages but not in the totality of the CD68+ elements (red staining); NOS2-CD68 colocalization is observed in very rare cells.
The functioning of the MyD88-dependent pathway was investigated by tracing the downstream activation of NF-κB in LPS-stimulated cells. Nuclear translocation of p65 NF-κB subunit is a prerequisite for NF-κB activation. Western blot detection for p65 NF-κB subunit in the nuclear extracts of LPS-stimulated PECs and TAMs showed significantly delayed time kinetics for the nuclear translocation of p65 in TAMs, which was indicative of a defective NF-κB activity in these cells (Figure 5C). This observation is in agreement with our previous report on defective NF-κB activation in TAMs.11
Transcriptional profiling of TAMs exposed to LPS. (A) The bar graph presents a quantitative account of the gene modulations in PEC and TAM transcriptomes upon activation with LPS (100 ng/mL) for 4 hours. The data are representative of the microarray experiments described in Table 2. (B) Real-time PCR and (C) ELISA validation of few LPS-induced cytokine/chemokine genes in PECs and TAMs. The same pool of 4-hour LPS-stimulated PEC and TAM RNA used in the array was subjected to real-time PCR analysis for the indicated genes. All data were normalized with regard to β-actin gene expression. Results are given as fold increase in mRNA levels with respect to the untreated PECs. For ELISA, the culture supernatants of untreated or LPS-treated PECs and TAMs were collected following overnight incubation and determined for the indicated proteins. ND indicates not determined; Unt, untreated. LPS (100 ng/mL). Data are mean ± SD, representative of 3 independent experiments done in triplicate (PECs vs TAMs, P < .05). *For IL-12 real-time PCR and ELISA, the values correspond to IFN-γ-primed LPS treatment for PECs and TAMs (see “Results”).
Transcriptional profiling of TAMs exposed to LPS. (A) The bar graph presents a quantitative account of the gene modulations in PEC and TAM transcriptomes upon activation with LPS (100 ng/mL) for 4 hours. The data are representative of the microarray experiments described in Table 2. (B) Real-time PCR and (C) ELISA validation of few LPS-induced cytokine/chemokine genes in PECs and TAMs. The same pool of 4-hour LPS-stimulated PEC and TAM RNA used in the array was subjected to real-time PCR analysis for the indicated genes. All data were normalized with regard to β-actin gene expression. Results are given as fold increase in mRNA levels with respect to the untreated PECs. For ELISA, the culture supernatants of untreated or LPS-treated PECs and TAMs were collected following overnight incubation and determined for the indicated proteins. ND indicates not determined; Unt, untreated. LPS (100 ng/mL). Data are mean ± SD, representative of 3 independent experiments done in triplicate (PECs vs TAMs, P < .05). *For IL-12 real-time PCR and ELISA, the values correspond to IFN-γ-primed LPS treatment for PECs and TAMs (see “Results”).
Taken together, these results suggest a defective MyD88-dependent NF-κB pathway but a functional MyD88-independent pathway in TAMs. Divergent regulation of NF-κB and IRF-3/STAT1 is likely to underlie the differential gene expression profile of TAMs.
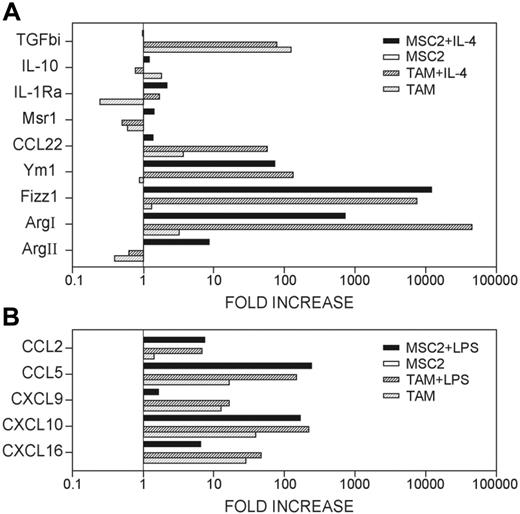
Profiling TAMs versus myeloid suppressor cells
Several reports have identified an M2-biased cell population in lymphoid organs of tumor-bearing hosts referred to as the myeloid suppressor cells (MSCs), which are suggested to contribute to the immunosuppressive phenotype.16 In an effort to put TAM profiling in a more general contest, we compared the TAMs with this cell population, with respect to their ability to express typical M2 genes, upon skewing by the M2 stimulus IL-4. As shown in Figure 6, resting TAMs expressed higher levels of Arg1, Fizz1, Ccl22, Il10, and Tgfb1 compared with MSCs. IL-4 treatment induced significant expression of M2 gene subset, Arg1, Fizz1, Ym1, Ccl22, Il10, and Tgfb1, which was comparable to (or even higher than) that of IL-4-treated myeloid suppressor cells (MSC2), a prototypic M2 population.16 This suggested that the TAMs were prone to M2 skewing compared with other M2 subpopulations, thus indicating their type II/M2 bias. However, interesting differences between the MSCs and TAMs were also evident. In particular, the expression of Ccl22 was significantly higher in the IL-4-treated TAMs in comparison with the M2-skewed MSC2 cells. Moreover, TGFβ expression was restricted to unstimulated TAMs and was not further increased by M2-biasing cytokines.
Due to the capability of TAMs to coexpress both M2-related genes and IFN-inducible M1 chemokines, LPS-activated TAMs and LPS-activated MSCs were compared for the expression of CCL2, CCL5, CXCL9, CXCL10, and CXCL16 (Figure 6B). As shown, following LPS activation, both TAMs and MSCs displayed appreciable levels of IFN-inducible chemokines.
Divergent regulation of the NF-κB and IRF-3/STAT1 pathway in TAMs. (A) Laser confocal microscopic representation of IRF-3 activation in TAMs. Untreated or 2-hour LPS-treated (100 ng/mL) PECs and TAMs were stained for IRF-3 (red) or with SYTO for nuclear counter-staining (green) and visualized by laser confocal microscopy. Panels represent IRF-3 staining, nuclear staining, and merge plus phase-contrast images (left to right). (B) STAT1 activation in TAMs. Western blot (WB) for phospho-STAT1 expression. The cell lysates from untreated or LPS-treated (100 ng/mL) PECs and TAMs for the indicated time points were probed first in WB with phospho-STAT1 antibody and reprobed with STAT1 antibody. Equal loading is visualized by actin expression. (C) Nuclear translocation of p65 NF-κB subunit. Western blot for p65 NF-κB subunit protein in the nuclear extracts of untreated or LPS-treated PECs and TAMs for the indicated time periods. Bottom panel shows cytoplasmic levels of the same. Results are representative of 3 independent experiments.
Divergent regulation of the NF-κB and IRF-3/STAT1 pathway in TAMs. (A) Laser confocal microscopic representation of IRF-3 activation in TAMs. Untreated or 2-hour LPS-treated (100 ng/mL) PECs and TAMs were stained for IRF-3 (red) or with SYTO for nuclear counter-staining (green) and visualized by laser confocal microscopy. Panels represent IRF-3 staining, nuclear staining, and merge plus phase-contrast images (left to right). (B) STAT1 activation in TAMs. Western blot (WB) for phospho-STAT1 expression. The cell lysates from untreated or LPS-treated (100 ng/mL) PECs and TAMs for the indicated time points were probed first in WB with phospho-STAT1 antibody and reprobed with STAT1 antibody. Equal loading is visualized by actin expression. (C) Nuclear translocation of p65 NF-κB subunit. Western blot for p65 NF-κB subunit protein in the nuclear extracts of untreated or LPS-treated PECs and TAMs for the indicated time periods. Bottom panel shows cytoplasmic levels of the same. Results are representative of 3 independent experiments.
Discussion
The results reported here show that TAMs from a murine sarcoma express a unique transcriptional profile. Resting TAMs showed higher expression of genes coding for immunosuppressive cytokines (Il10, Tgfbi), phagocytosis-related receptors/molecules (Msr2 and C1q), and inflammatory chemokines (Ccl2 and Ccl5), as expected, as well as, unexpectedly, IFN-inducible chemokines (Cxcl9, Cxcl10, Cxcl16). Compared with PECs, LPS-mediated activation of TAMs resulted in defective expression of several proinflammatory cytokines (eg, IL-1β, IL-6, TNF-α) and chemokines (eg, CCL3) and in the strong up-regulation of immunosuppressive cytokines (IL-10, TGFβ) and IFN-inducible chemokines (CCL5, CXCL9, CXCL10, and CXCL16). This functional profile was associated with defective activation of NF-κB and full activation of the MyD88-independent IRF-3 and STAT1 pathway.
The isolation procedure may have affected the profile and properties of TAMs; however, PECs that underwent the same isolation procedure did not show the same phenotype. Moreover, immunohistology has confirmed in this and other tumors key features of the TAM phenotype (eg, IL-12low, TNF-αlow, CCL2high, CCL5high, CXCL9high, CXCL10high, CXCL16high, Dectin-1high, IL-10high, MGL1high, NOS2low; Figure 3; Kataki et al32 and Kunz et al33 ). Therefore it is likely that the transcriptome of TAMs described in the present study is representative of cells in situ.
Several lines of evidence suggest that TNF-α and IL-1β can play a role in tumor progression.34 TAMs have a TNFlow phenotype in this and other studies in murine and human tumors.4 It is important to emphasize that TNF-α is generally not discriminatory between M1 and M2 macrophages, since at least one form of M2 polarization is characterized by a TNFhigh phenotype.2,6
Due to its capability to induce hemorrhagic necrosis and to stimulate antitumor immunity,35 locoregional administration of high doses of TNF-α is being used for the treatment of patients with locally advanced solid tumors.36 This therapeutic approach is apparently in contrast with preclinical findings suggesting that TNF-α may act as endogenous tumor promoter (eg, in ovarian and breast cancers).35 However, low-dose TNF-α was reported to promote the proliferation of some malignant cell lines.36,37 This evidence suggests a biphasic and dose-dependent effect of TNF-α on tumor progression and indicates that low TNF-α production by TAMs, as observed in the present and previous studies,4,32,35 may be optimal to promote tumor growth and metastasis. As the level of TNF-α production by TAMs may vary among tumors of different origin and different stages, it is likely to represent a determinant for the “balance” of protumoral versus antitumoral activities expressed by TAMs.
The expression profile of TAMs included genes not previously reported in this population. For instance, TAMs expressed Ly6a, e and CD81, which play a role in the interaction of antigen-presenting cells with T cells and in maintenance of T-helper 2 (Th2) phenotype.38,39 Therefore, these molecules may contribute to skewing the adaptive immune response at the tumor site in a Th2 direction.
Expression of M2- and M1-associated molecules by TAMs and MSCs. (A) RNA from untreated and IL-4-treated TAMs or untreated and IL-4-treated MSC-2 cells was used for real-time PCR analysis corresponding to the indicated genes. (B) RNA from untreated and LPS-treated TAMs or untreated and LPS-treated MSC-2 cells was analyzed by real-time PCR for the mRNA expression of the indicated genes. Results are given as fold increase in mRNA expression with respect to that in untreated MSC-2 cells. Data were normalized to expression of actin gene and representative of 3 independent experiments done in triplicate.
Expression of M2- and M1-associated molecules by TAMs and MSCs. (A) RNA from untreated and IL-4-treated TAMs or untreated and IL-4-treated MSC-2 cells was used for real-time PCR analysis corresponding to the indicated genes. (B) RNA from untreated and LPS-treated TAMs or untreated and LPS-treated MSC-2 cells was analyzed by real-time PCR for the mRNA expression of the indicated genes. Results are given as fold increase in mRNA expression with respect to that in untreated MSC-2 cells. Data were normalized to expression of actin gene and representative of 3 independent experiments done in triplicate.
A prominent set of genes expressed in TAMs were chemokines (Ccl2, Ccl5, Cxcl9, Cxcl10, Cxcl16), a finding confirmed at the protein level (Figure 2) and by immunohistochemistry (Figure 3). By producing chemokines such as CCL2, TAMs are likely to sustain and amplify tumor-elicited monocyte recruitment. CCL2 and CXCL10 have contrasting effects on angiogenesis.40-45 TAM production of these chemokines may tune new vessel formation and contribute to the irregular distribution and shape of tumor microvessels.
When LPS was used, an extensively characterized46-48 activation signal, TAMs showed quantitative and qualitative differences compared with PECs. The set of LPS-regulated genes was considerably smaller in TAMs compared with PECs. Moreover, a set of proinflammatory genes (chemokine/cytokine genes like Tnfa, Il1b, Ccl3, Ccl6, Ccl9; the oxidative burst-related genes Sod2, Lysz, Mt2; and several other LPS-inducible genes like Slpi, Timp1) were weakly induced in TAMs compared with PECs, whereas genes related to immunosuppression (Il10 and Tgfbi) showed a stronger up-regulation in TAMs versus PECs (Table 2). These results confirm and extend previous observation4,11 and are consistent with the view that TAMs are part of mechanisms of inhibition and diversion of specific immunity. LPS was used in the present study as a classic model of activation stimulus for TAMs. LPS is recognized by TLR4, which reportedly also interacts with components present in the tumor microenvironment (eg, hsp and derivatives of fibrinogen).18-21 In addition, and perhaps more interestingly, there is at present renewed interest for TLR agonists as immunotherapeutic agents in cancer.49,50 Hence, definition of the microenvironmental and signaling pathways responsible for the altered responsiveness of TAMs to TLR4 agonists may be relevant to the design of therapeutic strategies aimed at boosting innate and adaptive immunity against tumors.
Macrophage polarization into M1 and M2 cells provides a useful conceptual framework for the plasticity of mononuclear phagocytes.4 However, different versions of M2 cells have been described and the M1-M2 paradigm should be viewed as an operationally useful scheme.
The in vitro and in vivo results obtained in the present study, with resting and LPS-activated cells, are generally consistent with the view of TAMs as a unique and distinct polarized M2 population (IL-12low, TNF-αlow, NOS2low, IL-10high, Dectin-1high, MGL1high, TGF-βhigh, scavenger receptorhigh), characterized at variance with classic M2 cells by high levels of IFN-inducible chemokines. Scavenger receptors are generally up-regulated in M2-polarized macrophages. However, unexpectedly, TAMs showed high levels of CXCL10 and related chemokines, identified and characterized as IFN inducible.17
Thus, TAMs represent a unique macrophage population with key properties of M2 cells, which coexpress IFN-inducible chemokines. Interestingly, under M1- and M2-polarizing conditions (IL-4 versus LPS), MSCs also display the capability to express M1- and M2-associated molecules.
Signaling was investigated in an effort to define the molecular basis for the distinct phenotype of TAMs. NF-κB was not constitutively activated in TAMs and its activation in response to prototypic stimulus LPS was defective, in agreement with previous observations.4,11 p65 NF-κB translocation to the nucleus was delayed in LPS-stimulated TAMs. This finding, as well as delayed IκBα phosphorylation (data not shown), is reminiscent of the phenotype of MyD88-deficient macrophages.51 In contrast, TAMs showed appreciable levels of baseline STAT1 phosphorylation and, upon exposure to LPS, prominent IRF-3 translocation and STAT1 phosphorylation. Thus, TAMs show a unique dissociation of LPS signaling, with impaired MyD88-dependent NF-κB activation and full function of the MyD88-independent IRF-3 pathway. This molecular phenotype is consistent with the distinct transcriptional profile of LPS-activated TAMs, characterized by low inflammatory cytokine (eg, IL-12, TNF-α) induction as well as constitutive and inducible production of molecules such as CCL2, CCL5, CXCL9, CXCL10, and CXCL16.
Lack of constitutive NF-κB activity and defective NF-κB activation in response to LPS in TAMs is consistent with previous results in mouse and human tumors.4,11 These findings may seem at odds with recent results demonstrating a key role of the NF-κB pathway in liver and colon carcinogenesis.52,53 In particular, lineage-restricted inactivation of IKKβ revealed a nonredundant role of NF-κB activation in myeloid cells in colon carcinogenesis,53 an observation generally consistent with a protumor function of inflammatory reactions and TAMs in particular.34,35,54 This apparent discrepancy may reflect the different tumor systems investigated (spontaneous versus transplanted; carcinoma versus sarcomas), with differential involvement of different polarized inflammatory reactions. Alternatively, and more likely, vigorous NF-κB-dependent reactions underlie the overt inflammation that facilitates the early steps of colon carcinogenesis, but established neoplasia is propelled by a smoldering M2-polarized inflammatory milieu.
Prepublished online as Blood First Edition Paper, November 3, 2005; DOI 10.1182/blood-2005-01-0428.
Supported by Associazione Italiana Ricerca sul Cancro (AIRC), Italy; European Community; Ministero Istruzione Università Ricerca (MIUR), Italy; and Istituto Superiore Sanitá (ISS). Support from Alfredo Leonardi fellowship and G.L. Pfeiffer foundation to S.K.B. is acknowledged.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Prof S. Akira (Research Institute for Microbial Diseases, University of Osaka, Osaka, Japan) and Prof T. Fujita (Tokyo Metropolitan Institute of Medical Sciences, Tokyo, Japan) for the antibodies against IRF-3 and Dr M. Giannií for help with the phospho-specific antibodies. We thank Dr Pieter G. M. Leenen and Dr G. Raes for the kind donation of the anti-MGL1 antibody.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal