Mutations in LMAN1 (ERGIC-53) or MCFD2 cause combined deficiency of factor V and factor VIII (F5F8D). LMAN1 and MCFD2 form a protein complex that functions as a cargo receptor ferrying FV and FVIII from the endoplasmic reticulum to the Golgi. In this study, we analyzed 10 previously reported and 10 new F5F8D families. Mutations in the LMAN1 or MCFD2 genes accounted for 15 of these families, including 3 alleles resulting in no LMAN1 mRNA accumulation. Combined with our previous reports, we have identified LMAN1 or MCFD2 mutations as the causes of F5F8D in 71 of 76 families. Among the 5 families in which no mutations were identified, 3 were due to misdiagnosis, with the remaining 2 likely carrying LMAN1 or MCFD2 mutations that were missed by direct sequencing. Our results suggest that mutations in LMAN1 and MCFD2 may account for all cases of F5F8D. Immunoprecipitation and Western blot analysis detected a low level of LMAN1-MCFD2 complex in lymphoblasts derived from patients with missense mutations in LMAN1 (C475R) or MCFD2 (I136T), suggesting that complete loss of the complex may not be required for clinically significant reduction in FV and FVIII.
Introduction
Combined deficiency of factor V and factor VIII (F5F8D) was first described in a family by Oeri et al1 in 1954. Most patients have plasma levels of factor V (FV) and factor VIII (FVIII) (both antigen and activity) in the range of 5 to 30 U/dL and exhibit mild to moderate bleeding manifestations.2 The disease is inherited as an autosomal recessive disorder. Positional cloning efforts initially demonstrated that F5F8D results from null mutations in the gene encoding the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) marker protein ERGIC-53 (now called LMAN1).3 However, a significant subset (30%) of affected individuals exhibited evidence for mutations in at least one additional gene, and cells from these affected individuals had normal levels of LMAN1.4,5 We recently identified a second gene (MCFD2) that accounted for most of the remaining families without mutations in LMAN1.6 LMAN1 and MCFD2 form a stable, calcium-dependent complex that likely serves as a cargo receptor for efficient ER-to-Golgi transport of FV and FVIII. Cargo receptors selectively package secreted proteins into COPII-coated vesicles budding from the ER for trafficking to the Golgi compartment.7-9 Consistent with this idea, FVIII was shown to bind to both LMAN1 and MCFD2 through calcium-dependent protein-protein interactions in cultured cells.10,11 The B domain of FVIII appears to be the primary determinant of binding to the LMAN1-MCFD2 complex.6,12 In vitro data suggest that the other cargo proteins including the lysosomal proteins cathepsin Z13 and cathepsin C14 may also depend on LMAN1/MCFD2 function, though evidence for deficiency of these or other proteins in F5F8D patients has not been reported.
A total of 19 different mutations in the LMAN1 gene have been reported to date in 49 families,2,15,16 all of which appear to be null mutations that abolish protein expression. Seven MCFD2 mutations have been identified in 10 families.6,16 Five of the latter mutations disrupt the open reading frame, with the remaining 2 mutations resulting in single-amino acid substitutions in the second putative EF-hand domain (D129E and I136T). Both missense mutations were shown to abolish the interaction of MCFD2 with LMAN1.6 Mutant MCFD2 expressed in COS1 cells failed to coimmunoprecipitate with LMAN1 but appears to still retain the ability to bind FVIII.11
We now report the analysis of 20 F5F8D families, including 10 families previously reported to lack LMAN1 mutations.4,5 Novel mutations in LMAN1 and MCFD2 were identified, including evidence for regulatory mutations that abolish LMAN1 mRNA expression. In addition, we studied the interaction of LMAN1 and MCFD2 in lymphoblasts derived from a patient with the I136T mutation in MCFD2 and a patient with a novel missense mutation in LMAN1 (C475R).
Patients, materials, and methods
Patient samples
Peripheral blood (10-15 mL) was collected from the affected individuals, with informed consent from all individuals, after diagnosis of F5F8D. The study protocol was approved by the institutional review board at the University of Michigan. The levels of FV and FVIII activities are reported in Table 1. Epstein-Barr virus (EBV)-immortalized lymphoblast lines were derived from 3 affected individuals (family 9, Zhang et al6 ; B16; B17) as previously described.3
LMAN1 and MCFD2 mutation analyses, geographic origins, and FV/FVIII levels in F5F8D patients
Patient . | Origin . | FV/FVIII levels, U/dL . | LMAN1 mutation . | MCFD2 mutation . | Reference . |
|---|---|---|---|---|---|
| B2 | Japan* | 11/19 | None | None | 3 in Nichols et al5 |
| B3 | Italy* | 2.3/6.3 | None | 149+5G>A | 5 in Nichols et al5 |
| B4-1 | Italy | 7.5/6.0 | None | 149+5G>A | 6 in Nichols et al5 |
| B4-2 | Italy | 10/10 | None | 149+5G>A | 6 in Nichols et al5 |
| B5 | Italy | 11/23 | None | 149+5G>A | 8 in Nichols et al5 |
| B6 | Iran* | 10/6 | None | –6–IG>C | A14 in Neerman-Arbez et al4 |
| B7 | Italy | 8/24 | None | None | A22 in Neerman-Arbez et al4 |
| B8 | Italy | VWD | None | N/A | A33 in Neerman-Arbez et al4 |
| B9 | Kosovo | 4.5/1.5 | N/A | 407T>C (I136T) | This study |
| B10 | Serbia | 7/7 | N/A | 149+5G>A | This study |
| B11 | Italy | 13/20 | 2T>C (M1T) | None | This study |
| B12-1 | Austria | 15/12 | 780delT | None | This study |
| B12-2 | Austria | 24/13 | 780delT | None | This study |
| B13 | Iraq | 14/25 | G961>T (E321X) | None | This study |
| B14 | Poland* | 16/23 | 839-841delA | None | This study |
| B15 | Belgium | 11/35 | 822+1G>A | None | This study |
| B16 | USA | 23/24 | 822+33insGGTT†‡ | None | This study |
| B17 | Argentina | 18/23 | 1423T>C (C475R)‡ | None | This study |
| B18 | Greece | 7/5 | None | 266A>C (D89A) | This study |
Patient . | Origin . | FV/FVIII levels, U/dL . | LMAN1 mutation . | MCFD2 mutation . | Reference . |
|---|---|---|---|---|---|
| B2 | Japan* | 11/19 | None | None | 3 in Nichols et al5 |
| B3 | Italy* | 2.3/6.3 | None | 149+5G>A | 5 in Nichols et al5 |
| B4-1 | Italy | 7.5/6.0 | None | 149+5G>A | 6 in Nichols et al5 |
| B4-2 | Italy | 10/10 | None | 149+5G>A | 6 in Nichols et al5 |
| B5 | Italy | 11/23 | None | 149+5G>A | 8 in Nichols et al5 |
| B6 | Iran* | 10/6 | None | –6–IG>C | A14 in Neerman-Arbez et al4 |
| B7 | Italy | 8/24 | None | None | A22 in Neerman-Arbez et al4 |
| B8 | Italy | VWD | None | N/A | A33 in Neerman-Arbez et al4 |
| B9 | Kosovo | 4.5/1.5 | N/A | 407T>C (I136T) | This study |
| B10 | Serbia | 7/7 | N/A | 149+5G>A | This study |
| B11 | Italy | 13/20 | 2T>C (M1T) | None | This study |
| B12-1 | Austria | 15/12 | 780delT | None | This study |
| B12-2 | Austria | 24/13 | 780delT | None | This study |
| B13 | Iraq | 14/25 | G961>T (E321X) | None | This study |
| B14 | Poland* | 16/23 | 839-841delA | None | This study |
| B15 | Belgium | 11/35 | 822+1G>A | None | This study |
| B16 | USA | 23/24 | 822+33insGGTT†‡ | None | This study |
| B17 | Argentina | 18/23 | 1423T>C (C475R)‡ | None | This study |
| B18 | Greece | 7/5 | None | 266A>C (D89A) | This study |
None indicates no mutations detected; VWD, von Willebrand disease; and N/A, not analyzed.
Nucleotide numberings are relative to the initiation codon of GenBank sequences NM_005570 for LMAN1 and NM_139279 for MCFD2.
Known consanguinity
Patient is heterozygous for this mutation with no mutation detected on the other allele
Cause-effect relationship not established for this intronic mutation
Sequencing of the LMAN1 and MCFD2 genes in F5F8D patients
All coding exons and intron-exon junctions were amplified from genomic DNA prepared from peripheral blood as previously reported. Polymerase chain reactions (PCRs) were performed as previously described3,6 and DNA sequencing was completed in the University of Michigan DNA Sequencing Core. Total RNA was prepared from patient-derived lymphoblasts using the Trizol reagent (Invitrogen, Carlsbad, CA). Reverse transcriptase-PCRs (RT-PCRs) were performed as previously described,6 using primers listed in Table 2. Heterozygous mutations were confirmed by cloning PCR products and sequencing individual clones.
Primers used in RT-PCR and genotyping analyses
Primer . | Sequence 5′ → 3′ . |
|---|---|
| a | GAATTGGAGCTGATGGCCTA |
| b | CGCAACAAACCCTATCCTGT |
| c | ATCTGCTGCAACTGGAGGTC |
| d | GACCTCCAGTTGCAGCAGAT |
| e | TGAAATCAAGCAGCTGAACC |
| f | AGGCATTCCTGCTCCTCTTT |
| g | TGCAGGTGCTCTTTGATGTC |
| MCFD2F | GACCATGAGATCCCTGCTCAGA |
| MCFD2R | CTGCAGTGATTTTGCAAATTCAG |
| BZ1F | CCAGCAGCCAAAGAGAAAAC |
| BZ1R | GCTTGTCCCAGGAAAAAGAG |
| BZ20F | CATGTGCCACTGACAACAAAC |
| BZ20R | TCTCTGACACCCCTGAGGAC |
Primer . | Sequence 5′ → 3′ . |
|---|---|
| a | GAATTGGAGCTGATGGCCTA |
| b | CGCAACAAACCCTATCCTGT |
| c | ATCTGCTGCAACTGGAGGTC |
| d | GACCTCCAGTTGCAGCAGAT |
| e | TGAAATCAAGCAGCTGAACC |
| f | AGGCATTCCTGCTCCTCTTT |
| g | TGCAGGTGCTCTTTGATGTC |
| MCFD2F | GACCATGAGATCCCTGCTCAGA |
| MCFD2R | CTGCAGTGATTTTGCAAATTCAG |
| BZ1F | CCAGCAGCCAAAGAGAAAAC |
| BZ1R | GCTTGTCCCAGGAAAAAGAG |
| BZ20F | CATGTGCCACTGACAACAAAC |
| BZ20R | TCTCTGACACCCCTGAGGAC |
Immunoprecipitation and Western blot analysis
Patient-derived lymphoblasts were lysed and immunoprecipitated with rabbit anti-MCFD26 or monoclonal anti-LMAN1 antibodies (a gift from H.-P. Hauri, University of Basel, Switzerland) as previously described.6 The immunoprecipitates were further analyzed by Western blot with monoclonal anti-MCFD26 or rabbit anti-P58 (a gift from R. F. Pettersson, Karolinska Institute, Stockholm, Sweden).
Results
Further analysis of previously reported F5F8D patients
Previous LMAN1 gene sequence analysis failed to identify mutations in 17 of 54 F5F8D families.4,5 Ten of these families were studied in our recent report, which identified MCFD2 mutations in 7 families.6 No LMAN1 or MCFD2 mutations were identified in the remaining 3 families, raising the possibility of additional locus heterogeneity and the involvement of a third F5F8D gene. One of the 3 families (family 2, Zhang et al6 ) exhibited evidence for linkage to the MCFD2 locus,6 suggesting a mutation in a regulatory region of the gene that was missed by direct sequence analysis. The other 2 families (families 3 and 8, Zhang et al6 ) showed genetic evidence against linkage to either LMAN1 or MCFD2. Reanalysis of the FV and FVIII levels in probands from these 2 families now suggests errors in the initial F5F8D diagnosis. Reevaluation of the family 3 proband demonstrated a normal level of FVIII, with a FV level at 1% of normal, consistent with a diagnosis of simple FV deficiency. Indeed, linkage data from this family are consistent with a mutation on chromosome 1 in the vicinity of the F5 gene. Reanalysis of both male and female probands from family 8 (Zhang et al6 ) identified a FVIII level of approximately 10%, with a FV level of 40% to 50%. This family may have von Willebrand disease combined with a F5 null allele or a coincidental FV level at the lower end of normal.
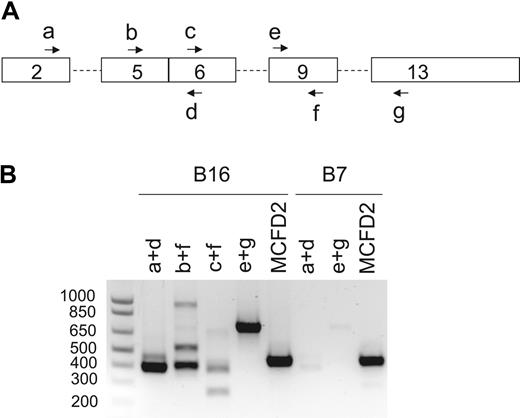
We sequenced the MCFD2 gene of the probands from the remaining 7 of the 17 previously reported F5F8D families4,5 (renumbered as B2-B8; Table 1) and identified mutations in 4 families. A novel homozygous mutation (-6-1G>C) that abolishes the consensus splice acceptor site of MCFD2 exon 2 was identified in family B6. Homozygosity for a previously identified splice donor site mutation (149 + 5G>A) was identified in 3 Italian families (B3-B5). No LMAN1 or MCFD2 mutations were identified in families B2, B7, and B8. Genotyping revealed that the proband of family B2, whose parents are first cousins, is homozygous at the LMAN1 locus and heterozygous at the MCFD2 locus (Table 3), suggesting an unidentified LMAN1 mutation. Although no mutation was identified in B7, Western blot analysis of the proband detected no LMAN1 protein.4 Immunoprecipitation of cell lysates from 5 × 106 lymphoblasts derived from the B7 proband also failed to detect LMAN1 (Figure 1C), suggesting a regulatory mutation in the LMAN1 gene that was missed by direct sequence analysis. Indeed, RT-PCR detected only a trace amount of LMAN1 mRNA and revealed an aberrantly spliced species of LMAN1 mRNA that skipped exon 4 (Figure 2). The low level of mRNA in this patient may be the result of nonsense-mediated decay. Reevaluation of the B8 proband revealed reduced von Willebrand factor and FVIII levels and a normal level of FV, consistent with a diagnosis of von Willebrand disease.
Haplotype analysis for 6 families with the 149+5G>A mutation and 2 families with the 407T>C (I136T) mutation
Family . | Haplotype . | Mutation . |
|---|---|---|
| 10 in Zhang et al6 | N/D-C-C-C-N/D-17 | 149+5G>A |
| 12 in Zhang et al6 | N/D-C-C-C-N/D-17 | 149+5G>A |
| B3 | N/D-A-C-C-N/D-19 | 149+5G>A |
| B4 | N/D-A-C-C-N/D-18 | 149+5G>A |
| B5 | N/D-A-C-C-N/D-19 | 149+5G>A |
| B10 | N/D-C-T-G-N/D-19 | 149+5G>A |
| 9 in Zhang et al6 | 19/22-C-C-C-19-20 | I136T |
| B9 | 19-C-C-C-18/19-18/19 | I136T |
Family . | Haplotype . | Mutation . |
|---|---|---|
| 10 in Zhang et al6 | N/D-C-C-C-N/D-17 | 149+5G>A |
| 12 in Zhang et al6 | N/D-C-C-C-N/D-17 | 149+5G>A |
| B3 | N/D-A-C-C-N/D-19 | 149+5G>A |
| B4 | N/D-A-C-C-N/D-18 | 149+5G>A |
| B5 | N/D-A-C-C-N/D-19 | 149+5G>A |
| B10 | N/D-C-T-G-N/D-19 | 149+5G>A |
| 9 in Zhang et al6 | 19/22-C-C-C-19-20 | I136T |
| B9 | 19-C-C-C-18/19-18/19 | I136T |
Three single-nucleotide polymorphisms located at the nucleotides 2895, 3099, and 3116 relative to the initiation codon of LMAN1 mRNA, as well as 2 CA repeat markers (BZ1 and D2S2227), were genotyped. Haplotypes are shown in the format BZ1 (CA repeat numbers)-C2895A-C3099T-C3116G-D2S2227 (CA repeat number)-BZ20 (GAAA repeat number).
N/D indicates genotyping not done for this marker.
Mutational analysis of additional F5F8D patients
We analyzed the LMAN1 and MCFD2 genes of 11 F5F8D patients from 10 previously unreported families (B9-B18; Table 1). These families included the first reported cases from the Balkans, Austria, Poland, Belgium, Argentina, and Greece. Homozygosity for LMAN1 null mutations were identified in 5 families, including 4 novel mutations (780delT in B12, 839-841delA in B14, E321X in B13, and 822 + 1G>A in B15) and 1 previously reported mutation (2T>C in B11). We also identified homozygous MCFD2 mutations in 2 families from the former Yugoslavia (B9-B10) and a family from Greece (B18). B9 carries a missense mutation (I136T) that has previously been reported in a Venezuelan family.6 B10 carries the 149 + 5G>A splicing mutation identified here in 3 Italian families and previously in US, Swiss, and Indian families.6,16 To examine the genetic origin of this mutation, we constructed haplotypes using 3 single-nucleotide polymorphisms in the 3′ untranslated region (UTR) of MCFD2 and 3 microsatellite markers (BZ1, located ∼ 570 kb upstream of MCFD2; D2S2227, located ∼ 123 kb downstream of MCFD2; and BZ20, located ∼1900 kb downstream of MCFD2). Among 6 families with the 149 + 5G>A mutation, 4 different haplotypes were identified, indicating at least 4 independent origins for this mutation (Table 3). These data are consistent with distinct geographic origins for many of these families and suggest a recurring mutation, perhaps due to a mutation hotspot at a CpG dinucleotide.17 The haplotypes of the 2 families (B9 and 9II-5, Zhang et al6 ) with the I136T missense mutation in MCFD2 diverge at both the BZ1 and D2S2227 markers (Table 3), suggesting that this mutation also arose independently. Lastly, B18 carries a novel homozygous missense mutation (D89A) that changes an amino acid at a position in the first EF-hand domain that is invariant in all MCFD2 orthologs.6 Although cells from family B18 are not available for study, we expect the D89A mutation to abolish binding of the mutant MCFD2 to LMAN1, similar to previously described D129E and I136T mutations.6
Immunoprecipitation and Western blot analysis of patient lymphoblasts. (A) Western blot analysis of LMAN1 in lymphoblasts from probands in families B16 and B17 compared with cells carrying the MCFD2 I136T mutation and wild-type (wt) cells. (B) A small amount of LMAN1-MCFD2 complex is detected in lymphoblasts with the MCFD2 I136T mutation. Lysates from 5 × 106 cells were immunoprecipitated (IP) with rabbit anti-MCFD2 followed by Western blot analysis with monoclonal anti-LMAN1 (top panel) or anti-MCFD2 antibodies (bottom panel). (C) Lymphoblasts from the B17 proband were untreated (N) or treated with the proteasome inhibitors MG132 (MG) or lactacystin (Lac) for 6 hours. Lysates from 5 × 106 cells of B17 and B7 lymphoblasts were immunoprecipitated with monoclonal anti-LMAN1 followed by Western blot analysis with polyclonal rabbit anti-LMAN1.
Immunoprecipitation and Western blot analysis of patient lymphoblasts. (A) Western blot analysis of LMAN1 in lymphoblasts from probands in families B16 and B17 compared with cells carrying the MCFD2 I136T mutation and wild-type (wt) cells. (B) A small amount of LMAN1-MCFD2 complex is detected in lymphoblasts with the MCFD2 I136T mutation. Lysates from 5 × 106 cells were immunoprecipitated (IP) with rabbit anti-MCFD2 followed by Western blot analysis with monoclonal anti-LMAN1 (top panel) or anti-MCFD2 antibodies (bottom panel). (C) Lymphoblasts from the B17 proband were untreated (N) or treated with the proteasome inhibitors MG132 (MG) or lactacystin (Lac) for 6 hours. Lysates from 5 × 106 cells of B17 and B7 lymphoblasts were immunoprecipitated with monoclonal anti-LMAN1 followed by Western blot analysis with polyclonal rabbit anti-LMAN1.
RT-PCR analysis of LMAN1 mRNA from probands of families B7 and B16. (A) Schematics of the LMAN1 cDNA with locations of PCR primers (Table 2, a-g) indicated. (B) Agarose gel electrophoresis of the RT-PCR products after amplification of LMAN1 mRNA with the various primer combinations indicated. The bottom bands in B16/b+f and B16/c+f lanes represent alternatively spliced species with exon 8 skipping, whereas the top bands represent the normally spliced products. The 900-bp band in the B16/b+f lane was a nonspecific PCR product according to sequence analysis. Two specific PCR products were observed in the B7/a+d lane. Sequencing of the latter PCR products further amplified by nested PCR revealed that the top band represents the normal spliced product, whereas the bottom band represents an alternatively spliced species with exon 4 skipping. Amplification of the coding region of MCFD2 mRNA served as a control for RNA quality.
RT-PCR analysis of LMAN1 mRNA from probands of families B7 and B16. (A) Schematics of the LMAN1 cDNA with locations of PCR primers (Table 2, a-g) indicated. (B) Agarose gel electrophoresis of the RT-PCR products after amplification of LMAN1 mRNA with the various primer combinations indicated. The bottom bands in B16/b+f and B16/c+f lanes represent alternatively spliced species with exon 8 skipping, whereas the top bands represent the normally spliced products. The 900-bp band in the B16/b+f lane was a nonspecific PCR product according to sequence analysis. Two specific PCR products were observed in the B7/a+d lane. Sequencing of the latter PCR products further amplified by nested PCR revealed that the top band represents the normal spliced product, whereas the bottom band represents an alternatively spliced species with exon 4 skipping. Amplification of the coding region of MCFD2 mRNA served as a control for RNA quality.
Western blot analysis of lymphoblasts derived from the proband of family B16 detected no LMAN1 (Figure 1A), indicating that loss of LMAN1 protein expression is the cause of F5F8D in this patient. Sequencing of the genomic DNA detected no mutations within the exons or splice junctions of the LMAN1 and MCFD2 genes. We isolated mRNA from lymphoblasts and performed RT-PCR analysis, identifying an alternatively spliced species of LMAN1 mRNA (Figure 2). Sequencing confirmed that the lower band corresponds to splicing of exon 7 to exon 9, with skipping of exon 8. The upper band corresponds to the wild-type LMAN1 product (exons 7-8-9). Sequencing of intron 7 demonstrated a heterozygous 4-nucleotide (GGTT) insertion 33 bp downstream of exon 7. The 4-nucleotide (nt) insertion may be responsible for this aberrant splicing, although an effect of another more distant mutation cannot be excluded. The wild-type product could represent residual normal splicing for the same allele. Alternatively, the second allele may carry an unidentified mutation that markedly reduces transcription or mRNA processing.
Missense mutations disrupt LMAN1-MCFD2 complex formation
We identified a heterozygous missense LMAN1 mutation (C475R) in the proband of an Argentinean family of Italian origin (B17). No other mutations were found in this patient. RT-PCR analysis of lymphoblast RNA revealed that only the missense allele is expressed at the mRNA level, indicating that the second LMAN1 allele in this patient carries an unidentified mutation that results in a cis-defect in transcription or mRNA stability. Cysteine 475 plays an important role in forming disulfide-linked homodimers and homohexamers of LMAN1. Substitution of C475 with alanine has previously been reported to abolish homohexamer formation without affecting homodimer formation.18,19 LMAN1C475A was shown to exhibit impaired transport out of the ER18 but retained a normal intracellular localization pattern.19 We analyzed cell lysates of lymphoblasts derived from the proband of B17. No endogenous LMAN1 was detectable by Western blot analysis (Figure 1A), though upon immunoprecipitation of lysates from 5 × 106 cells, a trace amount of LMAN1 was detected (Figure 1B). This is in contrast to the reported C475A substitution, which exhibited a turnover comparable to the wild-type protein.18 In addition to disrupting a disulfide bond that is critical for homohexamer formation, the C475R mutation presumably further compromises LMAN1 folding, leading to ER-associated protein degradation. However, treatment of cells with the potent proteasome inhibitors MG132 and lactacystin only slightly increased the amount of LMAN1 accumulation (Figure 1C), suggesting that degradation of LMAN1C475R is mediated largely through a proteasome-independent pathway.
We previously showed that the I136T mutation in MCFD2 identified in B9 and a Venezuelan family (family 9, Zhang et al6 ) abolished binding to LMAN1 in an in vitro transfection assay.6 To further study the endogenous behavior of this mutant, we performed coimmunoprecipitation analysis in lymphoblasts derived from a Venezuelan patient homozygous for the I136T mutation. Cell lysates were immunoprecipitated with anti-MCFD2 and analyzed by Western blotting with anti-MCFD2 or anti-LMAN1. A small amount of MCFD2I136T was detected at a level higher than that observed in LMAN1 mutant lymphoblasts (Figure 1B). A trace amount of LMAN1 also coimmunoprecipitated with MCFD2I136T (Figure 1B). Taken together, these data suggest that LMAN1 and MCFD2I136T retain a weak interaction. However, the residual amount of LMAN1-MCFD2 complex is apparently either functionally deficient or of insufficient quantity to support FV and FVIII secretion.
Discussion
We report the analysis of LMAN1 and MCFD2 mutations in 10 previously reported and 10 new F5F8D families. Homozygous MCFD2 and LMAN1 mutations were identified in 12 families. Heterozygous mutations were identified in only one LMAN1 allele in 3 families, all of which exhibit markedly reduced LMAN1 protein expression, indicating the existence of unidentified mutations in the second LMAN1 allele. Of the remaining 5 families, 3 can be explained by misdiagnosis of a similar bleeding disorder. Unidentified mutations in LMAN1 or MCFD2 are likely to explain F5F8D in the remaining 2 families that exhibit linkage to 1 of the 2 loci. Presumably regulatory or splicing mutations in unsequenced portions of the corresponding gene are responsible, though patient-derived cells were not available for further study. Thus, the locus heterogeneity identified in F5F8D appears to be limited to the LMAN1 and MCFD2 genes. These data are consistent with the failure of previous biochemical studies to identify additional components of the LMAN1-MCFD2 receptor complex or this receptor-mediated secretory pathway.11 Alternatively, mutations in other components of this pathway may exhibit a different phenotype distinct from F5F8D.
Combined with previous reports, at least 25 LMAN1 and 9 MCFD2 mutations have now been identified. Among these, 8 LMAN1 mutations and 3 MCFD2 mutations were identified in multiple families. In addition, 3 families carry unidentified regulatory mutations. No clear genotype/phenotype relationship can be identified to account for variations in FV and FVIII levels among F5F8D patients (Table 1). For example, even for patients homozygous for the same 149 + 5G>A mutation, levels of FV/FVIII varied from 2.3/1.2 to 11/23 (Table 1).16 Although variability in FV and FVIII assays could contribute, the range of observed values more likely reflects the combined effects of modifier genes for FV/FVIII and environmental factors. Although most reported F5F8D patients are from the Mediterranean region,3-6,20-22 an increasing number of cases have been identified in other geographic regions.4,5,15,16,23,24 A high percentage of F5F8D patients are associated with known consanguinity and/or are homozygous for a single mutation, suggesting that the geographic distribution of this disease is due at least in part to the varying prevalence of consanguineous marriages rather than specific positive selection for heterozygous carriers.
All previously reported LMAN1 mutations were predicted to be null, leading to the speculation that even a trace amount of functional LMAN1 expression may be sufficient to support normal FV and FVIII trafficking. However, we detected a low level of LMAN1-MCFD2 complex in patient lymphoblasts carrying either the LMAN1 C475R mutation (data not shown) or the MCFD2 I136T mutation (Figure 1). The LMAN1-MCFD2I136T complex may retain residual FVIII-binding activity, since MCFD2 carrying the similar D129E mutation was shown to bind FVIII.11 The latter observation suggests that a threshold level of LMAN1-MCFD2 activity above that observed for the LMAN1-MCFD2I136T complex may be required for the intracellular trafficking of FV and FVIII, though we cannot rule out the possibility that the LMAN1-MCFD2I136T interaction may be further weakened in more relevant cells such as hepatocytes in vivo. These results also suggest that targeting the LMAN1-MCFD2 complex as a therapeutic approach to anticoagulation is likely to require marked reduction of one or both components of this protein complex.
Prepublished online as Blood First Edition Paper, November 22, 2005; DOI 10.1182/blood-2005-09-3620.
Supported by grants PO1 HL057346, R37 HL039693, and RO1 HL52173 from the National Institutes of Health and a Career Development Award from the National Hemophilia Foundation (B.Z.). R.J.K. and D.G. are investigators of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal