GATA-2 is a zinc finger transcription factor essential for differentiation of immature hematopoietic cells. We analyzed the function of GATA-2 by a combined method of tetracycline-dependent conditional gene expression and in vitro hematopoietic differentiation from mouse embryonic stem (ES) cells using OP9 stroma cells (OP9 system). In the presence of macrophage colony-stimulating factor (M-CSF), the OP9 system induced macrophage differentiation. GATA-2 expression in this system inhibited macrophage differentiation and redirected the fate of hematopoietic differentiation to other hematopoietic lineages. GATA-2 expression commencing at day 5 or day 6 induced megakaryocytic or erythroid differentiation, respectively. Expression levels of PU.1, a hematopoietic transcription factor that interferes with GATA-2, appeared to play a critical role in differentiation to megakaryocytic or erythroid lineages. Transcription of PU.1 was affected by histone acetylation induced by binding of GATA-2 to the PU.1 promoter region. This study demonstrates that the function of GATA-2 is modified in a context-dependent manner by expression of PU.1, which in turn is regulated by GATA-2.
Introduction
Hematopoiesis is a well-coordinated, sequential process in which multipotential hematopoietic cells differentiate through intermediate progenitor cells to form the progenies of more than 8 distinct lineages.1 Hematopoietic differentiation is regulated by complex interactions among numerous transcription factors and cofactors.2 However, the functions of the individual genes involved in the process remain largely undefined. In addition, epigenetic regulation of gene expression has recently emerged as an important factor in hematopoiesis, as well as in other cell differentiation systems.3-5
The GATA family of transcriptional regulatory protein factors is comprised of 6 members, GATA-1 through GATA-6.6 Of the 6 mammalian GATA family members, 3 (GATA-1, -2, and -3) are indispensable for hematopoiesis. GATA-1 and GATA-3 are lineage-specific transcription factors. GATA-1 is expressed at high levels in erythroid cells, megakaryocytes, mast cells, and eosinophils.7-11 GATA-1-deficient erythroid precursors fail to differentiate beyond the proerythroblast stage.12-15 GATA-3 is expressed in the T-lymphoid lineage16 and is required for T-lymphocyte development.17,18
In contrast to GATA-1 and GATA-3, GATA-2 is broadly expressed in the hematopoietic system. It is expressed at particularly high levels in early hematopoietic cells as well as in megakaryocyte lineages.19-21 The function of GATA-2 has been examined by null mutation and overexpression studies. A GATA-2 knockout mouse exhibited panhematopoietic defects and embryonic lethality at day 12.5 postcoitum.22 In the in vivo and in vitro differentiation analyses using Gata2 null embryonic stem (ES) cells, GATA-2 was required for proliferation and survival of immature hematopoietic progenitors.23 Overexpression of GATA-2 induced proliferation of hematopoietic progenitors.24
Interactions between lineage-specific transcription factors have recently been shown to be important in determining the fate of hematopoietic cells.25 An archetypal example is provided by the interaction between PU.1 and GATA transcription factors.26-30 PU.1 is a hematopoietic cell-specific transcription factor of the Ets family that is required for development of some lymphoid and myeloid lineages.31 PU.1 can also function as an oncoprotein in erythroid precursors after proviral insertion or transgenesis.26,31 In some contexts, GATA factors and PU.1 have antagonistic functions in the development of distinct hematopoietic lineages. For example, GATA-1 blocks PU.1 transactivation by blocking binding of the PU.1 coactivator c-jun, and PU.1 inhibits GATA-1 transactivation by inhibiting its binding to DNA.26-28,32 On the other hand, generation of mast cells requires cooperated functions of both GATA-2 and PU.1.33
A research method combining in vitro ES cell differentiation with tetracycline-based conditional gene expression is a powerful tool for analyzing the functions of various genes at distinct stages of hematopoietic development and differentiation.24,34-36 In a previous in vitro study using mouse ES cells cocultured on the stromal cell line OP9, we demonstrated that GATA-2 increased the proliferation of immature hematopoietic cells.24
In the present study, we used the OP9 stroma cell-based differentiation method to examine the effects of GATA-2 on the development of macrophages from ES cells. We found that GATA-2 not only inhibited macrophage differentiation, but it also induced lineage distortion to megakaryocyte or erythroid cell differentiation. Lineage distortion was dependent on the timing of Gata2 expression. These results show that the function of GATA-2 in hematopoietic cells depends on differentiation status. Furthermore, we found that the interaction between PU.1 and GATA-2 plays a crucial role in cell fate determination, and epigenetic alteration of PU.1 (Sfpi1) gene expression could be involved in this process.
Materials and methods
Cell culture
Mouse ES cell lines that conditionally expressed mouse Gata2 on tetracycline induction were established and maintained as previously described.24 Although the data of this paper were obtained from one cell line, the other 2 lines gave essentially similar results. Hematopoietic induction of the ES cells was carried out by coculture with OP9 stromal cells.37,38 For macrophage differentiation induction, 10 ng/mL human macrophage colony-stimulating factor (M-CSF; a kind gift from Morinaga Milk, Tokyo, Japan) was added to the coculture at day 5. Following this, an inhibitor of HDAC, trichostatin A (TSA; Sigma, St Louis, MO), was added to the culture at 5 ng/mL at day 5. Cells were stained with May-Giemsa dye for morphologic analysis or with acetylcholinesterase (AchE) for detection of megakaryocyte differentiation.
A retrovirus-producing cell line, Plat-E, was a kind gift from Dr T. Kitamura (Tokyo University, Tokyo, Japan). Plat-E cells were maintained and transfected with retroviral vectors as described previously.39 ES cells were dissociated by trypsin treatment on day 5 after differentiation induction and were infected with the Plat-E retroviral supernatant by spinoculation (1100g, 2 hours). Microscopic images were obtained and captured using an Olympus Provis microscope with a UPlan APO 40 ×/0.85 numeric aperture objective lens, an Olympus DP70 camera, and DC controller software (all from Olympus, Tokyo, Japan). Images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
Plasmid construction
A retroviral plasmid vector, pMY.IRES.EGFP was a kind gift from Dr T. Kitamura (Tokyo University, Tokyo, Japan). The plasmid pMY.IRES.EYFP was constructed by replacing the EGFP with the EYFP gene, which had been excised from pEYFP (BD Biosciences; Franklin Lakes, NJ). The murine PU.1 cDNA, which was a kind gift from Dr T. Oikawa (Sasaki Institute, Tokyo, Japan), was inserted into pMY.IRES.EYFP to create pMY.PU.1.IRES.EYFP.
Flow cytometry
Hematopoietic cells cocultured with OP9 stromal cells were harvested by gentle pipetting. FcBlock (BD Biosciences) was added to the cell suspension to block nonspecific staining before the cells were stained with antibodies. The antibodies used were specific for biotin-Mac-1 (BD Biosciences), biotin-TER-119 (Dr. T. Kina, Kyoto University, Kyoto, Japan), biotin-platelet glycoprotein V (GPV) (Seikagaku, Tokyo, Japan), and PE-CD71 (eBioscience, San Diego, CA). The biotinylated antibodies were visualized with Cy-chrome-conjugated streptavidin (BD Biosciences). Surface marker expression and cell sorting were carried out using a FACSCalibur (BD Biosciences).
Western blotting
Cells were suspended in 62.5 mM Tris-HCl (pH 6.8) buffer containing 3% SDS, 10% glycerol, and 5% 2-mercaptoethanol. The cell lysates were electrophoresed and transferred to a PVDF membrane (Immobilon, Millipore, Bedford, MA). The membranes were probed with antibodies to GATA-1 (N6), GATA-2 (H-116), or PU.1 (T-21), all purchased from Santa Cruz Biotechnology (Santa Cruz, CA), followed by staining with HRP-conjugated antirat or antirabbit antibodies (Zymed, San Francisco, CA). Antibody staining was visualized using the enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia, Buckinghamshire, United Kingdom).
RT-PCR
Total RNA was purified using a RNeasy Mini Kit (Qiagen, Valencia, CA), and cDNA synthesis was carried out using the Thermoscript reverse transcription-polymerase chain reaction (RT-PCR) system (Gibco BRL, Rockville, MD) with 1 μg total RNA. cDNA samples were serially diluted by 3-fold and PCRs were subsequently performed. Primer sequences (5′→3′) were: Gata2 sense: cgg aat tcg aca cac cac ccg ata ccc acc tat; Gata2 antisense: cgg aat tcg cct acg cca tgg cag tca cca tgc t; PU.1 sense: tgg aag ggt ttt ccc tca cc; PU.1 antisense: tgc tgt cct tca tgt cgc cg; Gapdh sense: ggg tgg agc caa acg ggt cat c; Gapdh antisense: gcc agt gag ctt ccc gtt cag c.
Chromatin immunoprecipitation
For chromatin cross-linking, cells were treated with 1% formaldehyde for 10 minutes at 37°C and washed twice with PBS containing 1 mM PMSF. The cells were lysed in 50 mM Tris-HCl (pH 8.1) buffer containing 1% SDS, 10 mM EDTA, and 1 mM PMSF, and sonicated to shear the gDNA. The lysates were preincubated with protein-G Sepharose (Amersham Pharmacia) for 30 minutes with agitation at 4°C. The unbound fraction was collected and anti-GATA-2 antibody (H-116) or antiacetylated histone H4 antibody (Upstate Biotechnology, Lake Placid, NY) was added to the lysates. The protein-DNA complexes bound to the antibodies were precipitated with protein-G Sepharose, and the DNA was eluted by adding NaCl for 6 hours at 65°C followed by proteinase K treatment. The isolated DNA was recovered using the QIAEX II DNA purification system (Qiagen), and PCR analysis of the isolated DNA was carried out. The primer sequences for the PU.1 promoter region were: PU.1 sense: cag ctg tgg tac tta gct gg; PU.1 antisense: ctt gct gaa gta gtc cac tg.40
Reporter assay
The PU.1 genomic region containing -627 to +435 (transcriptional initiation site was referred to +1) was amplified from gDNA by PCR. The PCR product was cloned into pBluescript and DNA sequence was confirmed by DNA sequencer (ABI, Foster City, CA). The PU.1 promoter region (-627 to +150) was inserted into a promoterless luciferase vector, pGL3-basic (Promega, Madison, WI). The reporter construct and pRL-TK carrying control Renilla luciferase gene were cotransfected with PU.1 expression vector (pEF-PU.1) and GATA-2 expression vector (pOPI3-GATA-2) into CV-1 cells by calcium phosphate method as previously described.24 Transcriptional activities were measured by dual luciferase reporter kit (Promega) and transfection efficiency was normalized by RL-TK activity.
Results
Induction of megakaryocyte differentiation by GATA-2
In the course of hematopoietic differentiation from ES cells on the OP9 stromal layer, multipotential definitive hematopoietic progenitors appear at day 5 and continue to differentiate to various hematopoietic lineages in the absence of exogenous cytokines.37,38 The time course of this differentiation recapitulates early hematopoietic development. All hematopoietic cells at a specific time point are at the same stage of differentiation because differentiation is initiated only once and progresses synchronously.
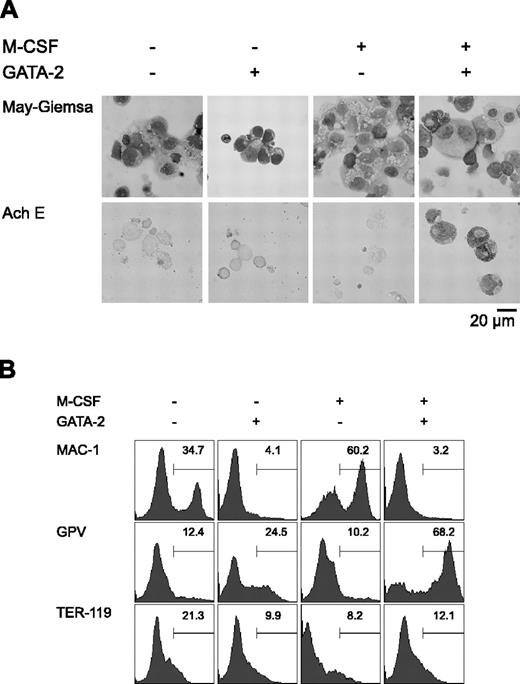
Initially, we carried out a morphologic analysis of day 9 differentiation-induced cells with or without overexpression of GATA-2 from days 5 to 9 (Figure 1A). In the absence of M-CSF, overexpression of GATA-2 did not significantly alter the morphology of the cells. In the presence of M-CSF, the majority of the cells were morphologically identifiable macrophages. Overexpression of GATA-2 from day 5 in the presence of M-CSF inhibited macrophage differentiation, and AchE+ megakaryocytic cells appeared. These morphologic changes corresponded closely to the expression of macrophage, megakaryocytic, and erythroid cell lineage markers (Mac-1, GPV, and TER-119, respectively; Figure 1B).
Redirection of differentiation from macrophage to megakaryocytic lineages induced by GATA-2. (A) May-Giemsa and AchE staining of cells at day 9. (B) Surface expression patterns of Mac-1, GPV, and TER-119 at day 9. Addition of M-CSF and induction of GATA-2 expression occurred at the times indicated.
Redirection of differentiation from macrophage to megakaryocytic lineages induced by GATA-2. (A) May-Giemsa and AchE staining of cells at day 9. (B) Surface expression patterns of Mac-1, GPV, and TER-119 at day 9. Addition of M-CSF and induction of GATA-2 expression occurred at the times indicated.
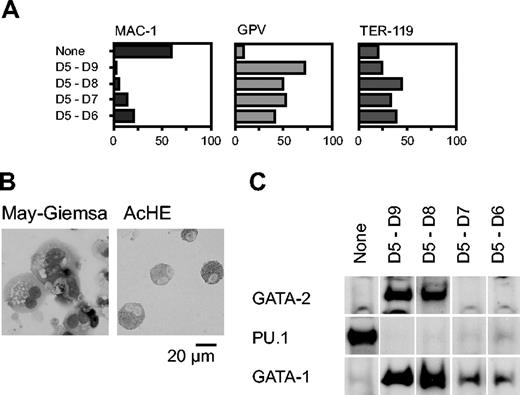
We next analyzed the effect of Gata2 when the gene was overexpressed for shorter periods, commencing with day 5. Percentages of cells positive for Mac-1, GPV, and TER-119 at day 9 are shown in Figure 2A. GPV expression analysis and AchE staining showed that differentiation to megakaryocytes occurred even when expression was induced only for 1 or 2 days (D5-D6 and D5-D7, respectively; Figure 2A-B). The effect of transient GATA-2 expression on the levels of GATA-1, GATA-2, and PU.1 proteins was analyzed by Western blotting (Figure 2C). When Gata2 was not overexpressed, neither GATA-2 nor GATA-1 was detectable, but PU.1 was detected, reflecting macrophage differentiation. In contrast, when GATA-2 was expressed from days 5 to 8 or 5 to 9, the expression pattern was completely altered: GATA-2 and GATA-1 were detectable but PU.1 was undetectable. When GATA-2 was expressed for shorter periods, from days 5 to 6 or 5 to 7, GATA-2 expression was undetectable at day 9. Thus, transient GATA-2 expression down-regulated PU.1 expression and up-regulated GATA-1 expression concomitantly.
Induction of erythroid differentiation by GATA-2
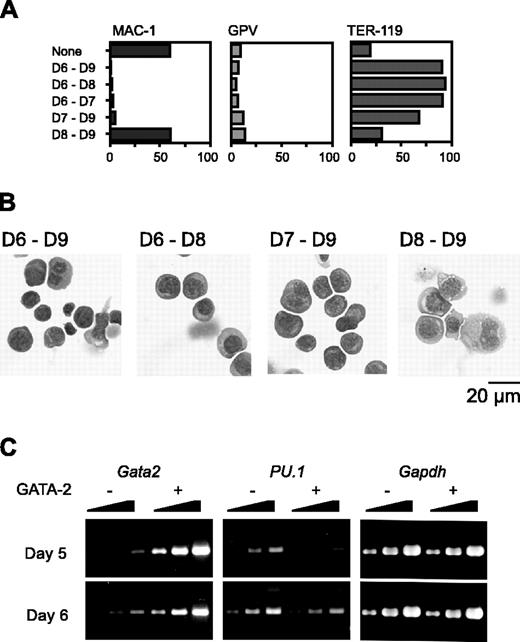
We next varied the day on which Gata2 expression was initiated. Because hematopoietic differentiation occurs only once and proceeds in a strictly synchronous manner, cells at different time points have different differentiation statuses in our experimental system. When Gata2 expression commenced on day 6 or 7, Mac-1 expression was suppressed. Expression of the megakaryocyte marker GPV was not up-regulated (Figure 3A); instead, the expression of the erythroid marker TER-119 was very high. Morphologic examination confirmed that erythroid differentiation had occurred (Figure 3B).
RT-PCR analysis showed that the PU.1 expression pattern was quite different when GATA-2 expression began at day 5 versus day 6. Expression levels of PU.1 at 1 day after initiation of Gata2 expression are shown in Figure 3C. When Gata2 expression was induced at day 5, PU.1 expression was significantly reduced to about 10% of the control. This decrease of PU.1 expression was presumably due to the down-regulation of PU.1 expression and was not attributable to the differentiation to macrophages because 1 day was not enough for the differentiation to macrophages. However, when Gata2 expression was induced at day 6, no significant alteration of PU.1 expression was detected. These data suggest that reduced or continuous expression of PU.1 promotes lineage skewing to megakaryocytes or erythroid cells, respectively.
Lineage determination from megakaryocyte to erythroid lineage by PU.1
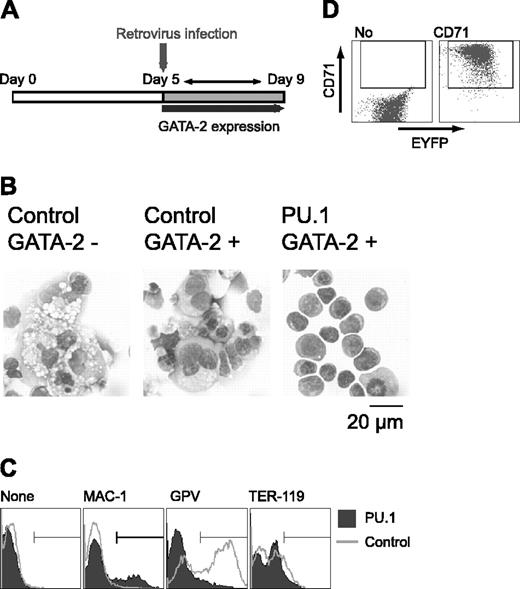
To clarify the effect of PU.1 expression on cell differentiation, Gata2 expression was induced at day 5, and PU.1 gene expression was enforced by retroviral infection at day 5 after induction in the presence of M-CSF. Retroviral infection served to prevent the down-regulation of PU.1 expression as described in Figure 3C (Figure 4A). Control EYFP retrovirus-infected cells differentiated megakaryocytes. In contrast, PU.1 retrovirus-infected cells exhibited erythroid morphology, like that of proerythroblasts and early basophilic erythroblasts (Figure 4B). GPV expression was detected in cells infected with the control retrovirus, but it was abolished by the enforced expression of PU.1. When the PU.1 retrovirus was transfected and no GATA-2 expression was induced, PU.1 had no effect on differentiation (data not shown), showing that the effect of PU.1 on ES cell lineage redirection from macrophage to erythroid cells results from its cooperative interaction with GATA-2.
Effects of GATA-2 expression on differentiation of hematopoietic cells and expression of transcription factors. GATA-2 expression commenced on day 5 and continued for the durations shown (A,C). (A) Percentages of Mac-1+, GPV+, and TER-119+ cells at day 9. Three independent experiments were carried out. Similar results were obtained from the experiments and representative data are shown. (B) May-Giemsa and AchE staining of cells at day 9. (C) GATA-2 was expressed from days 5 to 7. Western blotting analyses of GATA-2, PU.1, and GATA-1 proteins at day 9.
Effects of GATA-2 expression on differentiation of hematopoietic cells and expression of transcription factors. GATA-2 expression commenced on day 5 and continued for the durations shown (A,C). (A) Percentages of Mac-1+, GPV+, and TER-119+ cells at day 9. Three independent experiments were carried out. Similar results were obtained from the experiments and representative data are shown. (B) May-Giemsa and AchE staining of cells at day 9. (C) GATA-2 was expressed from days 5 to 7. Western blotting analyses of GATA-2, PU.1, and GATA-1 proteins at day 9.
Effects of GATA-2 expression commencing at different stages of differentiation on hematopoietic cells and expression of transcription factors. The duration of GATA-2 expression was as indicated (A-B). (A) Percentages of Mac-1+, GPV+, and TER-119+ cells at day 9. Three independent experiments were performed. Essentially similar results were obtained from the experiments and representative data are shown. (B) May-Giemsa staining of cells at day 9. (C) RT-PCR analyses of Gata2 and PU.1 1 day after induction of GATA-2 expression. Serially diluted cDNAs (1, 1:3, and 1:9) were used for PCR. GATA-2 expression was induced on day 5 or 6 as indicated.
Effects of GATA-2 expression commencing at different stages of differentiation on hematopoietic cells and expression of transcription factors. The duration of GATA-2 expression was as indicated (A-B). (A) Percentages of Mac-1+, GPV+, and TER-119+ cells at day 9. Three independent experiments were performed. Essentially similar results were obtained from the experiments and representative data are shown. (B) May-Giemsa staining of cells at day 9. (C) RT-PCR analyses of Gata2 and PU.1 1 day after induction of GATA-2 expression. Serially diluted cDNAs (1, 1:3, and 1:9) were used for PCR. GATA-2 expression was induced on day 5 or 6 as indicated.
Although cells expressing PU.1 and GATA-2 exhibited erythroid morphology, TER-119 expression was not detectable (Figure 4C). And the vast majority of the cells expressed neither Mac-1 nor GPV. PU.1 has been reported to interfere with GATA-1 function and inhibit erythroid differentiation.26 Because overexpression of PU.1 could have inhibited terminal erythroid differentiation, we analyzed the expression of the transferrin receptor CD71 in these cells. Although CD71 is not a specific marker for erythroid cells, strong CD71 expression is detected only in immature erythroid cells41 and not in megakaryocytes (data not shown). As shown in Figure 4D, the cells infected with PU.1 retrovirus expressed CD71 at very high levels, indicating that these cells were immature erythroid cells. Thus, PU.1 appears to be a key transcription factor in promoting the differentiation redirection to megakaryocytic or erythroid lineages in this experimental system.
Effects of histone deacetylase inhibitor on lineage determination and PU.1 expression
The effects of GATA-2 expression on PU.1 expression levels differed when expression was induced on day 5 versus day 6, as shown in Figure 3C. This result could have arisen from the different epigenetic status of the PU.1 locus at different stages of differentiation. Therefore, to analyze whether DNA methylation or histone modification influenced differentiation redirection, we carried out experiments using 5-azacytidine (5-AC), a DNA methylation inhibitor, and TSA, a histone deacetylase inhibitor.
Gata2 expression was induced on day 5 in the presence of M-CSF and in the presence or absence of 5-AC or TSA. Although 5-AC did not influence megakaryocytic differentiation (data not shown), TSA caused the differentiation to redirection from megakaryocytes to erythroid cells. GPV expression by Gata2 was abolished, and TER-119 expression was up-regulated (Figure 5A). In addition, TSA-treated cells exhibited erythroid morphology (Figure 5B). Expression of PU.1 was examined in the absence or presence of TSA after induction of Gata2 overexpression at day 5 (Figure 5C). PU.1 expression was significantly reduced at day 6 in the absence, but not the presence, of TSA. Meanwhile, expression levels of GATA-1 and Gata2 were not altered by the addition of TSA (Figure 5C). Taken together with our PU.1 overexpression data, these results suggest that PU.1 expression levels play a crucial role in determining megakaryocyte versus erythroid cell lineage commitments.
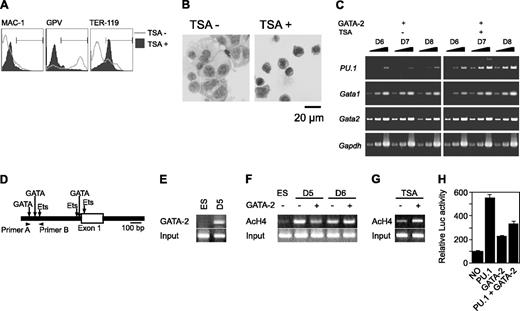
We next examined the GATA-2 binding and the acetylation status of histone H4 on the PU.1 promoter region using chromatin immunoprecipitation (ChIP) analysis. The murine PU.1 locus contains multiple GATA consensus sequences juxtaposed with Ets-binding sites at the promoter region (Figure 5D). We found that GATA-2 was bound to the site 8 hours after enforced expression at day 5 (Figure 5E). Subsequent to Gata2 induction at day 5, acetylated histone H4 at the PU.1 promoter region was reduced within 8 hours (Figure 5F). This reduced acetylation was reversed by the addition of TSA (Figure 5G). In contrast, acetylation was enhanced slightly when GATA-2 expression was induced at day 6 (Figure 5F). To analyze the effects of GATA-2 and PU.1 on the PU.1 promoter, we carried out the reporter expression analysis using the luciferase reporter gene containing approximately 0.7 kb of the PU.1 gene. PU.1 and GATA-2 increased the transcriptional activity by 5- and 2-fold, respectively. However, GATA-2 significantly reduced the transcriptional activation of PU.1.
Effects of retroviral expression of PU.1 on megakaryocytic differentiation in the presence of GATA-2. (A) Schematic diagram of the experiment. On day 5, cells were infected with control EYFP or PU.1-IRES-EYFP retrovirus, and expression of GATA-2 was induced. EYFP+ cells were sorted and examined on day 9. (B) May-Giemsa staining of the cells. Infected retroviruses and the expression of GATA-2 are as indicated. (C) Surface expression patterns of Mac-1, GPV, and TER-119. (D) Expression of CD71 in PU.1-IRES-EYFP-infected cells at day 9. Monoclonal antibodies used for staining are indicated (C-D).
Effects of retroviral expression of PU.1 on megakaryocytic differentiation in the presence of GATA-2. (A) Schematic diagram of the experiment. On day 5, cells were infected with control EYFP or PU.1-IRES-EYFP retrovirus, and expression of GATA-2 was induced. EYFP+ cells were sorted and examined on day 9. (B) May-Giemsa staining of the cells. Infected retroviruses and the expression of GATA-2 are as indicated. (C) Surface expression patterns of Mac-1, GPV, and TER-119. (D) Expression of CD71 in PU.1-IRES-EYFP-infected cells at day 9. Monoclonal antibodies used for staining are indicated (C-D).
Effects of GATA-2 expression on epigenetic modification and expression of PU.1. (A-C) On day 5, TSA was added and GATA-2 expression was induced. (A) Surface expression patterns of Mac-1, GPV, and TER-119 at day 9. (B) May-Giemsa staining at day 9. (C) RT-PCR analysis of PU.1, Gata1, and Gata2 expression at days 6 to 8. Serially diluted cDNAs (1, 1:3, and 1:9) were used for PCR. (D) Schematic PU.1 genomic locus and the primers used for ChIP analyses. GATA-2 expression was induced at day 5 or 6 as indicated, and ChIP analyses were carried out 8 hours later. (E) ChIP analysis using anti-GATA-2 antibody. (F) ChIP analysis of ES cells and differentiated cells using antiacetylated histone H4 antibody. (G) Effects of TSA on the amount of acetylated histone H4. GATA-2 expression commenced on day 5 and ChIP analyses were carried out 8 hours later. (H) Suppression of PU.1 promoter activity by GATA-2. The data are shown as the mean ± SD of 6 samples.
Effects of GATA-2 expression on epigenetic modification and expression of PU.1. (A-C) On day 5, TSA was added and GATA-2 expression was induced. (A) Surface expression patterns of Mac-1, GPV, and TER-119 at day 9. (B) May-Giemsa staining at day 9. (C) RT-PCR analysis of PU.1, Gata1, and Gata2 expression at days 6 to 8. Serially diluted cDNAs (1, 1:3, and 1:9) were used for PCR. (D) Schematic PU.1 genomic locus and the primers used for ChIP analyses. GATA-2 expression was induced at day 5 or 6 as indicated, and ChIP analyses were carried out 8 hours later. (E) ChIP analysis using anti-GATA-2 antibody. (F) ChIP analysis of ES cells and differentiated cells using antiacetylated histone H4 antibody. (G) Effects of TSA on the amount of acetylated histone H4. GATA-2 expression commenced on day 5 and ChIP analyses were carried out 8 hours later. (H) Suppression of PU.1 promoter activity by GATA-2. The data are shown as the mean ± SD of 6 samples.
Discussion
In this study, we examined the biologic function of GATA-2 in ES cells using conditional gene expression in combination with an OP9 differentiation induction system. Conditional expression of GATA-2 induced the cell differentiation redirection from macrophages to megakaryocytes and erythroid cells. Specific lineage determination was dependent on the timing of GATA-2 expression and was presumably controlled by the expression of another hematopoietic transcription factor, PU.1. Additionally, PU.1 expression was governed by epigenetic modification of the gene by GATA-2.
Dependence of GATA-2 function on the cell differentiation context
During the course of development of macrophages from mouse ES cells, enforced expression of Gata2 commencing on day 5 or day 6 of differentiation induction caused cell fate determination to megakaryocytes or erythrocytes, respectively. Considering the starting time points of conditional GATA-2 expression, 2 possibilities may explain this differentiation disturbance. One is the lineage switch from macrophages to megakaryocytic or erythroid lineages and the other is the preferential differentiation to erythro-megakaryocytic lineages rather than macrophages. In any case, the redirection to megakaryocytes and erythroid cells occurred dependent on the commencement of GATA-2 expression. It was unlikely that apoptotic cell death of macrophages was the cause of preferential differentiation to erythro-megakaryocytic lineages because no significant increase of apoptotic cell death was induced by GATA-2 expression. These data demonstrate that the biologic function of GATA-2 is dependent on the differentiation context of cells.
Previous reports concerning the effects of GATA-2 on hematopoietic cell differentiation and proliferation have yielded dissimilar results. Overexpression of Gata2 in hematopoietic stem cells was reported to block their proliferation.42 On the other hand, we previously reported that conditional overexpression of Gata2 in hematopoietic progenitor cells enhanced cell proliferation.24 These results suggest that GATA-2 has different functions in hematopoietic stem and progenitor cells. Recently, the effects of Gata2 gene dosage on generation and expansion of hematopoietic stem cells in the aorta-gonad-mesonephros (AGM) region, in which hematopoietic stem cells develop, and the other hematopoietic organs (yolk sac, fetal liver, and adult bone marrow) were examined.43 The results indicated that GATA-2 has different roles in the AGM region and other organs during ontogeny. Thus, GATA-2 functions in multiple ways in hematopoietic development and differentiation.
We previously demonstrated that the biologic function of GATA-2 opposes that of a GATA-2/estrogen receptor (ER) chimera.24 GATA-2 and GATA-2/ER exhibited different binding to other hematopoietic transcription factors, including c-Myb and PU.1. GATA-2 and GATA-2/ER also had different characteristics with regard to transcriptional interference with c-Myb and PU.1. These different characteristics presumably underlie the divergent biologic functions of GATA-2 and its chimeric GATA-2/ER counterpart. In other words, different GATA-2 complexes exhibit different biologic functions. In support of this idea, GATA-2 has been shown to interact with various nuclear factors, including p300/CBP,44 HDAC,45 c-Myb,24 PU.1,24,46 Fog-1,47 RARα,48 and PLZF.49 Taken together, these observations indicate that the diverse functions of GATA-2 are brought about by different transcriptional complexes, dependent on the cell differentiation context.
PU.1 expression and redirection of lineage distortion
Among the various hematopoietic transcription factors and cofactors, PU.1 is strongly implicated in redirecting the differentiation to megakaryocytes and erythroid cells in cooperation with GATA-2. Reciprocal inhibition between GATA factors and PU.1 has been reported.26-30 However, the functional interrelationships of GATA factors and PU.1 are complex. They function synergistically on some occasions but antagonistically on other occasions. The interplay between PU.1 and GATA-2 promotes mast cell differentiation,32 but an inhibitory interaction between GATA-1 and PU.1 determines the cell lineage commitment of myelo-erythroid progenitors.33
In our experiments, macrophage differentiation was blocked by conditional expression of GATA-2. The inhibitory effect persisted even when GATA-2 was expressed at the late stage of macrophage differentiation. Although the expression of Mac-1 was not reduced, morphologic analysis exhibited the inhibition of macrophage differentiation (Figure 3 A-B), when Gata2 was expressed since day 8. This inhibition may be explained by inhibition of the transcriptional activity of PU.1 by GATA-2 because high expression of PU.1 is essential for macrophage differentiation. However, our Western blotting data demonstrate that transcriptional interference is not the sole cause of differentiation inhibition by GATA-2. When Gata2 was expressed from days 5 to 9, PU.1 expression was severely inhibited, and up-regulation of GATA-1 was observed on day 9. This expression pattern was observed even when Gata2 was expressed for only 1 day (day 5). Concomitant expression of GATA-2 and PU.1 in macrophages appears to induce rapid degradation of PU.1 and subsequent inhibition of differentiation.
The erythroid and megakaryocytic lineages are derived from common bipotential erythrocyte-megakaryocyte progenitors and are believed to be closely related.21 On the other hand, macrophages belong to the myeloid lineage, which is distinct from the erythro-megakaryocytic lineages from the point of cell differentiation. GATA-2 overexpression redirected the differentiation from the myeloid to the erythro-megakaryocytic lineage, and GATA-2-modulated expression of PU.1 determined subsequent differentiation to erythroid versus megakaryocytic lineages in cooperation with GATA-2. Low levels of PU.1 led to differentiation to megakaryocytes, and high levels of PU.1 led to differentiation to erythroid cells. This effect mirrors the function of PU.1 in differentiation to macrophage and B cells in that high or low expression levels of PU.1 are essential for macrophage or B cell differentiation, respectively.50 Fine regulation of PU.1 expression appears to be essential in determining the fate of hematopoietic cells.
Effects of epigenetic status
The roles of transcription factors are often examined by means of overexpression studies. However, our present results serve as a warning that the results of simple overexpression experiments should not be overinterpreted because cell differentiation conditions may also play a role. In the present experiment, expression of PU.1 was affected by conditional expression of Gata2, and the results of Gata2 overexpression should be interpreted as the combinatorial effect of expression of both GATA-2 and PU.1. Two mutually nonexclusive explanations are possible for the complex biologic effects of GATA-2. One is simple transcriptional interference between GATA-2 and PU.1 as shown in Figure 5H. Although GATA-2 repressed the autoregulation of PU.1 promoter by the PU.1, this simple transcriptional interference could not be sufficient for the cell differentiation context-dependent function of GATA-2 and some epigenetic mechanisms should be involved.51
DNA methylation and histone modification are the major mechanisms for epigenetic control of gene regulation. The methylation status of CpG sites flanking exon 1 of the PU.1 gene was reported to be important for transcriptional regulation of PU.1.52 However, DNA methylation does not seem to be involved in erythro-megakaryocytic differentiation determination or PU.1 gene regulation by GATA-2 in our study because 5-AC did not influence differentiation or gene expression.
In contrast, histone acetylation modification plays important roles in both differentiation and gene expression, as revealed by the experiment using TSA. TSA inhibited the down-regulation of PU.1 expression caused by Gata2 expression at day 5, and it induced erythroid, rather than megakaryocytic, differentiation. Taken with the results of retroviral PU.1 overexpression, the correspondence between these results appears significant. Presumably, inhibition of PU.1 down-regulation induced by TSA was the cause of erythroid differentiation. When Gata2 was expressed from day 5, inhibition of PU.1 expression was caused by binding of GATA-2 to the PU.1 promoter region, where it presumably recruited the HDAC corepressor complex, which could be associated with GATA-2,45 and brought about deacetylation of histone H4. On the other hand, histone H4 acetylation was not affected by Gata2 expression commencing on day 6. The reason for the differences in the effects of GATA-2 on histone H4 acetylation, depending on whether Gata2 expression commenced on day 5 or day 6, remains elusive. However, a difference in susceptibility to epigenetic modification could be the key to the context-dependent functions of GATA-2.
Prepublished online as Blood First Edition Paper, October 27, 2005; DOI 10.1182/blood-2005-06-2527.
Supported in part by grants from the Ministry of Education, Science, Sports, and Culture; the Support Program for Technology Development on the Basis of Academic Findings (NEDO); the Uehara Memorial Foundation; and the Osaka Cancer Foundation.
K.K., M.T., J.Z., H.Y., A.S., D.S., H.U., and E.S. performed research on both molecular and cell biology; T.N. designed this research and wrote this manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs T. Kitamura (Tokyo University), T. Oikawa (Sasaki Institute), and T. Kina (Kyoto University) and Morinaga Milk Co for providing materials. We also thank Ms M. Ikeuchi, Y. Fujita, T. Asada, and A. Mizokami for their assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal