Infection with human immunodeficiency virus type 1 (HIV-1) is characterized by dysfunction of HIV-1–specific T cells. To control the virus, antigen-loaded dendritic cells (DCs) might be useful to boost and broaden HIV-specific T-cell responses. In the present study, monocyte-derived DCs from nontreated HIV-1–seropositive patients were electroporated with codon-optimized (“humanized”) mRNA encoding consensus HxB-2 (hHXB-2) Gag protein. These DCs elicited a strong HIV-1 Gag-specific interferon-γ (IFN-γ) response by an HLA-A2–restricted CD8+ T-cell line. Moreover, hHXB-2 gag mRNA-electroporated DCs also triggered IFN-γ secretion by autologous peripheral blood mononuclear cells (PBMCs), CD4+ T cells, and CD8+ T cells from all patients tested. Next, a novel strategy was developed using autologous virus sequences. Significant specific IFN-γ T-cell responses were induced in all patients tested by DCs electroporated with patients' autologous polymerase chain reaction (PCR)–amplified and in vitro–transcribed proviral and plasma viral mRNA encoding either Gag or Env. The stimulatory effect was seen on PBMCs, CD8+ T cells, and CD4+ T cells, demonstrating both major histocompatibility complex (MHC) class I and MHC class II antigen presentation. Moreover, a significant interleukin-2 (IL-2) T-cell response was induced by DCs electroporated with hHxB-2 or proviral gag mRNA. These findings open a major perspective for the development of patient-specific immunotherapy for HIV-1 disease.
Introduction
During the past 20 years, human immunodeficiency virus (HIV) infection has become a pandemic, with more than 40 million people infected and already more than 20 million deaths from acquired immunodeficiency syndrome (AIDS).1 Data from exposed uninfected and from long-term nonprogressors strongly suggest that both HIV-specific CD8+ and CD4+ T-cell functions are essential for protective immunity.2 In all infected persons, HIV-specific CD8+ cytotoxic T-lymphocyte (CTL) immune responses are activated following primary infection, and they can be quite effective in lowering the viral load. However, HIV tends to escape from immune control by mutation of critical epitopes. T-cell responses against the new epitopes are induced, but HIV escapes again. During this process of viral adaptation, all the previous variants are stored as proviral DNA,3 constituting a “latent reservoir.” In the chronic progressive stage of the disease, CD4+ and CD8+ T cells become progressively more dysfunctional, and CTLs against new and previously targeted epitopes do not fully mature to effector stage,4 resulting in increasing viral load and clinical immunodeficiency. Highly active antiretroviral therapy (HAART) can lower the plasma viral load to undetectable levels but is unable to eliminate the proviral latent reservoir.5-9 Clearly, if we want to develop an immunotherapy to complement the effect of HAART, it is not sufficient that the T-cell responses against the circulating virus are enhanced. To minimize the risk of escape, it is equally important that immune responses against the entire latent reservoir are activated.10
Dendritic cells (DCs) are professional antigen-capturing and -presenting cells that are able to stimulate effective immune responses both in vitro and in vivo.11-13 In the context of DC-based immunotherapy, it has been shown by Larsson et al14 that DCs passively pulsed with immunogenic HIV peptides or loaded with HIV proteins using recombinant vaccinia virus specifically stimulate HIV-specific interferon-γ (IFN-γ)–producing T cells in vitro. Other teams have also successfully used DCs expressing HIV antigens (eg, pulsed with peptides, transduced by different vectors, or loaded with apoptotic infected cells) to stimulate memory15-18 or even primary19-24 CD8+ T-cell responses in vitro. In vivo, simian immunodeficiency virus (SIV)–specific CD8+ and CD4+ T-cell responses were induced in monkeys using SIV antigen-expressing DCs.22 Importantly, viral control was obtained in HIV-infected humanized severe combined immunodeficient (SCID) mice23,24 and in SIV-infected rhesus macaques after DC immunization.25 Finally, Lu et al26 showed that DCs pulsed with chemically inactivated autologous virus specifically stimulated HIV-specific immune responses in vitro and in vivo in HIV-1–seropositive patients.
During the past few years, we27-29 and others12,30-32 have developed a novel antigen-loading strategy for DCs based on transfection of antigen-encoding mRNA. This technology represents a safe (eg, noninfectious) and clinically applicable protocol with many advantages over peptide pulsing and viral transduction.33-34 Following on our previously published data on the use of mRNA-electroporated DCs, we now demonstrate in the first part of this study that DCs cultured from HIV-1–seropositive patients specifically stimulate HIV-1 Gag-specific autologous T cells in vitro after electroporation with a codon-optimized mRNA encoding a consensus HxB2 HIV-1 Gag protein. In the second part, we develop a strategy aiming to stimulate a broad CTL response ideally directed against the totality of the “autologous” variants. To this end, DCs from HIV-1–seropositive patients were electroporated with mRNA derived from autologous proviral and plasma viral sequences and used to specifically trigger effector memory T cells.
Patients, materials, and methods
Study population
Peripheral blood samples (100 mL) were obtained from 28 HIV-1–seropositive patients (designated as P1 to P28) recruited at the Clinical Department of the Institute of Tropical Medicine of Antwerp according to institutional guidelines and after obtaining informed consent. Demographic and clinical information (designated number, age, sex, years of documented HIV-1 seropositivity, therapy, CD4+ T-cell count, viral load, and presence or absence of HLA*0201 major histocompatibility antigen) of these patients is summarized in Table 1. Only antiviral therapy–naive patients with a CD4+ T-cell count above 0.40 × 109/L (400/μL) were included. Peripheral blood samples (buffy coats) from 5 healthy HIV-1–seronegative controls (designated as C1 to C5) were provided by the Antwerp Blood Transfusion Center (Red Cross Flanders, Belgium). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque gradient separation (Amersham Biosciences, Freiburg, Germany).
Demographic and clinical information of HIV-1–seropositive individuals
Patients . | Age, y . | Sex . | Time HIV positive, y . | CD4+ cells/μL . | HIV viral load, copies/mL . | HLA*0201 . |
|---|---|---|---|---|---|---|
| P1 | 55 | M | 5 | 558 | < 50 | – |
| P2 | 36 | M | 2 | 537 | < 400 | + |
| P3 | 41 | M | 1 | 572 | 4 180 | – |
| P4 | 36 | M | 1 | 571 | 52 900 | + |
| P5 | 37 | M | 3 | 803 | 9 740 | + |
| P6 | 49 | M | 9 | 669 | 37 800 | – |
| P7 | 44 | M | 3 | 414 | 159 000 | – |
| P8 | 40 | M | 4 | 539 | 4 710 | + |
| P9 | 43 | M | 3 | 755 | 2 790 | + |
| P10 | 56 | F | 3 | 621 | < 400 | – |
| P11 | 45 | M | 1 | 871 | 25 400 | + |
| P12 | 30 | M | 4 | 495 | 30 500 | + |
| P13 | 33 | M | 1 | 900 | < 500 | – |
| P14 | 40 | M | 1 | 543 | 18 000 | – |
| P15 | 23 | F | 3 | 874 | 1 300 | + |
| P16 | 55 | F | 2 | 532 | 25 100 | – |
| P17 | 36 | M | 1 | 497 | 19 700 | – |
| P18 | 35 | M | 8 | 546 | < 50 | + |
| P19 | 46 | M | 1 | 1552 | 26 500 | – |
| P20 | 40 | M | 0.5 | 679 | 461 000 | + |
| P21 | 51 | M | 0.5 | 542 | 140 000 | – |
| P22 | 39 | M | 2 | 763 | 30 000 | – |
| P23 | 29 | M | 0.25 | 470 | 215 000 | – |
| P24 | 28 | M | 1 | 610 | 28 800 | – |
| P25 | 27 | M | 2 | 486 | 57 600 | + |
| P26 | 42 | M | 3 | 390 | 43 500 | – |
| P27 | 30 | M | 1 | 575 | 24 400 | + |
| P28 | 31 | M | 2 | 547 | 77 000 | + |
Patients . | Age, y . | Sex . | Time HIV positive, y . | CD4+ cells/μL . | HIV viral load, copies/mL . | HLA*0201 . |
|---|---|---|---|---|---|---|
| P1 | 55 | M | 5 | 558 | < 50 | – |
| P2 | 36 | M | 2 | 537 | < 400 | + |
| P3 | 41 | M | 1 | 572 | 4 180 | – |
| P4 | 36 | M | 1 | 571 | 52 900 | + |
| P5 | 37 | M | 3 | 803 | 9 740 | + |
| P6 | 49 | M | 9 | 669 | 37 800 | – |
| P7 | 44 | M | 3 | 414 | 159 000 | – |
| P8 | 40 | M | 4 | 539 | 4 710 | + |
| P9 | 43 | M | 3 | 755 | 2 790 | + |
| P10 | 56 | F | 3 | 621 | < 400 | – |
| P11 | 45 | M | 1 | 871 | 25 400 | + |
| P12 | 30 | M | 4 | 495 | 30 500 | + |
| P13 | 33 | M | 1 | 900 | < 500 | – |
| P14 | 40 | M | 1 | 543 | 18 000 | – |
| P15 | 23 | F | 3 | 874 | 1 300 | + |
| P16 | 55 | F | 2 | 532 | 25 100 | – |
| P17 | 36 | M | 1 | 497 | 19 700 | – |
| P18 | 35 | M | 8 | 546 | < 50 | + |
| P19 | 46 | M | 1 | 1552 | 26 500 | – |
| P20 | 40 | M | 0.5 | 679 | 461 000 | + |
| P21 | 51 | M | 0.5 | 542 | 140 000 | – |
| P22 | 39 | M | 2 | 763 | 30 000 | – |
| P23 | 29 | M | 0.25 | 470 | 215 000 | – |
| P24 | 28 | M | 1 | 610 | 28 800 | – |
| P25 | 27 | M | 2 | 486 | 57 600 | + |
| P26 | 42 | M | 3 | 390 | 43 500 | – |
| P27 | 30 | M | 1 | 575 | 24 400 | + |
| P28 | 31 | M | 2 | 547 | 77 000 | + |
Gag-specific CD8+ T-cell line
An HLA-A2–restricted CD8+ T-cell line directed against the consensus SLYNTVATL (77 to 85) peptide of the HIV-1 Gag protein was generated by the Antigen Presentation by Dendritic Cell Group in Paris.18 One day before use, a frozen vial of the CD8+ T-cell line was thawed, and cells were allowed to recover for 24 hours at 37°C in RPMI 1640 medium (Cambrex, Verviers, Belgium) supplemented with 5% pooled human serum (PHS) and 1 U/mL interleukin-2 (IL-2) (Roche Molecular Biochemicals, Mannheim, Germany).
In vitro generation of monocyte-derived DCs
Monocytes were isolated from PBMCs by magnetic isolation using CD14 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. About 8 × 106 to 10 × 106 monocytes were obtained starting from 100 × 106 PBMCs with purity levels of more than 90%. Monocyte-derived immature DCs (iDCs) were generated as described before35 in 2.5% PHS. In some experiments, maturation of iDCs was induced on day 6 of culture using a mixture of IL-1, IL-6, prostaglandin E2 (PGE2), and tumor necrosis factor-α (TNF-α) as described.36 Immunophenotyping of DCs was performed as described before.37 The monocyte-depleted PBMCs were cryopreserved.36
Plasmid DNA constructs
The pGEM4Z/h-gag/A64 plasmid (pGEMhgag), generated by the Laboratory of Physiology and Immunology in Brussels, was used to prepare a humanized (codon-optimized) mRNA encoding the HxB-2 HIV-1 Gag protein (hHxB-2 gag).38 The original humanized cDNA was provided by B.V.
Amplification of autologous gag or env mRNA derived from PBMC proviral sequences
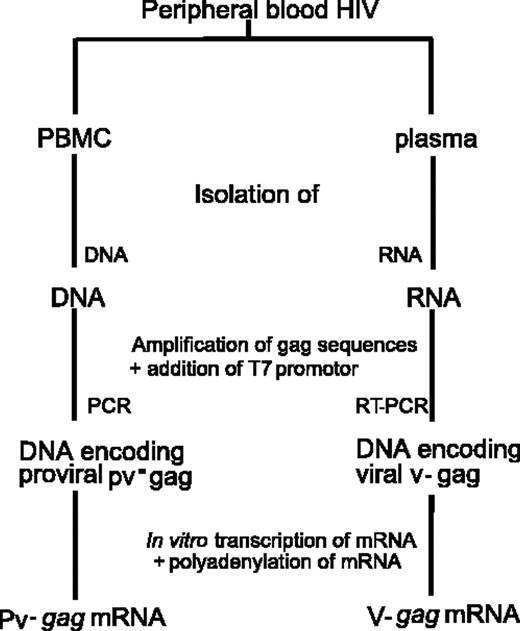
For DNA extraction, 10 × 106 PBMCs from HIV-1–seropositive blood donors (Figure 1) were resuspended in 200 μL lysis buffer (Merck, Darmstadt, Germany) and treated with 100 μg/mL proteinase K (Qiagen, Chatsworth, CA) overnight at 56°C, followed by phenol chloroform extraction to purify the DNA. Proviral gag (pv-gag) DNA sequences were amplified by nested polymerase chain reaction (PCR) using the GeneAmp XL PCR kit (Applied Biosystems, Lennik, Belgium). In the first-round PCR, 4 primers (final concentration 0.25 μM) were used. Next, 5 μL aliquots of the amplified products were subjected to a second-round PCR with an inner sense primer containing the 23-nucleotide T7 bacteriophage promoter sequence at the 5′ end (the underlined sequence represents the T7 promoter sequence): forward “H1G787-T7” primer (5′-TAATACGACTCACTATAGGGAGGATGGGTGCGAGAGCGTCAGTATT-3′) and reverse “H1G2307” primer (5′-ATAAGCGGCCGCTTATTGTGACAATGGGTCGTTGCCA-3′). The cycle conditions for the first round PCR were 1 cycle of 94°C for 120 seconds, 16 cycles of 94°C for 15 seconds, 50°C for 30 seconds, 68°C for 90 seconds, followed by 12 cycles of 94°C for 15 seconds, 68°C for 90 seconds, with an increment of 5 seconds per cycle and a final extension step of 68°C for 7 minutes. For the second-round PCR reaction, the conditions were the same except that the annealing temperature was increased to 55°C. Amplified DNA products were purified using a QiaQuick PCR purification kit (Qiagen). Amplified DNA was then in vitro transcribed using a T7 Message Machine in vitro transcription kit followed by polyadenylation using a poly(A) Tailing kit (Ambion, Cambridgeshire, United Kingdom), according to manufacturer's instructions (Figure 1). RNA was stored at -80°C in small aliquots. For the amplification of proviral env (pv-env) (gp120) the same protocol was followed with an inner sense primer “T7-JFES” (5′-TAATACGACTCACTATAGGGAGGAGAGCAGAAGACAGTGGCAATG-3′) and reverse primer “ED12 Stop” (5′-AGGCTAAGTGCTTCCTGCTGCTCCCAAGAACCCAAGGA-3′).
Amplification of autologous gag or env mRNA derived from plasma virus
Total RNA was extracted out of plasma virus from HIV-1–seropositive patients (Figure 1) using the QIAamp viral RNA mini kit (Qiagen), followed by reverse transcription using Expand RT (Roche Molecular Biochemicals), as described by manufacturer's instructions. The cDNA obtained was then used for nested PCR (Expand High Fidelity PCR System; Roche Molecular Biochemicals) using the same primers as described in “Amplification of autologous gag or env mRNA derived from PBMC proviral sequences,” resulting in full gag or env cDNA fragments containing a T7 promoter sequence at the 5′ end. Afterward, the amplified DNA was in vitro transcribed using a T7 Message Machine in vitro transcription kit followed by polyadenylation using a poly(A) Tailing kit (Ambion). The resulting viral-gag (v-gag) or viral-env (v-env) (gp120) mRNA was stored at -80°C in small aliquots.
Amplification procedure for autologous proviral pv-gag and pv-env and plasma viral v-gag and v-env mRNA.
Amplification procedure for autologous proviral pv-gag and pv-env and plasma viral v-gag and v-env mRNA.
DC electroporation
Electroporation of in vitro–transcribed mRNA was performed as described previously.28 After electroporation, cells were either directly used for DC/T-cell coculture experiments or further cultured alone for in vitro characterization experiments in fresh complete medium (including 2.5% PHS and cytokines for DC culture/maturation).
Peptide pulsing of DCs
An HxB-2 Gag peptide pool (NIH AIDS Research & Reference Reagent Program, Germantown, MD) was used for the detection of Gag-specific effector memory T cells. Pulsing of the peptide was performed as described previously.27 Briefly, 106 iDCs were washed twice with RPMI 1640 medium (Cambrex) and incubated for 1 hour with 20 μg/mL peptide in serum-free medium supplemented with 2.5 μg/mL β2-microglobulin (Sigma, Bornem, Belgium). Afterward, DCs were matured, washed, and used as stimulators for autologous T cells.
Intracellular staining for HIV-1 Gag protein
Immature DCs (iDCs) were electroporated with hHxB-2 gag, autologous pv-gag, or v-gag mRNA and analyzed for intracellular Gag expression 24 hours after electroporation. mRNA-electroporated and mock-electroporated (control) cells were washed and fixed for 15 minutes with fixation reagent A (Serotec, Oxford, United Kingdom) at room temperature. Next, cells were permeabilized and stained for 30 minutes at 4°C in reagent B (Serotec) with 5 μL anti-Gag HIV-1 RD-1–conjugated monoclonal antibody (Beckman Coulter, Fullerton, France) and analyzed by flow cytometry (FacsCalibur; Becton Dickinson, Erembodegem, Belgium).
Gag p24 ELISA
Immature DCs, mock electroporated or electroporated with various gag mRNA, were cultured in RPMI 1640 medium supplemented with 2.5% PHS at 2 × 105 cells per 200 μL in 96-well-plates. After 24 hours, supernatant samples were collected, and p24 Gag secretion was measured by ELISA.39
Stimulation of a Gag-specific CD8+ T-cell line using mRNA-electroporated DCs
Immature DCs from HIV-1–seropositive patients were electroporated on day 6 of culture with hHxB-2 gag mRNA, pv-gag mRNA, or v-gag mRNA and matured for 24 hours. As stimulators, mock-electroporated (control) and mDCs from the same donor were used. A total of 20 000 mDCs were cocultured in quadruplicate with 6 × 104 CD8+ HIV-1 Gag-specific T cells in RPMI medium supplemented with 5% PHS for 24 hours in an anti–IFN-γ antibody–coated enzyme-linked immunospot (ELISPOT) plate (Diaclone, Besançon, France). As positive and negative controls, T cells were cocultured with, respectively, 1 μM HIV-1 Gag77-85 peptide (SLYNTVATL) or with 1 μM Tax11-19 peptide (LLFGYPVYV), a negative control peptide derived from human T-cell leukemia lymphoma virus type I (HTLV-I).18
Triggering of autologous HIV-1 Gag effector memory-specific T cells
Immature DCs were electroporated with hHxB-2 gag mRNA, pv-gag mRNA, or v-gag mRNA on day 6 of the culture. In some experiments, these iDCs were directly used for stimulation of autologous PBMCs, purified CD4+ T cells (using CD4 microbeads according to manufacturer's instructions; Miltenyi Biotec), or purified CD8+ T cells (using CD8 microbeads; Miltenyi Biotec). In other experiments, the electroporated iDCs were matured for 24 hours before stimulation of autologous “effector cells” (ie, either PBMCs, CD4+ T cells, or CD8+ T cells). Briefly, 5 × 104 mRNA- or mock-electroporated DCs were cocultured with 5 × 105 effector cells in an anti–IFN-γ antibody–coated ELISPOT plate (Diaclone) in RPMI 1640 medium supplemented with 2.5% PHS.14 Spot numbers were counted using an automated ELISPOT reader (AID, Strassberg, Germany).
Triggering of autologous HIV-1 Env effector memory-specific T cells
Immature DCs were electroporated with pv-env or v-env mRNA on day 6 of the culture and matured. Afterward, env mRNA-electroporated mDCs were cocultured with autologous PBMCs, CD4+ cells, or CD8+ T cells in an IFN-γ antibody–coated ELISPOT plate (Diaclone) in RPMI 1640 medium supplemented with 2.5% PHS.
Heteroduplex mobility assay (HMA) and sequencing
HMA was largely performed as described before.40-41 The PCR template used for the in vitro transcription to mRNA was cloned in TA vector (Invitrogen, Leek, The Netherlands). Recombinant plasmids were used to transform competent Escherichia coli cells according to manufacturer's protocol, and transformants were grown on ampicillin plates. Afterward, the positive clones were amplified by PCR, and a nested PCR was used to amplify a region of 750 base pairs. Then heteroduplex molecules were obtained by mixing 5 μL PCR product with 1.1 μL annealing buffer (1 M NaCl, 100 mM Tris-HCl, 20 mM EDTA). The DNA fragments were denatured at 94°C for 2 minutes and reannealed by rapid cooling on wet ice. Electrophoresis was performed on a 5% polyacrylamide gel that included 20% urea at 250 V for 2.5 hours. Detection of heteroduplexes was done by staining with ethidium bromide and visualization under UV light. Clonal analysis of the PCR products was performed for 4 patients. Only those clones that showed a different migration pattern on HMA were further analyzed. Plasmid DNA containing the gag or env inserts were purified using Qiagen PCR purification kit (Qiagen). The electrophorogram was analyzed by Chromas,42 and the autologous proviral or viral gag or env sequences were aligned using CLUSTALW.43 The diversity was calculated by BioEdit.44
Statistics
Because individual responses were sometimes widely variable, we summarized data using the geometric mean (GM) and the standard error of the mean (SE). Two sets of data were compared using the unpaired t test. Correlations were calculated using the Spearman rank correlation test. Differences or correlations between sets of data were considered significant if P equaled .05.
Results
Monocyte-derived DCs from HIV-1–seropositive individuals can be efficiently loaded with antigen by electroporation with hHxB-2 gag mRNA
Cultured iDCs from HIV-1–seropositive patients were CD13+, CD14-, HLA-DR+, DC-SIGN+, CD80+, CD86+, and CD83+/- (data not shown). These iDCs were electroporated with codon-optimized hHxB-2 gag mRNA. Twenty-four hours later, both intracellular Gag protein expression (Figure 2A) and secretion of HIV-1 Gag protein (Figure 2B) were consistently demonstrated, whereas mock-electroporated DCs failed to express or secrete measurable p24 protein. Next, we investigated to what extent these hHxB-2 gag mRNA-electroporated DCs could process and present antigenic epitopes to an HLA-A*0201–restricted HIV-1 Gag peptide-specific CD8+ T-cell line using an IFN-γ ELISPOT assay as a read-out. As shown in Figure 2C, IFN-γ production by the HIV-1 Gag peptide-specific CD8+ T-cell line was induced after stimulation with the HIV-1 Gag77-85 peptide, but also by hHxB-2 gag mRNA-electroporated DCs, indicating efficient major histocompatibility complex (MHC) class I–restricted peptide processing and presentation starting from consensus gag mRNA.
Electroporation of monocyte-derived DCs from HIV-1–seropositive individuals with various gag mRNA results in efficient expression, secretion, and presentation of Gag protein and peptides. (A) iDCs from an HIV-1–seropositive individual (patient P1) were mock electroporated or electroporated with hHxB-2 gag mRNA and further matured for 24 hours. Afterward they were analyzed for intracellular Gag protein. Histogram overlay showing anti-Gag fluorescence of mock-electroporated control mDCs (open histogram) and hHxB-2 gag mRNA-electroporated mDCs (filled histogram). The example shown was representative for patients P1, P3, P4, and P10. (B) Gag protein secretion was measured in the supernatant 24 hours after electroporation and maturation. This experiment was performed and is shown for patients P1, P3, P5, P7, and P8. mDC-mock indicates mock-electroporated mDCs; mDC-hHxB-2 gag, mDCs electroporated with hHxB-2 gag mRNA. (C) iDCs from patient P15 were electroporated with hHxB-2 gag mRNA or mock electroporated, further matured for 24 hours, and the antigen-processing and -presenting capacity was investigated in IFN-γ ELISPOT using a CD8+ Gag-specific T-cell line. The HIV-1 Gag77-85 peptide was used as a positive control and the HTLV-I Tax11-19 peptide as a negative control. The numbers are GMs ± SE (quadruplicate wells) of IFN-γ SFCs per 60 000 responder cells. (D) iDCs from an HIV-1–seropositive individual (patient P14) were mock electroporated or electroporated with autologous proviral gag mRNA and further matured for 24 hours. Afterward they were analyzed for intracellular Gag protein. Histogram overlay showing anti-Gag fluorescence of mock-electroporated control mDCs (open histogram) and pv-gag mRNA-electroporated mDCs (filled histogram). (E) Gag p24 protein secretion was measured by ELISA in the supernatant of pv-gag mRNA-electroporated mDCs (DC-pv-gag) and v-gag mRNA-electroporated mDCs (DC-v-gag). This experiment was performed and is shown for patients P9, P12, P13, P21, and P22. Gag p24 protein secretion of mock-electroporated mDCs was below background (data not shown). (F) iDCs from an HIV-1–seropositive patient (P12) were mock electroporated, electroporated with autologous pv-gag or v-gag mRNA, and further matured. After 24 hours, these mDCs were cocultured in an ELISPOT assay with a Gag peptide-specific CD8+ T-cell line. Peptide controls and SFCs are the same as in panel C.
Electroporation of monocyte-derived DCs from HIV-1–seropositive individuals with various gag mRNA results in efficient expression, secretion, and presentation of Gag protein and peptides. (A) iDCs from an HIV-1–seropositive individual (patient P1) were mock electroporated or electroporated with hHxB-2 gag mRNA and further matured for 24 hours. Afterward they were analyzed for intracellular Gag protein. Histogram overlay showing anti-Gag fluorescence of mock-electroporated control mDCs (open histogram) and hHxB-2 gag mRNA-electroporated mDCs (filled histogram). The example shown was representative for patients P1, P3, P4, and P10. (B) Gag protein secretion was measured in the supernatant 24 hours after electroporation and maturation. This experiment was performed and is shown for patients P1, P3, P5, P7, and P8. mDC-mock indicates mock-electroporated mDCs; mDC-hHxB-2 gag, mDCs electroporated with hHxB-2 gag mRNA. (C) iDCs from patient P15 were electroporated with hHxB-2 gag mRNA or mock electroporated, further matured for 24 hours, and the antigen-processing and -presenting capacity was investigated in IFN-γ ELISPOT using a CD8+ Gag-specific T-cell line. The HIV-1 Gag77-85 peptide was used as a positive control and the HTLV-I Tax11-19 peptide as a negative control. The numbers are GMs ± SE (quadruplicate wells) of IFN-γ SFCs per 60 000 responder cells. (D) iDCs from an HIV-1–seropositive individual (patient P14) were mock electroporated or electroporated with autologous proviral gag mRNA and further matured for 24 hours. Afterward they were analyzed for intracellular Gag protein. Histogram overlay showing anti-Gag fluorescence of mock-electroporated control mDCs (open histogram) and pv-gag mRNA-electroporated mDCs (filled histogram). (E) Gag p24 protein secretion was measured by ELISA in the supernatant of pv-gag mRNA-electroporated mDCs (DC-pv-gag) and v-gag mRNA-electroporated mDCs (DC-v-gag). This experiment was performed and is shown for patients P9, P12, P13, P21, and P22. Gag p24 protein secretion of mock-electroporated mDCs was below background (data not shown). (F) iDCs from an HIV-1–seropositive patient (P12) were mock electroporated, electroporated with autologous pv-gag or v-gag mRNA, and further matured. After 24 hours, these mDCs were cocultured in an ELISPOT assay with a Gag peptide-specific CD8+ T-cell line. Peptide controls and SFCs are the same as in panel C.
Stimulatory capacity of hHxB-2 gag mRNA-electroporated DCs from HIV-1–seropositive individuals toward autologous T cells
To investigate if DCs from HIV-1–seropositive individuals could efficiently trigger autologous effector memory T cells, we used hHxB-2 gag mRNA-electroporated iDCs in 14 independent experiments. As compared with mock-electroporated iDCs, hHxB-2 gag mRNA-electroporated iDCs induced significantly (P < .005) more IFN-γ spot-forming cells (SFCs) in all cocultures with autologous PBMCs, CD4+ cells, or CD8+ T cells (Table 2). As compared with whole PBMCs and on a per million cells basis, CD8+ T cells were similarly responsive and CD4+ T cells tended to be less responsive. We did not find any significant correlation between viral load and IFN-γ SFCs (r = 0.244, P = .40) or between CD4+ T-cell count and IFN-γ SFCs (r = 0.147, P = .62).
Stimulatory capacity of codon-optimized HxB-2 HIV-1 gag mRNA-electroporated DCs from HIV-1–seropositive individuals
Patients or controls* . | PBMCs . | . | CD4+ T cells . | . | CD8+ T cells . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | DC-mock . | DC-h-gag . | DC-mock . | DC-h-gag . | DC-mock . | DC-h-gag . | |||
| P1 | 5 ± 3 | 561 ± 33* | 6 ± 5 | 279 ± 33 | 0 ± 0 | 428 ± 13 | |||
| P2 | 34 ± 16 | 1258 ± 48 | — | — | — | — | |||
| P3 | 12 ± 6 | 473 ± 56 | 8 ± 8 | 122 ± 26 | — | — | |||
| P4 | 438 ± 85 | 1024 ± 178 | — | — | — | — | |||
| P5 | 35 ± 12 | 707 ± 125 | 55 ± 19 | 631 ± 36 | 29 ± 2 | 1248 ± 203 | |||
| P6 | 173 ± 33 | 762 ± 69 | — | — | — | — | |||
| P7 | 28 ± 8 | 172 ± 70 | 16 ± 5 | 82 ± 9 | — | — | |||
| P8 | 86 ± 11 | 262 ± 26 | — | — | — | — | |||
| P9 | |||||||||
| iDCs | 2 ± 1 | 923 ± 67 | — | — | 2 ± 0 | 2400 ± 94 | |||
| mDCs | 11 ± 16 | 1012 ± 51 | — | — | 48 ± 23 | 2668 ± 55 | |||
| P10 | |||||||||
| iDCs | 4 ± 1 | 816 ± 162 | — | — | — | — | |||
| mDCs | 5 ± 4 | 1115 ± 205 | — | — | — | — | |||
| P11 | |||||||||
| iDCs | 6 ± 0 | 98 ± 13 | — | — | — | — | |||
| mDCs | 8 ± 7 | 398 ± 61 | — | — | 5 ± 6 | 468 ± 136 | |||
| P26 | |||||||||
| iDCs | 35 ± 1 | 994 ± 12 | — | — | — | — | |||
| mDCs | 156 ± 83 | 1465 ± 375 | — | — | — | — | |||
| P27 | |||||||||
| iDCs | 84 ± 14 | 418 ± 10 | — | — | — | — | |||
| mDCs | 88 ± 20 | 676 ± 14 | — | — | — | — | |||
| P28 | |||||||||
| iDCs | 24 ± 6 | 72 ± 16 | — | — | — | — | |||
| mDCs | 6 ± 2 | 312 ± 44 | — | — | — | — | |||
| GM ± SE | |||||||||
| iDCs | 11 ± 31 | 201 ± 100 | 14 ± 11 | 205 ± 125 | 8 ± 11 | 1086 ± 572 | |||
| mDCs | 20 ± 30 | 427 ± 237 | |||||||
| C1 | 3 ± 2 | 3 ± 3 | 39 ± 10 | 38 ± 9 | — | — | |||
| C2 | 11 ± 2 | 22 ± 9 | 6 ± 4 | 6 ± 4 | 48 ± 10 | 76 ± 14 | |||
| C3 | |||||||||
| iDCs | 37 ± 3 | 41 ± 6 | — | — | — | — | |||
| mDCs | 29 ± 5 | 30 ± 6 | — | — | — | — | |||
Patients or controls* . | PBMCs . | . | CD4+ T cells . | . | CD8+ T cells . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | DC-mock . | DC-h-gag . | DC-mock . | DC-h-gag . | DC-mock . | DC-h-gag . | |||
| P1 | 5 ± 3 | 561 ± 33* | 6 ± 5 | 279 ± 33 | 0 ± 0 | 428 ± 13 | |||
| P2 | 34 ± 16 | 1258 ± 48 | — | — | — | — | |||
| P3 | 12 ± 6 | 473 ± 56 | 8 ± 8 | 122 ± 26 | — | — | |||
| P4 | 438 ± 85 | 1024 ± 178 | — | — | — | — | |||
| P5 | 35 ± 12 | 707 ± 125 | 55 ± 19 | 631 ± 36 | 29 ± 2 | 1248 ± 203 | |||
| P6 | 173 ± 33 | 762 ± 69 | — | — | — | — | |||
| P7 | 28 ± 8 | 172 ± 70 | 16 ± 5 | 82 ± 9 | — | — | |||
| P8 | 86 ± 11 | 262 ± 26 | — | — | — | — | |||
| P9 | |||||||||
| iDCs | 2 ± 1 | 923 ± 67 | — | — | 2 ± 0 | 2400 ± 94 | |||
| mDCs | 11 ± 16 | 1012 ± 51 | — | — | 48 ± 23 | 2668 ± 55 | |||
| P10 | |||||||||
| iDCs | 4 ± 1 | 816 ± 162 | — | — | — | — | |||
| mDCs | 5 ± 4 | 1115 ± 205 | — | — | — | — | |||
| P11 | |||||||||
| iDCs | 6 ± 0 | 98 ± 13 | — | — | — | — | |||
| mDCs | 8 ± 7 | 398 ± 61 | — | — | 5 ± 6 | 468 ± 136 | |||
| P26 | |||||||||
| iDCs | 35 ± 1 | 994 ± 12 | — | — | — | — | |||
| mDCs | 156 ± 83 | 1465 ± 375 | — | — | — | — | |||
| P27 | |||||||||
| iDCs | 84 ± 14 | 418 ± 10 | — | — | — | — | |||
| mDCs | 88 ± 20 | 676 ± 14 | — | — | — | — | |||
| P28 | |||||||||
| iDCs | 24 ± 6 | 72 ± 16 | — | — | — | — | |||
| mDCs | 6 ± 2 | 312 ± 44 | — | — | — | — | |||
| GM ± SE | |||||||||
| iDCs | 11 ± 31 | 201 ± 100 | 14 ± 11 | 205 ± 125 | 8 ± 11 | 1086 ± 572 | |||
| mDCs | 20 ± 30 | 427 ± 237 | |||||||
| C1 | 3 ± 2 | 3 ± 3 | 39 ± 10 | 38 ± 9 | — | — | |||
| C2 | 11 ± 2 | 22 ± 9 | 6 ± 4 | 6 ± 4 | 48 ± 10 | 76 ± 14 | |||
| C3 | |||||||||
| iDCs | 37 ± 3 | 41 ± 6 | — | — | — | — | |||
| mDCs | 29 ± 5 | 30 ± 6 | — | — | — | — | |||
All samples were iDCs unless otherwise noted.
Monocyte-derived immature dendritic cells (iDCs) or monocyte-derived mature dendritic cells (mDCs) from HIV-1–seropositive patients were electroporated with hHxB-2 gag mRNA (DC-h-gag) or mock electroporated (DC-mock) and cocultured with autologous PBMCs or purified CD4+ or CD8+ T cells in ELISPOT plates for 24 hours. The resulting levels of IFN-γ SFCs per 106 responder cells (triplicate wells) are shown as geometric mean ± standard error.
— indicates not done.
The same experiment was done with dendritic cells from HIV-1–seronegative controls (C1-C3) to assay the specificity of the response. The combined results of the different seropositive donors are presented as the overall GM ± SE
In 6 experiments (P9 to P11 and P26 to P28), mRNA-electroporated iDCs were allowed to mature before use as stimulators in the ELISPOT assay. As shown in Table 2, mDCs triggered significantly more IFN-γ SFCs per 106 T cells (427 ± 237) as compared with iDCs (201 ± 100 SFCs; P = .003). Therefore, in most of the following experiments iDCs were first electroporated and then matured before being used as stimulators.
To examine whether the observed immune response is a reflection of preexisting HIV-1 Gag-specific T-cell pool in HIV-1–seropositive individuals, similar experiments were done in HIV-1–seronegative control blood donors (C1 to C3). Low numbers of SFCs were consistently found regardless of whether mock- or hHxB-2 gag-electroporated iDCs or mDCs were used.
DCs from HIV-1–seropositive individuals are efficiently loaded by electroporation with autologous gag mRNA and present a Gag epitope toward a specific CD8+ T-cell line
Whereas the codon-optimized hHxB-2 gag mRNA encodes a consensus HIV-1 Gag protein, amplification of proviral gag (pv-gag) and viral gag (v-gag) provides the opportunity to obtain mRNA encoding autologous HIV-1 Gag proteins. The procedure to obtain pv-gag and v-gag mRNA is schematically represented in Figure 1. As compared with hHxB-2 gag mRNA-electroporated DCs, translation efficiency of pv-gag and v-gag was lower as measured with intracellular staining (Figure 2D), and HIV-1 Gag p24 secretion also tended to be lower (Figure 2E).
Next we investigated to what extent these pv-gag or v-gag mRNA-electroporated DCs could process antigen and present the MHC class I–restricted Gag77-85 epitope to the peptide-specific CD8+ T-cell line. As shown in Figure 2F, both pv-gag and v-gag mRNA-electroporated DCs induced similar numbers of IFN-γ SFCs in the T-cell line, indicating antigenic presentation of Gag77-85.
DCs electroporated with autologous gag mRNA efficiently stimulate autologous T cells
Viral and/or proviral gag mRNA were prepared from 5 HIV-1–seropositive patients and used to electroporate autologous DCs. The DCs were matured and cocultured with autologous PBMCs, CD4+ cells, or CD8+ T cells during a 24-hour ELISPOT assay. As shown in Table 3, IFN-γ secretion was efficiently triggered in all cases (P < .05 as compared with mock-electroporated DCs in PBMCs, CD4+ cells, and CD8+ T cells). Similar to hHxB-2 gag, autologous v-gag or pv-gag induced fewer SFCs per 106 CD4+ T cells as compared with CD8+ T cells (P < .05), and no significant correlation could be found between IFN-γ SFCs and either viral load or peripheral blood CD4+ counts. The amount of SFCs induced by pv-gag or v-gag mRNA-electroporated DCs was comparable.
Stimulatory capacity of autologous gag mRNA-electroporated mature DCs from HIV-1–seropositive individuals
Cells and patient no. . | mDC-mock . | mDC-pv-gag . | mDC-v-gag . |
|---|---|---|---|
| PBMCs | |||
| P14 | 4 ± 2 | 236 ± 60 | 384 ± 96 |
| P15 | 4 ± 9 | 58 ± 10 | 252 ± 54 |
| P20 | 34 ± 10 | 424 ± 234 | 356 ± 56 |
| P21 | 70 ± 55 | 906 ± 112 | 290 ± 28 |
| P22 | 78 ± 18 | 116 ± 38 | 98 ± 44 |
| GM ± SE | 12 ± 9 | 225 ± 194 | 225 ± 55 |
| CD4+ T cells | |||
| P15 | 306 ± 26 | 158 ± 62 | 686 ± 18 |
| P20 | 48 ± 0 | 536 ± 0 | 328 ± 3 |
| P21 | 58 ± 0 | 280 ± 3 | 224 ± 4 |
| P22 | 36 ± 14 | 160 ± 42 | 128 ± 12 |
| GM ± SE | 74 ± 65 | 248 ± 89 | 283 ± 122 |
| CD8+ T cells | |||
| P15 | 204 ± 66 | 662 ± 72 | 908 ± 78 |
| P20 | 40 ± 8 | 1008 ± 32 | 604 ± 34 |
| P21 | 133 ± 0 | 1307 ± 3 | 1107 ± 4 |
| P22 | 130 ± 26 | 576 ± 104 | 604 ± 16 |
| GM ± SE | 109 ± 34 | 842 ± 168 | 778 ± 123 |
Cells and patient no. . | mDC-mock . | mDC-pv-gag . | mDC-v-gag . |
|---|---|---|---|
| PBMCs | |||
| P14 | 4 ± 2 | 236 ± 60 | 384 ± 96 |
| P15 | 4 ± 9 | 58 ± 10 | 252 ± 54 |
| P20 | 34 ± 10 | 424 ± 234 | 356 ± 56 |
| P21 | 70 ± 55 | 906 ± 112 | 290 ± 28 |
| P22 | 78 ± 18 | 116 ± 38 | 98 ± 44 |
| GM ± SE | 12 ± 9 | 225 ± 194 | 225 ± 55 |
| CD4+ T cells | |||
| P15 | 306 ± 26 | 158 ± 62 | 686 ± 18 |
| P20 | 48 ± 0 | 536 ± 0 | 328 ± 3 |
| P21 | 58 ± 0 | 280 ± 3 | 224 ± 4 |
| P22 | 36 ± 14 | 160 ± 42 | 128 ± 12 |
| GM ± SE | 74 ± 65 | 248 ± 89 | 283 ± 122 |
| CD8+ T cells | |||
| P15 | 204 ± 66 | 662 ± 72 | 908 ± 78 |
| P20 | 40 ± 8 | 1008 ± 32 | 604 ± 34 |
| P21 | 133 ± 0 | 1307 ± 3 | 1107 ± 4 |
| P22 | 130 ± 26 | 576 ± 104 | 604 ± 16 |
| GM ± SE | 109 ± 34 | 842 ± 168 | 778 ± 123 |
Immature DCs were electroporated with autologous proviral gag (mDC-pv-gag), viral gag (mDC-v-gag) mRNA, or mock electroporated (mDC-mock) and matured. Afterward these mDCs were cocultured with autologous PBMCs, purified CD4+ cells, or CD8+ T cells in ELISPOT plates. The amount of IFN-γ SFCs per 106 responder cells (triplicate wells) is shown as individual GM ± SE. The results from patients P15, P20, P21, and P22 are complete for all parameters. For comparative reasons only, the results of these 4 patients were summarized as overall GM ± SE.
DCs electroporated with gag mRNA also trigger IL-2–secreting T cells
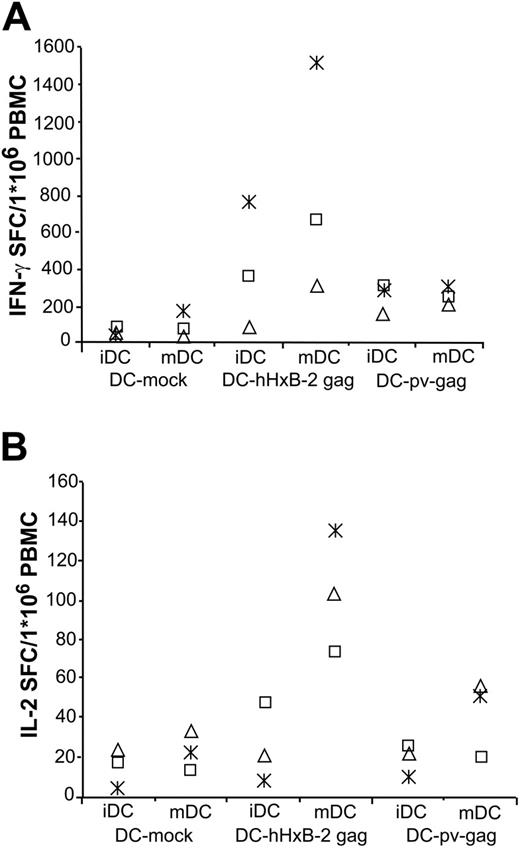
Because IFN-γ alone is not a good parameter for protection,45 we also measured IL-2 secretion. The same ELISPOT approach as described before was used. As shown in Figure 3, IFN-γ and IL-2 secretion could be triggered upon stimulation with hHxB-2 gag or pv-gag mRNA-electroporated DCs in cocultures with autologous PBMCs. The number of IL-2 SFCs was on average about 10 times less than that of IFN-γ SFCs. Significantly more IFN-γ (685 ± 364 SFCs for hHxB-2 gag and 249 ± 27 SFC for pv-gag) and IL-2 SFCs (101 ± 18 SFCs for hHxB-2 gag and 40 ± 11 SFCs for pv-gag) were induced in PMBCs stimulated with gag mRNA-electroporated and mDCs as compared with stimulation with mock-electroporated mDCs (47 ± 43 IFN-γ SFCs and 22 ± 6 IL-2 SFCs). For the IFN-γ SFCs, the statistical comparison yielded the following numbers: mDC-mock versus mDCs pv-gag: P < .01; mDC-mock versus mDC hHxB-2 gag: P < .05. For the IL-2 SFCs the P values were as follows: mDC-mock versus mDC pv-gag: P < .05; mDC-mock versus mDC hHxB-2 gag: P < .005. On the other hand, whereas iDCs significantly induced IFN-γ SFCs (iDC-mock versus iDC pv-gag: P < .01; iDC-mock versus iDC hHxB-2 gag: P < .05), they failed to significantly induce IL-2 SFCs (iDC-mock versus iDC pv-gag: P > .2; iDC-mock versus iDC hHxB-2 gag: P > .25). The GM ± SE of SFCs in the IFN-γ ELISPOT were the following: 49 ± 16 SFCs if mock-electroporated iDCs were used; 289 ± 199 SFCs and 238 ± 45 SFCs if hHxB-2 gag or pv-gag were used as stimulator cells. In the IL-2 assay, 13 ± 5 SFCs were measured if mock-electroporated iDCs were used and 20 ± 12 and 19 ± 5 if, respectively, hHxB-2 gag- and pv-gag–electroporated DCs were used as stimulator cells.
DCs electroporated with gag mRNA trigger autologous T cells to secrete IFN-γ and IL-2. (A) iDCs from an HIV-1–seropositive individual electroporated with hHxB-2 or proviral gag mRNA were used as stimulators for autologous PBMCs in IFN-γ ELISPOT assay or were further matured. Twenty-four hours later, mDCs were used as stimulators for autologous PBMCs during a 24-hour IFN-γ ELISPOT assay. (B) The same protocol was followed in parallel for the IL-2 ELISPOT assay. IL-2 SFCs were significantly induced in cultures with gag mRNA-electroporated mDCs in comparison with mock-electroporated mDCs. The difference between mock-electroporated iDCs and gag mRNA-electroporated iDCs was not significant (see “Stimulatory capacity of DCs electroporated with mRNA of autologous amplified env”). Both experiments were done for patients P26 (asterisk), P27 (□), and P28 (▵).
DCs electroporated with gag mRNA trigger autologous T cells to secrete IFN-γ and IL-2. (A) iDCs from an HIV-1–seropositive individual electroporated with hHxB-2 or proviral gag mRNA were used as stimulators for autologous PBMCs in IFN-γ ELISPOT assay or were further matured. Twenty-four hours later, mDCs were used as stimulators for autologous PBMCs during a 24-hour IFN-γ ELISPOT assay. (B) The same protocol was followed in parallel for the IL-2 ELISPOT assay. IL-2 SFCs were significantly induced in cultures with gag mRNA-electroporated mDCs in comparison with mock-electroporated mDCs. The difference between mock-electroporated iDCs and gag mRNA-electroporated iDCs was not significant (see “Stimulatory capacity of DCs electroporated with mRNA of autologous amplified env”). Both experiments were done for patients P26 (asterisk), P27 (□), and P28 (▵).
Stimulatory capacity of DCs electroporated with gag mRNA versus DCs pulsed with HxB-2 Gag peptide pool
To compare the mRNA approach with a standard antigen-loading strategy, peptide pulsing and mRNA electroporation were performed in parallel. In 3 experiments (patients P16, P17, and P19), DCs pulsed with the HxB-2 Gag peptide pool triggered more IFN-γ SFCs than DCs electroporated with hHxB-2 gag mRNA (Table 4), but this difference was significant for PBMCs (P < .05) only and not for CD4+ (P = .08) or CD8+ (P = .47) T cells. In 4 other experiments (patients P15, P20, P21, and P22), pv-gag or v-gag mRNA-electroporated DCs were compared with peptide pulsing. Peptide-pulsed DCs triggered more IFN-γ SFCs, the difference being significant for CD8+ T cells and CD4+ T cells (P < .05) but not for PBMCs (P = .09). In summary, HxB-2 Gag peptides generally induced higher SFC responses as compared with DCs transfected with gag mRNA derived from either consensus sequence or from autologous virus.
Stimulatory capacity of mDCs electroporated with gag mRNA versus DCs pulsed with HxB-2 Gag peptide pool
Patients or controls . | mDC-mock . | mDC-hHxB-2 gag . | mDC-pv-gag . | mDC-v-gag . | mDC-pulsed HxB-2 Gag . |
|---|---|---|---|---|---|
| IFN-γ SFCs per 106 PBMCs | |||||
| P16 | 6 ± 1 | 64 ± 18 | — | — | 742 ± 56 |
| P17 | 68 ± 26 | 482 ± 70 | — | — | 722 ± 56 |
| P19 | 67 ± 32 | 118 ± 62 | — | — | 740 ± 0 |
| GM ± SE | 30 ± 21 | 154 ± 31 | — | — | 735 ± 6 |
| P15 | 4 ± 8 | — | 58 ± 5 | 252 ± 54 | 774 ± 60 |
| P20 | 30 ± 16 | — | 620 ± 331 | 327 ± 79 | 2369 ± 81 |
| P21 | 94 ± 112 | — | 771 ± 101 | 219 ± 40 | 572 ± 240 |
| P22 | 78 ± 18 | — | 116 ± 38 | 98 ± 44 | 574 ± 120 |
| GM ± SE | 31 ± 21 | — | 238 ± 179 | 205 ± 48 | 881 ± 435 |
| IFN-γ SFCs per 106 CD4+ T cells | |||||
| P16 | 50 ± 10 | 156 ± 11 | — | — | 1154 ± 2 |
| P17 | 46 ± 4 | 56 ± 8 | — | — | 424 ± 64 |
| P19 | 30 ± 10 | 26 ± 4 | — | — | 186 ± 36 |
| GM ± SE | 41 ± 6 | 61 ± 39 | — | — | 450 ± 292 |
| P15 | 306 ± 26 | — | 158 ± 10 | 686 ± 18 | 1782 ± 224 |
| P20 | 48 ± 0 | — | 536 ± 0 | 328 ± 3 | 2642 ± 0 |
| P21 | 58 ± 0 | — | 280 ± 3 | 224 ± 4 | 672 ± 2 |
| P22 | 36 ± 14 | — | 160 ± 48 | 128 ± 12 | 356 ± 32 |
| GM ± SE | 74 ± 65 | — | 248 ± 89 | 283 ± 122 | 1030 ± 525 |
| IFN-γ SFCs per 106 CD8+ T cells | |||||
| P16 | 41 ± 4 | 253 ± 4 | — | — | 441 ± 15 |
| P17 | 120 ± 10 | 1210 ± 22 | — | — | 490 ± 270 |
| P19 | 26 ± 17 | 112 ± 4 | — | — | 720 ± 9 |
| GM ± SE | 50 ± 29 | 325 ± 345 | — | — | 538 ± 86 |
| P15 | 204 ± 66 | — | 662 ± 36 | 908 ± 72 | 1536 ± 28 |
| P20 | 40 ± 4 | — | 1018 ± 32 | 604 ± 34 | 2554 ± 36 |
| P21 | 133 ± 0 | — | 1307 ± 3 | 1107 ± 4 | 1251 ± 2 |
| P22 | 130 ± 26 | — | 576 ± 104 | 604 ± 16 | 1428 ± 32 |
| GM ± SE | 109 ± 34 | — | 844 ± 169 | 778 ± 123 | 1627 ± 293 |
| IFN-γ SFCs per 106 PBMCs | |||||
| C4 | 11 ± 4 | 17 ± 2 | — | — | 248 ± 13 |
| C5 | 22 ± 5 | 46 ± 8 | — | — | 291 ± 37 |
Patients or controls . | mDC-mock . | mDC-hHxB-2 gag . | mDC-pv-gag . | mDC-v-gag . | mDC-pulsed HxB-2 Gag . |
|---|---|---|---|---|---|
| IFN-γ SFCs per 106 PBMCs | |||||
| P16 | 6 ± 1 | 64 ± 18 | — | — | 742 ± 56 |
| P17 | 68 ± 26 | 482 ± 70 | — | — | 722 ± 56 |
| P19 | 67 ± 32 | 118 ± 62 | — | — | 740 ± 0 |
| GM ± SE | 30 ± 21 | 154 ± 31 | — | — | 735 ± 6 |
| P15 | 4 ± 8 | — | 58 ± 5 | 252 ± 54 | 774 ± 60 |
| P20 | 30 ± 16 | — | 620 ± 331 | 327 ± 79 | 2369 ± 81 |
| P21 | 94 ± 112 | — | 771 ± 101 | 219 ± 40 | 572 ± 240 |
| P22 | 78 ± 18 | — | 116 ± 38 | 98 ± 44 | 574 ± 120 |
| GM ± SE | 31 ± 21 | — | 238 ± 179 | 205 ± 48 | 881 ± 435 |
| IFN-γ SFCs per 106 CD4+ T cells | |||||
| P16 | 50 ± 10 | 156 ± 11 | — | — | 1154 ± 2 |
| P17 | 46 ± 4 | 56 ± 8 | — | — | 424 ± 64 |
| P19 | 30 ± 10 | 26 ± 4 | — | — | 186 ± 36 |
| GM ± SE | 41 ± 6 | 61 ± 39 | — | — | 450 ± 292 |
| P15 | 306 ± 26 | — | 158 ± 10 | 686 ± 18 | 1782 ± 224 |
| P20 | 48 ± 0 | — | 536 ± 0 | 328 ± 3 | 2642 ± 0 |
| P21 | 58 ± 0 | — | 280 ± 3 | 224 ± 4 | 672 ± 2 |
| P22 | 36 ± 14 | — | 160 ± 48 | 128 ± 12 | 356 ± 32 |
| GM ± SE | 74 ± 65 | — | 248 ± 89 | 283 ± 122 | 1030 ± 525 |
| IFN-γ SFCs per 106 CD8+ T cells | |||||
| P16 | 41 ± 4 | 253 ± 4 | — | — | 441 ± 15 |
| P17 | 120 ± 10 | 1210 ± 22 | — | — | 490 ± 270 |
| P19 | 26 ± 17 | 112 ± 4 | — | — | 720 ± 9 |
| GM ± SE | 50 ± 29 | 325 ± 345 | — | — | 538 ± 86 |
| P15 | 204 ± 66 | — | 662 ± 36 | 908 ± 72 | 1536 ± 28 |
| P20 | 40 ± 4 | — | 1018 ± 32 | 604 ± 34 | 2554 ± 36 |
| P21 | 133 ± 0 | — | 1307 ± 3 | 1107 ± 4 | 1251 ± 2 |
| P22 | 130 ± 26 | — | 576 ± 104 | 604 ± 16 | 1428 ± 32 |
| GM ± SE | 109 ± 34 | — | 844 ± 169 | 778 ± 123 | 1627 ± 293 |
| IFN-γ SFCs per 106 PBMCs | |||||
| C4 | 11 ± 4 | 17 ± 2 | — | — | 248 ± 13 |
| C5 | 22 ± 5 | 46 ± 8 | — | — | 291 ± 37 |
Immature DCs were pulsed with HxB-2 Gag peptide pool (mDC-pulsed HxB-2 Gag) or electroporated with codon-optimized gag (mDC-hHxB-2 Gag, autologous proviral (mDC-pv-gag), or viral (mDC-v-gag) gag mRNA or mock electroporated (mDC-mock). After maturation, these cells were cocultured with autologous PBMCs, purified CD4+ cells, or CD8+ T cells in ELISPOT plates. The amounts of IFN-γ SFCs per 106 responder cells (triplicate wells) are shown as GM ± SE. GM ± SE was calculated from the data of 2 groups of HIV-1—seropositive patients, where different comparisons were made: patients P16, P17, and P19 on one hand and patients P15, P20, P21, and P22 on the other hand.
Controls (C4 and C5) were HIV-1—seronegative individuals.
— indicates not done.
To examine whether the observed responses to peptides were HIV specific, similar experiments were performed in HIV-1–seronegative donors (C4 and C5). It is clear that DCs pulsed with HxB-2 peptide pool trigger a nonspecific stimulation of T cells from HIV-1–seronegative donors (Table 4), thus questioning if the peptide response in HIV-1 seropositives is entirely specific.
Stimulatory capacity of DCs electroporated with mRNA of autologous amplified env
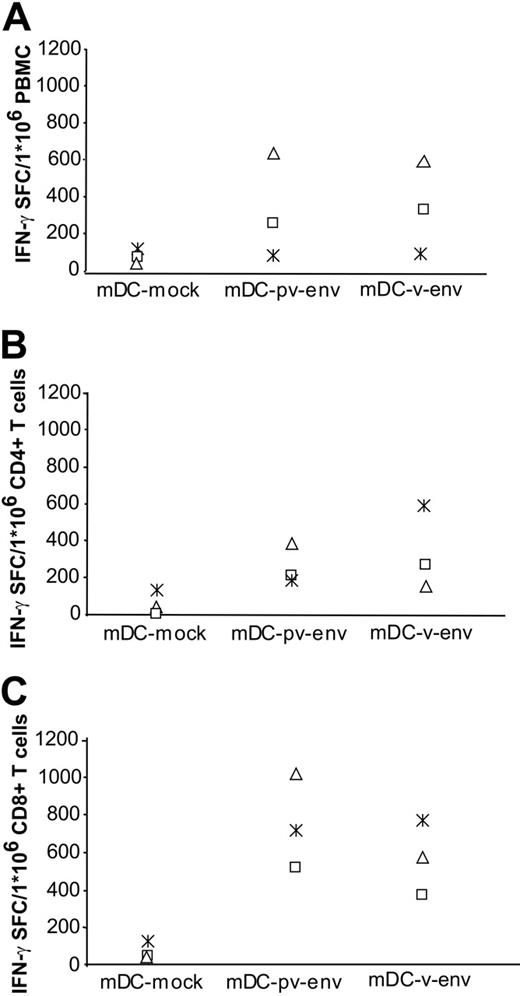
To show that mRNA coding for another HIV antigen can also efficiently be used for antigen presentation, we tested a similar strategy for pv-env and v-env mRNA electroporation. As shown in Figure 4, Env-specific effector memory T cells are significantly triggered by mDCs electroporated with pv-env (236 ± 163 SFCs for PBMCs as effectors; 269 ± 39 and 696 ± 140 SFCs for, respectively, CD4+ T cells and CD8+ T cells as effectors) or v-env mRNA (230 ± 132 SFCs for PBMCs as effectors; 271 ± 136 and 524 ± 100 SFCs for, respectively, CD4+ T cells and CD8+ T cells as effectors). The stimulatory effect of pv-env and v-env mRNA-electroporated DCs was evident in autologous PBMCs (mock versus pv-env, P = .05; mock versus v-env, P = .05) but also in CD4+ (mock versus pv-env, P = .007; mock versus v-env, P = .05) and CD8+ T cells (mock versus pv-env, P = .004; mock versus v-env, P = .004). The amount of IFN-γ SFCs triggered by pv-env mRNA-electroporated mDCs was not significantly different from that induced by v-env mRNA-electroporated mDCs (P = .4 for PBMCs as effectors; P = .34 and P = .17 if CD4+ or CD8+ T cells were used as effectors, respectively). Overall ELISPOT Env responses are in the same order of magnitude as the responses for Gag.
Sequencing
Sequence analysis was done as a first rough estimate of the heterogeneity within and between the proviral and viral forms of gag and env used in our strategy. PCR templates of gag (patients P12 and P14) or env (patients P24 and P25) from the plasma virus as well as from PBMC-derived DNA provirus were cloned and amplified. Eighteen to 44 clones per patient were obtained and prepared for heteroduplex mobility assay. Table 5 shows that 15% to 30% of env clones and approximately 50% of gag clones displayed a different mobility. Remarkably, it turned out that each of the viruses from these 4 randomly chosen patients belonged to 4 different subtypes. Analysis of sequence diversity within virus and provirus showed a diversity of 0.91% to 2.59% for gag and of 1.36% to 4.57% for env. Sequence diversity between viral and proviral sequences was always higher than within each of them separately. Importantly, both pv-gag and v-gag sequences from both patients tested showed mutations in the immunodominant SLYNTVATL Gag epitope. In contrast, we found no mutations in the immunodominant Env epitopes VSFEPIPIH and RGPGRAFVT, but multiple deletions and mutations throughout env were observed.
Heterogeneity of pv- and v-gag and -env
. | . | . | Differences in mobility on HMA . | . | Diversity within provirus and virus, % . | . | Diversity between pv and v, % . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients . | Sequenced gene . | Subtype . | Provirus . | Virus . | Provirus . | Virus . | . | Changes in epitopes . | ||
| P12 | gag | CRF01-AE | 11/25 | 13/25 | 2.59 | 1.69 | 2.83 | SLYNTVATL | ||
| SLFNTIATL* | ||||||||||
| SLFNTVATL† | ||||||||||
| P14 | gag | CRF02-AG | 14/25 | 13/25 | 0.91 | 1.69 | 1.90 | SLYNTVATL | ||
| SLFNTLATL‡ | ||||||||||
| P24 | env | CRF03-AB | 6/18 | 12/42 | 1.36 | 1.40 | 1.50 | Mutations in nonimmunodominant epitopes | ||
| P25 | env | B | 6/36 | 8/44 | 3.8 | 4.57 | 4.68 | Deletions and mutations in nonimmunodominant epitopes | ||
. | . | . | Differences in mobility on HMA . | . | Diversity within provirus and virus, % . | . | Diversity between pv and v, % . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients . | Sequenced gene . | Subtype . | Provirus . | Virus . | Provirus . | Virus . | . | Changes in epitopes . | ||
| P12 | gag | CRF01-AE | 11/25 | 13/25 | 2.59 | 1.69 | 2.83 | SLYNTVATL | ||
| SLFNTIATL* | ||||||||||
| SLFNTVATL† | ||||||||||
| P14 | gag | CRF02-AG | 14/25 | 13/25 | 0.91 | 1.69 | 1.90 | SLYNTVATL | ||
| SLFNTLATL‡ | ||||||||||
| P24 | env | CRF03-AB | 6/18 | 12/42 | 1.36 | 1.40 | 1.50 | Mutations in nonimmunodominant epitopes | ||
| P25 | env | B | 6/36 | 8/44 | 3.8 | 4.57 | 4.68 | Deletions and mutations in nonimmunodominant epitopes | ||
PCR templates of gag (patients P12 and P14) or env (patients P24 and P25) from plasma virus or from provirus were analyzed for differences in mobility on HMA (values indicate number of clones of the provirus tested on HMA showing difference in mobility out of total number of clones) and for differences in sequence.
Underlined letters represent mutations in the epitope.
These mutations were found in 7 of 11 proviruses and 1 of 13 viruses
This mutation was found in 4 of 11 proviruses and 12 of 13 viruses
These mutations were found in all proviruses and viruses
Stimulatory capacity of env mRNA-electroporated DCs from HIV-1–seropositive individuals. iDCs from HIV-1–seropositive individuals were electroporated on day 6 of culture with pv-env or v-env mRNA. After maturation, these DCs were used as stimulators for autologous PBMCs (A), purified CD4+ cells (B), or purified CD8+ T cells (C). The experiment was done for patients P23 (□), P24 (*), and P25 (▵).
Stimulatory capacity of env mRNA-electroporated DCs from HIV-1–seropositive individuals. iDCs from HIV-1–seropositive individuals were electroporated on day 6 of culture with pv-env or v-env mRNA. After maturation, these DCs were used as stimulators for autologous PBMCs (A), purified CD4+ cells (B), or purified CD8+ T cells (C). The experiment was done for patients P23 (□), P24 (*), and P25 (▵).
Discussion
The development of a DC–based immunotherapy against HIV has gained major interest because of the superior ability of antigen-loaded DCs to stimulate T-cell responses.2,11,14 We previously developed an antigen-loading method based on electroporation of in vitro–transcribed mRNA27 and demonstrated that influenza matrix protein mRNA-electroporated DCs stimulated specific memory CD8+ T-cell responses in vitro.28 In the present study, we first showed that DCs electroporated with codon-optimized hHxB-2 gag mRNA expressed high levels of intracellular Gag, secreted the protein, and induced IFN-γ secretion by a specific CD8+ T-cell line in an HLA-A*0201–restricted way, pointing to efficient processing and MHC class I presentation.
Using the same hHXB-2 gag mRNA, DCs also triggered autologous PBMCs to produce IFN-γ. Importantly, also, noncodon-optimized gag mRNA from a large number of patients' viruses was shown to be efficient in transfecting autologous DCs. Although these autologous virus sequences resulted in lower Gag protein expression levels, as compared with hHXB-2 gag mRNA transfection, DCs electroporated with patient-derived gag mRNA efficiently stimulated IFN-γ secretion by autologous PBMCs. The higher responses induced by consensus as compared with autologous gag might be related to the codon optimization of the former, resulting in a more efficient translation, as also reflected in the higher protein expression. However, we suggest that covering the spectrum of autologous viral sequences is important to induce an individually adapted response to avoid immune escape.
Further analysis revealed that both CD8+ T cells and CD4+ T cells from the patients responded in this setting, pointing to efficient MHC class I and class II presentation. In view of future therapeutic applications, it is encouraging that purified CD4+ T cells could efficiently and specifically be triggered, even in the absence of MHC class II targeting signals, because it is well known that memory CD4+ T cells are needed to induce virus-specific CTLs.45
In addition, we were also able to trigger ex vivo Env-specific T cells by in vitro stimulation with autologous DCs transfected with proviral or viral env mRNA. The IFN-γ ELISPOT responses induced by env mRNA were in the same range as the responses to Gag. This observation seems to be in contrast with some previous studies showing higher IFN-γ ELISPOT responses to Gag than to Env.46-55 In those studies, however, PBMCs were stimulated with peptides. Clearly, our results suggest that any of the multiple HIV genes could be used as mRNA to load DCs and stimulate effector functions of autologous T cells. This may be important, because it is likely that multiple, if not all, HIV gene products will have to be included in an immunotherapeutic strategy.
Previously, most studies on virus-specific T cells were predominantly focused on measuring IFN-γ. Recently, a series of studies have shown the importance of IL-2 secretion.50-52 Using intracellular cytokine staining, Zimmerli et al53 and Harari et al54 compared the percentage of CD8+ and CD4+ T cells that secrete IL-2 after stimulation with Gag peptide pool between progressors and long-term nonprogressors. In both studies the CD8+ or CD4+ T cells from the progressors did not secrete IL-2, but those from long-term nonprogressors did. Remarkably, in our present study on 3 random patients (who seemed to be typical progressors), we showed that IL-2 could readily and significantly be triggered, although the amount was 10 times less than IFN-γ. Most probably, both the efficient antigen processing and presentation by DCs and the highly sensitive ELISPOT have contributed to this encouraging result. Nevertheless, in a future immunotherapeutic setting, it might be important to further enhance this IL-2 response. As expected from previous studies,11,56 we observed that mDCs induce higher responses as compared with iDCs, thus strongly supporting the use of mDCs in future immunotherapeutic approaches.
HIV-1 is characterized by an impressive variability within the infected population and within individual patients.3-4,10 The latter was illustrated in 4 randomly chosen patients from our cohort who apparently were infected by 4 different subtypes. HMA analysis revealed that up to 50% of the clones showed a differential migration pattern, and sequencing showed a high diversity, especially between clones from the proviral and the viral reservoir, with mutations in an immunodominant epitope (Gag) or mutations and deletions in nonimmunodominant epitopes (Env). Clearly, therapeutic vaccination with a consensus B subtype sequence, such as HxB-2, is likely to be insufficient to completely control both the prevalent plasma variants and the latent variants of patients with various subtypes.46 Admittedly, with our present strategy of DNA amplification not only the archived latent and replication-competent virus is amplified but also nonintegrated DNA.47 Recently Monie et al47 and Chun et al48 showed that resting CD4+ T cells are the major reservoir of provirus, and we have isolated DNA from PBMCs that contain both the latent reservoir and the virus dominant on that moment, which might also be present as a labile DNA virus in activated CD4+ T cells.6,49 Covering the entire intrapatient variability is a prerequisite for a successful DC-mediated therapy, and it seems that our mRNA strategy indeed has the potential of stimulating the patients' T cells against a broad range of variants from their own virus and provirus.
In view of these multiple indications of the validity of using the patients' own virus mRNA to load autologous DCs and stimulate autologous T cells, it was surprising that pulsing the patients' DCs with overlapping peptides derived from the consensus gag HxB-2 tended to induce more SFCs as compared with DCs electroporated with gag mRNA derived from HXB-2 autologous virus or provirus. At least part of this seemingly higher peptide response may be due to nonspecific stimulation by the HxB-2 Gag peptide pool, because some responses were also seen in HIV-1–seronegative controls. On the other hand, a slightly higher peptide response may indicate that processing of mRNA, although efficient, remains suboptimal in nontreated HIV-infected subjects. In any case, ex vivo peptide pulsing of DCs in the context of an immunotherapy for HIV-1 is not a realistic option, because it would imply the production of all possible peptides from all the antigenic variants in the patients' plasma and latent viral quasispecies.
In conclusion, we have shown for the first time that HIV-1–seropositive patients' DCs electroporated with consensus or autologous amplified viral and proviral gag or env mRNA are able to activate ex vivo IFN-γ production by autologous PBMCs, CD4+ cells, and CD8+ T cells as well as IL-2 production by autologous PBMCs. This mRNA-based strategy provides a major perspective for the development of a patient-specific immunotherapy directed against the entire autologous HIV quasispecies.
Prepublished online as Blood First Edition Paper, November 1, 2005; DOI 10.1182/blood-2005-01-0339.
Supported by the following grants: G.O.456.03 from the Fund for Scientific Research Flanders (FWO), Agreement 2002/210 with Agence de Recherche sur le Sida (ANRS), and GOA Fa20000/802 of the University of Antwerp. E.R.A.V.G. is a predoctoral fellow of the Institute for Science and Technology (IWT); G.V.d.B. and S.A. are predoctoral fellows, and V.F.I.V.T. a postdoctoral fellow, of the FWO; and G.H. and C.M. are predoctoral and postdoctoral fellows, respectively, of the ANRS.
E.R.A.V.G. performed all the culture experiments, including ELISPOT, and wrote the paper; P.P. contributed to study design, data analysis, and writing of the manuscript; L.H. contributed to study design, analyzed the heteroduplex mobility assays and sequences, and developed the method to produce proviral and viral mRNA; K.V., a technician, made proviral and viral mRNA and did all the heteroduplex mobility assays; F.M. contributed to the research (clinical part) and corevised the paper; A.D.R. contributed to the research (clinical part) and reviewed the manuscript; R.C. reviewed the protocol and paper and was the physician responsible for care of the patients studied; G.V.d.B. contributed to the organization of patient recruitment and to study design; D.R.V.B., senior researcher, supervised the flow cytometric and cell culture laboratory at the Antwerp University Hospital (UZA) and participated in the experimental set-up, result, discussion, and article proofreading; V.F.I.V.T. contributed to concept outline, project writing, and follow-up of the results; S.A. participated in cDNA cloning and article proofreading; B.V. contributed vital new reagents; C.M. and G.H. contributed to the experimental design for experiments with the Gag-specific T-cell line, execution and analysis, and participated in the interpretation and manuscript writing; A.H. contributed to study design, execution and analysis, and participated in the interpretation and manuscript writing; Z.N.B., coinitiator and copromotor, contributed to funding, study design, conceptual and technical ideas, analysis of the results, and writing and revision of the manuscript; and G.V., supervisor and promotor, contributed to study design, project writing, analyses of experiments, and writing and revision of the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Sandra Coppens (Virology Unit, ITMA) for the skillful help with the heteroduplex mobility assays. We acknowledge the skillful help of Anne-Claire Ripoche (Antigen Presentation by Dendritic Cell Group, Institute Cochin) and Luc Kestens (Laboratory of Immunology, ITMA) for the use of their culture facilities. We thank all members of the AC19 Dendritic Cell Work Group from the ANRS for helpful discussions.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal