Mutations in codon D816 of the KIT gene represent a recurrent genetic alteration in acute myeloid leukemia (AML). To clarify the biologic implication of activation loop mutations of the KIT gene, 1940 randomly selected AML patients were analyzed. In total, 33 (1.7%) of 1940 patients were positive for D816 mutations. Of these 33 patients, 8 (24.2%) had a t(8;21), which was significantly higher compared with the subgroup without D816 mutations. Analyses of genetic subgroups showed that KIT-D816 mutations were associated with t(8;21)/AML1-ETO and other rare AML1 translocations. In contrast, other activating mutations like FLT3 and NRAS mutations were very rarely detected in AML1-rearranged leukemia. KIT mutations had an independent negative impact on overall (median 304 vs 1836 days; P = .006) and event-free survival (median 244 vs 744 days; P = .003) in patients with t(8;21) but not in patients with a normal karyotype. The KIT-D816V receptor expressed in Ba/F3 cells was resistant to growth inhibition by the selective PTK inhibitors imatinib and SU5614 but fully sensitive to PKC412. Our findings clearly indicate that activating mutations of receptor tyrosine kinases are associated with distinct genetic subtypes in AML. The KIT-D816 mutations confer a poor prognosis to AML1-ETO-positive AML and should therefore be included in the diagnostic workup. Patients with KIT-D816-positive/AML1-ETO-positive AML might benefit from early intensification of treatment or combination of conventional chemotherapy with KIT PTK inhibitors.
Introduction
The KIT protooncogene encodes for a class III transmembrane receptor tyrosine kinase that is composed of an extracellular portion containing 5 immunoglobulin-like domains and an intracellular portion consisting of a juxtamembrane and 2 protein tyrosine kinase domains (PTK1 and PTK2) split up by an interkinase domain.1,2 Activating mutations in KIT have been reported in acute myeloid leukemia (AML) and are confined to either the extracellular (exon 8 mutations)3-5 or the PTK2 domain (D816 mutations).6 Both classes of KIT mutations have been identified predominantly in specific genetic subgroups of AML: exon 8 mutations in patients with inv(16) and D816 mutations in patients with t(8;21). In both AML subgroups, one of 2 parts of the transcription factor CBF is targeted: AML1 (CBFα) in t(8;21) and CBFB (CBFβ) in inv(16), leading to the common nomenclature of “core binding factor leukemia” for inv(16)/t(16;16)- and t(8;21)-positive AML. The clinical and prognostic significance of KIT mutations in AML is unclear. Preliminary data suggest that KIT exon 8 mutations are associated with an increased relapse rate4 and KIT-D816 mutations were correlated to a higher white blood cell (WBC) count in AML patients.7,8
On the molecular level the translocation (8;21) leads to the generation of the AML1-ETO fusion gene,9 which has been shown to influence differentiation, proliferation, and apoptosis in both in vitro and in vivo models and promotes self-renewal in retrovirally transduced primary human CD34+ cells.10 The t(8;21)/AML1-ETO-positive AML is commonly associated with a favorable prognosis.11-14
Recently, it was postulated that fusion genes involving transcription factors are not sufficient to induce leukemia and require additional “cooperative” mutations that trigger proliferation. In line with this hypothesis, it has been shown that activating mutations of the gene for tyrosine kinase FLT3 cooperate with PML-RARA to induce leukemia in mice.15 In contrast, the significance of additional genetic alterations in patients with t(8;21)/AML1-ETO-positive AML are poorly studied.
In the present study, we report that KIT-D816 mutations define an unfavorable subgroup in patients with AML1-ETO-positive AML. Thus, KIT-D816 mutations represent a reliable molecular marker identifying patients with a poor prognosis in an otherwise prognostically favorable AML group. As implied by our in vitro experiments, patients that are positive for AML1-ETO as well as for KIT-D816 mutations might benefit from new innovative therapeutic strategies (eg, KIT-selective PTK inhibitors).
Patients, materials, and methods
Patients
Patient samples were referred to the Laboratory for Leukemia Diagnostics between 1998 and 2004. All samples underwent a standardized processing including central sample registration, preparation, and evaluation by cytomorphology, cytochemistry, multiparameter immunophenotyping, cytogenetics, fluorescence in situ hybridization (FISH), and molecular genetics.16-18
Prior to therapy, all patients gave their informed consent for participation in the Acute Myeloid Leukemia Cooperative Group (AMLCG) studies. All samples investigated in this study were obtained at the time of diagnosis. Bone marrow samples were used when available; otherwise, peripheral blood was used. Selection criteria were limited to the availability of sufficient material and participation in the above-mentioned clinical trials. Thus, de novo as well as secondary and therapy-related AML were included. Patient characteristics are summarized in Table 1.
Presenting features of patients included in this analysis
Feature . | No. of cases in each group . |
|---|---|
| Male/female (ratio) | 1034/906 (1.14/1.00) |
| Etiology | |
| De novo AML | 1614 |
| s-AML | 201 |
| t-AML | 125 |
| FAB classification | |
| M0 | 80 |
| M1 | 332 |
| M2 | 693 |
| M3/M3v | 87 |
| M4 | 327 |
| M4eo | 97 |
| M5 | 156 |
| M6 | 78 |
| M7 | 10 |
| N/A | 80 |
| Karyotype | |
| Normal | 908 |
| t(15;17) | 84 |
| t(8;21) | 76 |
| inv(16)/t(16;16) | 97 |
| t(11q23)/MLL | 65 |
| inv(3)/t(3;3) | 39 |
| t(6;9) | 5 |
| –5/del(5q) | 20 |
| –7/del(7q) | 35 |
| Trisomy 8 | 81 |
| Trisomy 11 | 12 |
| Complex aberrant | 236 |
| All others | 255 |
| Not analyzed | 27 |
Feature . | No. of cases in each group . |
|---|---|
| Male/female (ratio) | 1034/906 (1.14/1.00) |
| Etiology | |
| De novo AML | 1614 |
| s-AML | 201 |
| t-AML | 125 |
| FAB classification | |
| M0 | 80 |
| M1 | 332 |
| M2 | 693 |
| M3/M3v | 87 |
| M4 | 327 |
| M4eo | 97 |
| M5 | 156 |
| M6 | 78 |
| M7 | 10 |
| N/A | 80 |
| Karyotype | |
| Normal | 908 |
| t(15;17) | 84 |
| t(8;21) | 76 |
| inv(16)/t(16;16) | 97 |
| t(11q23)/MLL | 65 |
| inv(3)/t(3;3) | 39 |
| t(6;9) | 5 |
| –5/del(5q) | 20 |
| –7/del(7q) | 35 |
| Trisomy 8 | 81 |
| Trisomy 11 | 12 |
| Complex aberrant | 236 |
| All others | 255 |
| Not analyzed | 27 |
N = 1940. Median age was 60 years (range, 15-90 years).
Treatment protocol of the German AMLCG Study
Eighty percent of the analyzed patients were treated according to the AMLCG studies. Treatment comprised the randomized comparisons of 9 days thioguanine, cytosine arabinoside, daunorubicin (TAD9)/TAD9 versus TAD9/high-dose cytosine arabinoside, miloxantrone (HAM) (AMLCG1992), and of TAD9/HAM versus HAM/HAM (AMLCG1999) double-induction therapy followed by TAD9 consolidation. Patients with acute promyelocytic leukemia (APL) were treated according to the respective APL protocol of the AMLCG.19 Patients in complete remission after TAD9 consolidation were subsequently randomized for monthly maintenance therapy or S-HAM as a second course of consolidation (AMLCG1992) or for autologous stem cell transplantation (AMLCG1999). The study design adhered to the Declaration of Helsinki and was approved by the ethics committees of the participating institutions prior to the study's initiation.
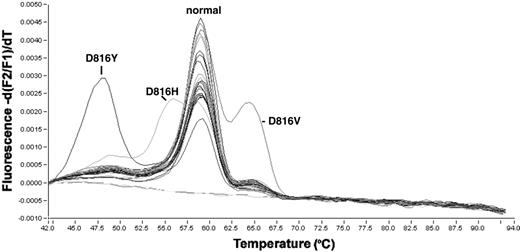
Detection of KIT-D816 mutations in patient samples by melting curve analysis. The y-axis represents fluorescence intensity and the x-axis the temperature. Mutations lead to different melting temperatures of the hybridization probes from the amplification product. Each individual peak indicates a different D816 mutation, whereas the center peak is composed of patient samples with nonmutated Asp816.
Detection of KIT-D816 mutations in patient samples by melting curve analysis. The y-axis represents fluorescence intensity and the x-axis the temperature. Mutations lead to different melting temperatures of the hybridization probes from the amplification product. Each individual peak indicates a different D816 mutation, whereas the center peak is composed of patient samples with nonmutated Asp816.
PCRs at diagnosis
Polymerase chain reactions (PCRs) for AML1-ETO fusion transcript were performed as has been described.20 For each sample an ABL-specific reverse transcriptase-PCR (RT-PCR) was performed to control the integrity of RNA as has been described.21 Strict precautions were taken to prevent contamination. Water instead of cDNA was included as a blank sample in each experiment. Amplification products were analyzed on 1.5% agarose gels stained with ethidium bromide.
Screening assay
Mononucleated cells were isolated by standard Ficoll-Hypaque density gradient centrifugation. Nucleic acid isolation and cDNA synthesis was performed as described before.21 Screening for KIT-D816 mutations was performed using a melting curve-based LightCycler assay (Roche Diagnostics, Mannheim, Germany) with forward primer KIT816F, CAGCCAGAAATATCCTCCTTACT; reverse primer KIT816R, TGTCAAGCAGAGAATGGGTACTC; and hybridization probes KIT816-FL sensor AGCCAGAGTCATCAAGAATGATTCTA-FL and KIT816-anchor LC-Red640-ATGTGGTTAAAGGAAACGTGAGTACCCA-P. The PCR reaction was carried out in a 20-μL reaction volume with 0.5 μM each of forward and reverse primer, 0.75 μM Hyb-Probes, 4 mM MgCl2, and 2 μL LightCycler-FastStart DNA Master Hybridization Probes (Roche Diagnostics, Mannheim, Germany). LightCycler data were analyzed using the LightCycler 3.0 software (Roche Diagnostics) and the second derivative maximum method. Each 20-μL reaction contains 2 μL cDNA, an equivalent of about 3000 cells. Amplification was performed with 45 cycles using 50°C annealing temperature. Final melting curve analysis was started at 40°C up to 95°C with slope of 0.2°C/second and continuous detection with channel F2/F1 (Figure 1).
Sequence analysis
All cases that were found to be positive were confirmed by sequence analysis. Approximately 100 ng of purified PCR products were directly sequenced with 3.3 pmol of each forward and reverse primer using the Big Dye Terminator Cycle Sequencing Kit (Applera, Darmstadt, Germany). After initial denaturation at 95°C for 5 minutes, 25 cycles at 94°C for 15 seconds and 60°C for 4 minutes were performed. Sequence analysis was performed on an ABI 310 or 3100 Avant sequence detection system (Applied Biosystems, Foster City, CA).
Cell proliferation of Ba/F3 cells and application of specific PTK inhibitors
IL-3-dependent Ba/F3 cells stably expressing either KIT-wild type (KIT-WT) or KIT-D816V were seeded at a concentration of 0.05 × 106/mL in the presence or absence of IL-3 and SCF, as described previously.5 The cells were initially treated with the indicated PTK inhibitors imatinib, PKC412 (both kindly provided by Novartis, Basel, Switzerland), and SU5614 (purchased from Calbiochem-Novabiochem, Bad Soden, Germany).At 72 hours, viable cells were counted in a standard hemacytometer after staining with trypan blue. Figures show mean values and standard deviations (SDs) of 3 independent experiments.
Transient transfection, preparation of whole-cell extracts, immunoprecipitation, and Western blotting
The 293 cells were seeded and transiently transfected, as described previously.5,23 Cells were starved for 12 hours and treated with the indicated concentrations of imatinib, PKC412, and SU5614 for 2 hours at 37°C, 5% CO2. After cell harvest and lysis, 300 μg of the lysates was immunoprecipitated with polyclonal rabbit α KIT antibody (c-19; Santa Cruz, Heidelberg, Germany). Immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with mouse monoclonal α phosphotyrosine (PY99; Santa Cruz) antibody and reprobed with α KIT antibody.
Cell cycle and apoptosis analyses
Cell cycle and apoptosis analyses were performed by measuring the DNA content of cell nuclei by staining with propidium iodide (PI) as described previously.23,24 Cells (0.2 × 106) were seeded and grown under conditions as named in “Cell proliferation of Ba/F3 cells and application of specific PTK inhibitors.” KIT-WT-expressing Ba/F3 cells were cultured in the presence of 100 ng/mL SCF. After 24 hours, cells were stained and analyzed by flow cytometry using standard methods. Cell nuclei showing a hypodiploid DNA content were considered to be apoptotic. For analysis of cell cycle distribution, sub-G1 cells were excluded by gating.
Statistical analysis
A calculation of overall and event-free survival was done only for patients included in the AMLCG study. Overall survival (OS) and event-free survival (EFS) were calculated according to Kaplan-Meier,25 their correlation with other parameters by Cox regression. OS was calculated from time of diagnosis to death and EFS from time of diagnosis to death, documentation of persistent leukemia, or relapse. Survival curves were compared using double-sided log rank test. Comparisons of dichotomous variables between different groups were performed by use of 2-sided Fisher exact test. For all explorations results were significant at a level of P less than .05 at both sides. SPSS (version 12.1.4) software (SPSS, Chicago, IL) was used for statistical analysis.
Results
Frequency of KIT-D816 mutations in AML samples
A total of 1940 patients were screened for KIT mutations in exon 17 of the tyrosine kinase domain 2. Patient characteristics are summarized in Table 1. Of the total cohort, 1614 patients had de novo AML, 201 had secondary AML (s-AML) after myelodysplastic syndrome (MDS), and 125 had therapy-related AML (t-AML). All patients were screened for mutations in exon 17 at and around codon Asp816 in the KIT receptor. In 33 patients (1.7%), a mutation affecting codon 816 of the KIT gene was detected (Figure 1). Characteristics of all cases with KIT-D816 mutations are given in Table 2. Sequencing of the mutations revealed a gac>gtc exchange (Asp→Val) in 21 patients, a gac>tac exchange (Asp→Tyr) in 2, and a gac>cac exchange (Asp→His) in 1 sample. The mutation of 9 patients that were found positive for a D816 mutation by melting curve analysis could not be sequenced due to the presence of less than 10% of mutated cells in the sample.
Characteristics of patients with KIT-D816 mutations
Case no. . | Sex/age, y . | Diagnosis . | BM blasts, % . | D816 mutation . | Additional mutations . | Karyotype . |
|---|---|---|---|---|---|---|
| 1 | M/32 | AML M4 | 35 | gac>gtc; Asp→ Val | — | 45,X,–Y [18] 46,XY [2] |
| 2 | M/27 | AML M2, mastocytosis of spleen | 30 | gac>gtc; Asp→ Val | — | 45,X,–Y [20] |
| 3 | M/34 | AML M2 | ND | gtg>cac; Asp→ His | FLT3-LM | 45,X,–Y,t(8;21)(q22;q22) [19] 46,XY [1] |
| 4 | M/56 | AML M2 | 80 | NA > 10% | — | 45,X,–Y,t(8;21)(q22;q22) [2] |
| 45,X,–Y,del(4)(q25q31),t(8;21)(q22;q22) [13] | ||||||
| 45,X,–Y,del(4)(q25q31),t(8;21)(q22;q22),del(9)(q22q34) [5] | ||||||
| 5 | F/62 | AML M2 | 85 | gac>gtc; Asp→ Val | FLT3-LM | 46,XX [25] |
| 6 | F/48 | AML M2 | 70 | NA > 10% | FLT3-LM | 46,XX [25] |
| 7 | F/46 | AML M4 | 60 | gac>gtc; Asp→ Val | — | 46,XX [25] |
| 8 | F/68 | AML M4eo | ND | gac>gtc; Asp→ Val | NRAS | 46,XX,inv(16)(p13q22)[9] 46,XX,t(3;21)(q26;q22),inv(16)(p13q22) [4] |
| 46,XX [5] | ||||||
| 9 | F/51 | AML M2 | 60 | NA > 10% | FLT3-TKD | 46,XX,t(3;21)(q26;q22) [15] 46,XX [5] |
| 10 | F/43 | AML M2, mastocytosis | ND | gac>gtc; Asp→ Val | — | 46,XX,t(8;21)(q22;q22) [1] 45,X,–X,t(8;21)(q22;q22),del(9)(q22) [2] |
| 11 | M/45 | AML M2 | 80 | gac>gtc; Asp→ Val | — | 46,XY [20] |
| 12 | M/66 | AML M2 | 30 | NA > 10% | — | 46,XY [23] |
| 13 | M/28 | AML M1 | ND | gac>gtc; Asp→ Val | FLT3-LM | 46,XY [25] |
| 14 | M/46 | AML M2 | ND | gac>gtc; Asp→ Val | — | 46,XY [25] |
| 15 | M/54 | AML M4 | 70 | gac>gtc; Asp→ Val | FLT3-LM | 46,XY [25] |
| 16 | M/37 | AML M2 | ND | NA > 10% | FLT3-LM | 46,XY [26] |
| 17 | M/60 | AML M4 | 80 | NA > 10% | — | 46,XY [28] |
| 18 | M/72 | s-AML M2 | 65 | gac>gtc; Asp→ Val | — | 46,XY,del(7)(q22),t(12;21)(q24.3;q22) [4] 46,XY [4] |
| 19 | M/72 | AML M4eo | 70 | gac>gtc; Asp→ Val | — | 46,XY,inv(16)(p13q22)[3] 48,XY, +4, +8,inv(16)(p13q22) [17] |
| 20 | M/73 | AML M2 | 20 | gac>gtc; Asp→ Val | — | 46,XY,t(17;21)(p13;q22) [12] 46,XY [9] |
| 21 | M/51 | t-AML M1 | 95 | gac>gtc; Asp→ Val | — | 46,XY,t(8;21)(q22;q22) [18] |
| 22 | M/64 | AML M1 | 90 | gac>gtc; Asp→ Val | — | 46,XY,t(8;21)(q22;q22) [2] 45,X,–Y,t(8;21)(q22;q22) [18] |
| 23 | M/23 | AML M2 | 80 | gac>gtc; Asp→ Val | — | 46,XY,t(8;21)(q22;q22) [4] 45,X,–Y,t(8;21)(q22;q22) [11] 46,XY [1] |
| 24 | M/51 | AML M2 | ND | NA > 10% | — | 46,XY,t(8;21)(q22;q22) [9] 45,X,–Y,t(8;21)(q22;q22) [6] |
| 25 | F/75 | AML M2 | 95 | NA > 10% | — | 47,XX, +4 [2] 47,XX, +4,der(13)t(8;13)(p11;p11) [7] 46,XX [11] |
| 26 | F/64 | AML M2 | ND | gac>gtc; Asp→ Val | — | 47,XX, +4 [21] 46,XX [4] |
| 27 | F/70 | s-AML M2 | 85 | gta>tac; Asp→ Tyr | NRAS, MLL-PTD | 47,XX, +8 [3] 46,XX [17] |
| 28 | M/76 | AML M2 | 70 | gta>tac; Asp→ Tyr | — | 47,XY, +4,t(8;21)(q22;q22) [20] |
| 29 | M/62 | AML M4 | 25 | gac>gtc; Asp→ Val | — | 47,XY, +8 [21] |
| 30 | M/80 | AML M2 | 50 | gac>gtc; Asp→ Val | — | 47,XY, +9 [14] 46,XY [6] |
| 31 | M/61 | s-AML M2 | ND | gac>gtc; Asp→ Val | — | 47,XY,inv(3)(q21q26), +6 [20] |
| 32 | M/79 | s-AML M2 | 60 | gac>gtc; Asp→ Val | — | 48,XY, +4, +8 [12] 46,XY [8] |
| 33 | M/75 | AML M4 | 80 | NA > 10% | — | 53,XY, +3, +6, +7, +8, +8, +12, der(13)t(3;13)(p23;p10), +18 [14] 46,XY [1] |
Case no. . | Sex/age, y . | Diagnosis . | BM blasts, % . | D816 mutation . | Additional mutations . | Karyotype . |
|---|---|---|---|---|---|---|
| 1 | M/32 | AML M4 | 35 | gac>gtc; Asp→ Val | — | 45,X,–Y [18] 46,XY [2] |
| 2 | M/27 | AML M2, mastocytosis of spleen | 30 | gac>gtc; Asp→ Val | — | 45,X,–Y [20] |
| 3 | M/34 | AML M2 | ND | gtg>cac; Asp→ His | FLT3-LM | 45,X,–Y,t(8;21)(q22;q22) [19] 46,XY [1] |
| 4 | M/56 | AML M2 | 80 | NA > 10% | — | 45,X,–Y,t(8;21)(q22;q22) [2] |
| 45,X,–Y,del(4)(q25q31),t(8;21)(q22;q22) [13] | ||||||
| 45,X,–Y,del(4)(q25q31),t(8;21)(q22;q22),del(9)(q22q34) [5] | ||||||
| 5 | F/62 | AML M2 | 85 | gac>gtc; Asp→ Val | FLT3-LM | 46,XX [25] |
| 6 | F/48 | AML M2 | 70 | NA > 10% | FLT3-LM | 46,XX [25] |
| 7 | F/46 | AML M4 | 60 | gac>gtc; Asp→ Val | — | 46,XX [25] |
| 8 | F/68 | AML M4eo | ND | gac>gtc; Asp→ Val | NRAS | 46,XX,inv(16)(p13q22)[9] 46,XX,t(3;21)(q26;q22),inv(16)(p13q22) [4] |
| 46,XX [5] | ||||||
| 9 | F/51 | AML M2 | 60 | NA > 10% | FLT3-TKD | 46,XX,t(3;21)(q26;q22) [15] 46,XX [5] |
| 10 | F/43 | AML M2, mastocytosis | ND | gac>gtc; Asp→ Val | — | 46,XX,t(8;21)(q22;q22) [1] 45,X,–X,t(8;21)(q22;q22),del(9)(q22) [2] |
| 11 | M/45 | AML M2 | 80 | gac>gtc; Asp→ Val | — | 46,XY [20] |
| 12 | M/66 | AML M2 | 30 | NA > 10% | — | 46,XY [23] |
| 13 | M/28 | AML M1 | ND | gac>gtc; Asp→ Val | FLT3-LM | 46,XY [25] |
| 14 | M/46 | AML M2 | ND | gac>gtc; Asp→ Val | — | 46,XY [25] |
| 15 | M/54 | AML M4 | 70 | gac>gtc; Asp→ Val | FLT3-LM | 46,XY [25] |
| 16 | M/37 | AML M2 | ND | NA > 10% | FLT3-LM | 46,XY [26] |
| 17 | M/60 | AML M4 | 80 | NA > 10% | — | 46,XY [28] |
| 18 | M/72 | s-AML M2 | 65 | gac>gtc; Asp→ Val | — | 46,XY,del(7)(q22),t(12;21)(q24.3;q22) [4] 46,XY [4] |
| 19 | M/72 | AML M4eo | 70 | gac>gtc; Asp→ Val | — | 46,XY,inv(16)(p13q22)[3] 48,XY, +4, +8,inv(16)(p13q22) [17] |
| 20 | M/73 | AML M2 | 20 | gac>gtc; Asp→ Val | — | 46,XY,t(17;21)(p13;q22) [12] 46,XY [9] |
| 21 | M/51 | t-AML M1 | 95 | gac>gtc; Asp→ Val | — | 46,XY,t(8;21)(q22;q22) [18] |
| 22 | M/64 | AML M1 | 90 | gac>gtc; Asp→ Val | — | 46,XY,t(8;21)(q22;q22) [2] 45,X,–Y,t(8;21)(q22;q22) [18] |
| 23 | M/23 | AML M2 | 80 | gac>gtc; Asp→ Val | — | 46,XY,t(8;21)(q22;q22) [4] 45,X,–Y,t(8;21)(q22;q22) [11] 46,XY [1] |
| 24 | M/51 | AML M2 | ND | NA > 10% | — | 46,XY,t(8;21)(q22;q22) [9] 45,X,–Y,t(8;21)(q22;q22) [6] |
| 25 | F/75 | AML M2 | 95 | NA > 10% | — | 47,XX, +4 [2] 47,XX, +4,der(13)t(8;13)(p11;p11) [7] 46,XX [11] |
| 26 | F/64 | AML M2 | ND | gac>gtc; Asp→ Val | — | 47,XX, +4 [21] 46,XX [4] |
| 27 | F/70 | s-AML M2 | 85 | gta>tac; Asp→ Tyr | NRAS, MLL-PTD | 47,XX, +8 [3] 46,XX [17] |
| 28 | M/76 | AML M2 | 70 | gta>tac; Asp→ Tyr | — | 47,XY, +4,t(8;21)(q22;q22) [20] |
| 29 | M/62 | AML M4 | 25 | gac>gtc; Asp→ Val | — | 47,XY, +8 [21] |
| 30 | M/80 | AML M2 | 50 | gac>gtc; Asp→ Val | — | 47,XY, +9 [14] 46,XY [6] |
| 31 | M/61 | s-AML M2 | ND | gac>gtc; Asp→ Val | — | 47,XY,inv(3)(q21q26), +6 [20] |
| 32 | M/79 | s-AML M2 | 60 | gac>gtc; Asp→ Val | — | 48,XY, +4, +8 [12] 46,XY [8] |
| 33 | M/75 | AML M4 | 80 | NA > 10% | — | 53,XY, +3, +6, +7, +8, +8, +12, der(13)t(3;13)(p23;p10), +18 [14] 46,XY [1] |
— indicates not present; ND, not done; NA, not applicable.
There was no significant difference with respect to de novo AML, AML after MDS prephase, or t-AML after preceding malignancy.
Frequency of KIT-D816 mutations in cytogenetic and morphologic AML subtypes
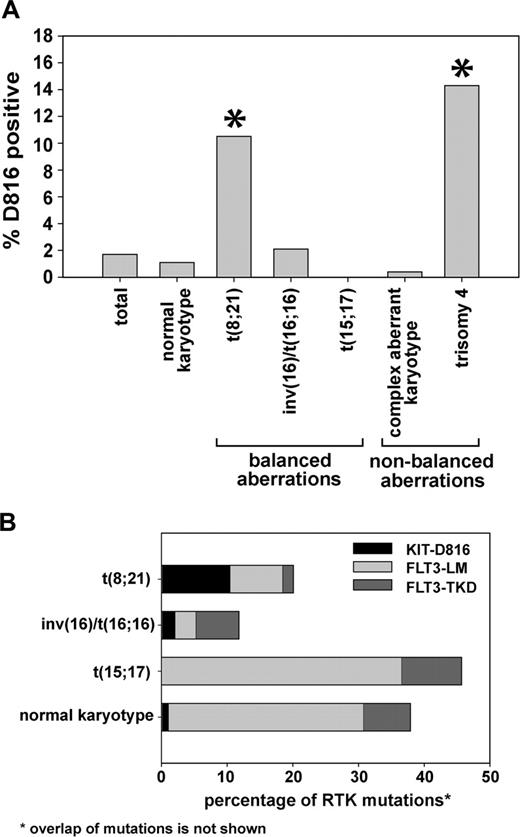
Cytogenetic analyses were available from 1913 (98.6%) of 1940 analyzed patients. Chi-square analysis showed that KIT-D816 mutations are not randomly distributed within cytogenetic subgroups (P < .001). Patients with a t(8;21) and a trisomy 4 that was present as sole cytogenetic aberration as well as concomitantly with other aberrations showed a significantly higher frequency of KIT-D816 mutations (10.5% and 14.3%, respectively; Figure 2A). In contrast, the frequency of KIT-D816 mutations was significantly lower in patients with a t(15;17) (0/84 patients = 0%) and a complex karyotype (1/236 = 0.4%).
We next analyzed additional cytogenetic alterations that were found in KIT-D816-positive patients. Of 33 patients with a KIT-D816 mutation, 10 (30.3%) had a normal karyotype, 8 (24.2%) had a t(8;21), 2 (6.1%) had an inv(16), 1 (3.0%) had a complex aberrant karyotype, 4 (12.1%) had a trisomy 4, and 8 (24.2%) had other rare aberrations (Table 2). The frequency of KIT-D816 mutations was significantly associated with t(8;21) and with trisomy 4, as assessed by Fisher exact test. This finding is in line with previous reports on the association of KIT mutations in CBF leukemias with these cytogenetic aberrations.26,27
Based on the tight association of t(8;21) and KIT-D816 mutations we further examined the frequency of other translocations involving the AML1 gene [rare t(AML1)] in patients with a KIT-D816 mutation: 4 patients had a translocation with AML1 other than t(8;21) (4/33; 12.1%). In detail, these were 2 cases with a t(3;21)/AML1-EVI1 (cases 8 and 9 in Table 2), one case with a t(12;21)(q24.3;q22) (case 18), and one case with a t(17;21)(p13; q22) (case 20). In cases 18 and 20, the AML1 rearrangement was confirmed by FISH, but the fusion partner was not yet further identified. Thus, a total of 36% of all KIT-D816-mutated cases had AML1 rearrangements.
Furthermore, 2 patients had a loss of the Y chromosome as sole chromosomal aberration (cases 1 and 2 in Table 2) and 2 additional patients showed a t(8;21) and a loss of the Y chromosome concomitantly (patients 3 and 4). A regression analysis showed that t(8;21), loss of Y, and trisomy 4 were independently associated with KIT-D816 mutations (P = .009, P = .024, and P < .001, respectively).
Cytomorphologic analyses that could be conducted with 1860 patients revealed that KIT-D816 mutations were not equally distributed among French-American-British (FAB) subgroups and were exclusively found in the M1 (n = 3), M2 (n = 22), M4 (n = 6), and M4eo (n = 2) cohorts. A total of 22 (67.0%) of 33 D816-positive patients were classified as M2.
Thus, these data suggest that KIT mutations are not only specifically associated with t(8;21), but with trisomy 4 or loss of Y chromosome. The latter is also a very common event in t(8;21) AML.
Activating mutations of receptor tyrosine kinases are associated with distinct cytogenetic subgroups
Experimental data from mouse models have suggested that certain mutations of RTK cooperate with distinct leukemic fusion genes to induce leukemia in mice.15,28 To validate this hypothesis in patients with AML, we analyzed the frequency of activating mutations in the FLT3 and KIT genes in different cytogenetic subgroups. As shown in Figure 2A, the frequency of KIT-D816 mutations in patients with AML1-ETO-positive leukemias was higher than in patients with normal and complex aberrant karyotypes. In contrast, mutations in the tyrosine kinase domain and length mutations in the FLT3 gene (FLT3-TKD and FLT3-LMs, respectively) were found at a much higher frequency in patients with a normal karyotype and a t(15;17) translocation. These data clearly show that activating mutations of FLT3 and KIT are associated with certain cytogenetic AML subtypes.
Cytogenetics of patients with D816 mutations. (A) Cytogenetic analyses were available in 1913 patients. The figure shows the frequency of D816 mutations within different cytogenetic subgroups. The overall frequency of this mutation was 1.7%. *Higher percentage of D816 mutations compared with the whole cohort as assessed by Fisher exact test (P < .05). The trisomy 4 was present as sole cytogenetic aberration as well as concomitantly with other aberrations. (B) The percentage of mutations in KIT and FLT3 in the indicated subgroups is shown. The overlap of RTK mutations is not shown in the figure. Thus, patients showing 2 different mutations are rated twice.
Cytogenetics of patients with D816 mutations. (A) Cytogenetic analyses were available in 1913 patients. The figure shows the frequency of D816 mutations within different cytogenetic subgroups. The overall frequency of this mutation was 1.7%. *Higher percentage of D816 mutations compared with the whole cohort as assessed by Fisher exact test (P < .05). The trisomy 4 was present as sole cytogenetic aberration as well as concomitantly with other aberrations. (B) The percentage of mutations in KIT and FLT3 in the indicated subgroups is shown. The overlap of RTK mutations is not shown in the figure. Thus, patients showing 2 different mutations are rated twice.
Prognostic significance of KIT-D816 mutations
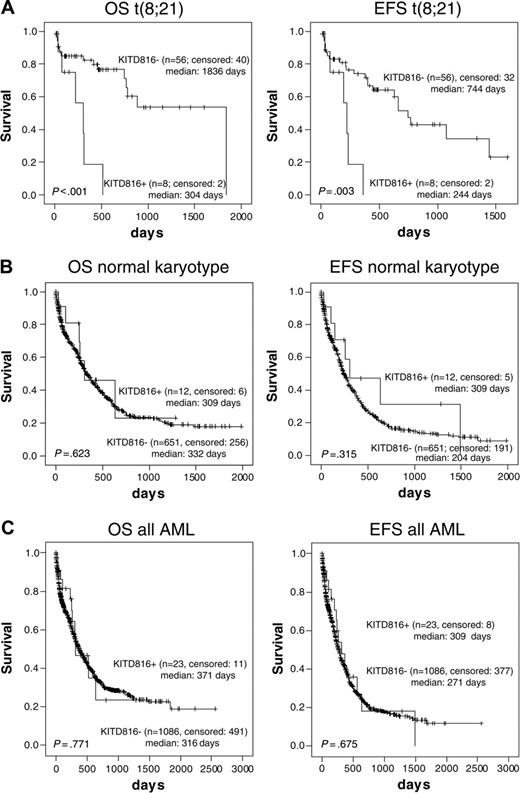
Analysis of the prognostic significance of KIT-D816 mutations was carried out (a) for 64 patients with t(8;21) and (b) for 663 patients that were classified to the subgroup with intermediate prognostic karyotypes (Figure 3). The median OS of t(8;21) patients without KIT-D816 mutations was 1836 days in contrast to 304 days for the KIT-D816-positive subgroup (Figure 3A; P < .001). Regarding EFS, the median was 244 days for the KIT-D816 mutation-positive patients and 744 days for the nonmutated subgroup (P = .003; Figure 3A). Age (P = .021) was another significant parameter on survival in the group with t(8;21) but FAB subtype, etiology, and leukocyte counts were not. Thus, the negative impact of KIT-D816 mutations was found to be independent of age (P < .001).
The same analysis was conducted for patients with a prognostically intermediate karyotype (Figure 3B) and for all unselected de novo AML (Figure 3C). In contrast to the t(8;21) subgroup, no significant differences in OS and EFS were observed (P = .623 and P = .315; P = .771 and P = .675, respectively).
Overall and event-free survival of patients with D816 mutations in subgroups with t(8;21) or with intermediate karyotype. OS and EFS for patients with mutated or nonmutated KIT-D816. (A) Only t(8;21) subgroup. (B) Patients with intermediate karyotype. (C) All AML. Figure shows Kaplan-Meier analyses for the patient numbers indicated. “Median” indicates median survival time; survival curves were compared using double-sided log rank test.
Overall and event-free survival of patients with D816 mutations in subgroups with t(8;21) or with intermediate karyotype. OS and EFS for patients with mutated or nonmutated KIT-D816. (A) Only t(8;21) subgroup. (B) Patients with intermediate karyotype. (C) All AML. Figure shows Kaplan-Meier analyses for the patient numbers indicated. “Median” indicates median survival time; survival curves were compared using double-sided log rank test.
A prognostic relevance of +4 alone in comparison to other intermediate risk group AML could not be shown (n = 8 vs 663 patients; median OS 320 vs 335 days, P = .509). Case numbers for sole -Y were too small for a separate analysis and thus subsequently -Y was combined with +4 to 1 group. No prognostic effect could be shown when +4 and -Y were combined (n = 14 vs 657; median OS 320 vs 335 days, P = .396). Thus, the unfavorable prognostic effect of KIT-D816 mutations in t(8,21) was independent of trisomy 4 or Y-chromosome loss (P < .001).
Sensitivity of KIT-D816V and KIT-WT to different PTK inhibitors
The activating KIT-D816V mutation is reported to be resistant to the selective PTK inhibitor imatinib that inhibits wild-type and juxtamembrane mutants of KIT.29 To test the sensitivity of the KIT-D816 mutant to different selective PTK inhibitors, the cDNAs for KIT-WT and KIT-D816V subcloned in pMSCV-IRES-eYFP mammalian expression vector were stably transfected in Ba/F3 cells as described before.5
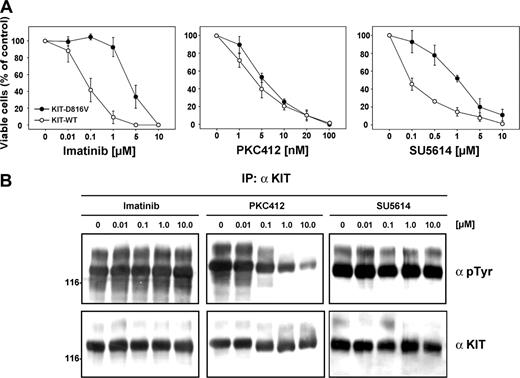
In addition to imatinib, we used PKC412 and SU5614, which have been reported to inhibit KIT-WT.23,30 We carried out proliferation assays in the presence and absence of these 3 inhibitors (Figure 4A). As described previously,29 the KIT-D816V mutant was resistant to imatinib (50% inhibitory concentration [IC50] = 3 μM) and SU5614 (IC50 = 1 μM) when compared with KIT-WT (IC50 = 0.05 μM and 0.1 μM, respectively). In contrast, PKC412 inhibited both KIT-WT and KIT-D816V with high similar efficiency in the nanomolar range (IC50 = 3 and 5 nM, respectively). To confirm these findings, we analyzed the effects of the PTK inhibitors on the autophosphorylation of the KIT-D816V receptor biochemically after transient transfection of 293 cells (Figure 4B). We could demonstrate by immunoprecipitation and Western blot analysis that only PKC412 treatment showed significant effects on the autophosphorylation of the KIT-D816V mutant. The IC50 of PKC412-induced receptor dephosphorylation was 240 nM as assessed by 3 independent experiments. This result is in line with the reported IC50 of 528 nM for the FLT3 receptor,31 another class III receptor tyrosine kinase and target of PKC412. In contrast, no significant dephosphorylation could be observed after incubation with imatinib or SU5614, even at high concentrations.
Effects of specific PTK inhibitors on KIT-transfected Ba/F3 cells. (A) Dose-response curves of the inhibitory activity of the PTK inhibitors imatinib, PKC412, and SU5614 in Ba/F3 KIT-WT and Ba/F3 KIT-D816V cells after 72 hours of incubation. Ba/F3 cells were seeded at a density of 0.05 × 106 cells/mL in the absence or presence of varying concentrations of imatinib, PKC412, and SU5614 and in the presence of 100 ng/mL recombinant human stem cell factor (SCF) in the case of KIT-WT. Viable cells were counted after 72 hours by trypan blue exclusion. The growth of untreated cells was defined as 100%. All values represent means and standard deviations from 3 independent experiments. (B) The 293 cells transfected with KIT-D816V were starved for 12 hours; treated with the indicated concentrations of imatinib, PKC412, and SU5614 for 2 hours; and lysed. Three hundred micrograms of each lysate was immunoprecipitated (IP) with α KIT antibody (α KIT) and immunoprecipitates were analyzed by Western blotting using antiphosphotyrosine antibody (αpTyr) and αKIT antibody.
Effects of specific PTK inhibitors on KIT-transfected Ba/F3 cells. (A) Dose-response curves of the inhibitory activity of the PTK inhibitors imatinib, PKC412, and SU5614 in Ba/F3 KIT-WT and Ba/F3 KIT-D816V cells after 72 hours of incubation. Ba/F3 cells were seeded at a density of 0.05 × 106 cells/mL in the absence or presence of varying concentrations of imatinib, PKC412, and SU5614 and in the presence of 100 ng/mL recombinant human stem cell factor (SCF) in the case of KIT-WT. Viable cells were counted after 72 hours by trypan blue exclusion. The growth of untreated cells was defined as 100%. All values represent means and standard deviations from 3 independent experiments. (B) The 293 cells transfected with KIT-D816V were starved for 12 hours; treated with the indicated concentrations of imatinib, PKC412, and SU5614 for 2 hours; and lysed. Three hundred micrograms of each lysate was immunoprecipitated (IP) with α KIT antibody (α KIT) and immunoprecipitates were analyzed by Western blotting using antiphosphotyrosine antibody (αpTyr) and αKIT antibody.
In order to further investigate the mechanism by which the compounds used in our study inhibit the proliferation of transformed Ba/F3 cells, we conducted cell cycle and apoptosis analyses using PI staining (Figure 5). After treatment for 24 hours with PKC412, both SCF-stimulated KIT-WT cells as well as KIT-D816V-transduced cells underwent a rapid increase in the percentage of apoptotic cells that was dose dependent (Figure 5A,C). Conversely, treatment with imatinib only led to a rise in the number of apoptotic nuclei in KIT-WT cells, whereas cells with KIT-D816V were absolutely resistant to this compound up to high doses. In both cell lines, PKC412 induced a significant accumulation of cells in G1/G0 phase (Figure 5B; data not shown). Surprisingly, no G1/G0 arrest was observed in SCF-stimulated KIT-WT cells that were treated with increasing doses of imatinib (data not shown), although this compound significantly increased the number of apoptotic nuclei (Figure 5A). This result, however, is congruent with the reported fact that some small cell lung cancer cell lines responding to imatinib are slowed in G2/M phase but are not arrested in G1/S phase.32
These data indicate that PKC412 has significant inhibitory activity against KIT receptors carrying a point mutation at Asp816 and might be of therapeutic benefit in patients with inferior-prognosis AML1-ETO/KIT-D816V-positive AML.
Discussion
Mutations or rearrangements of genes encoding transcription factors and tyrosine kinases represent 2 classes of the most frequent genetic aberrations in leukemia. Recently, it has been postulated that one of each class of these mutations cooperates in a 2-hit model to initiate a leukemic phenotype.33 Mutations of these 2 types seem to be of different efficacy in different combinations. For instance, FLT3-length mutations (FLT3-LMs) frequently occur together with t(15;17)/PML-RARA and t(6;9) DEK/CAN but are extremely rare in t(8;21)/AML1-ETO or inv(16)/CBFB-MYH11.21,34 Thus, FLT3-LMs seem to cooperate most effectively with PML-RARA or DEK-CAN and some experimental evidence already supports this hypothesis.15 In contrast, core binding factor leukemias have been described to frequently have KIT mutations.6
Cell cycle analyses of Ba/F3 KIT-D816V and KIT-WT cells after inhibitor treatment. (A) Ba/F3 cells expressing KIT-WT and KIT-D816V were grown for 24 hours in presence or absence of the indicated doses of PKC412 and imatinib and subsequently analyzed by propidium iodide (PI) staining of cell nuclei. Sub-G1 nuclei that had lost parts of their DNA by fragmentation were defined as apoptotic. KIT-WT cells were cultured in the presence of 100 ng/mL SCF. All values represent means and standard deviations from 3 independent experiments. (B) KIT-D816V cells treated with the indicated concentrations of PKC412 and imatinib were analyzed for cell cycle distribution. All values represent means and SDs from 3 independent experiments. (C) Representative histograms from flow cytometric analysis of PI-stained nuclei of Ba/F3 cells expressing KIT-D816V.
Cell cycle analyses of Ba/F3 KIT-D816V and KIT-WT cells after inhibitor treatment. (A) Ba/F3 cells expressing KIT-WT and KIT-D816V were grown for 24 hours in presence or absence of the indicated doses of PKC412 and imatinib and subsequently analyzed by propidium iodide (PI) staining of cell nuclei. Sub-G1 nuclei that had lost parts of their DNA by fragmentation were defined as apoptotic. KIT-WT cells were cultured in the presence of 100 ng/mL SCF. All values represent means and standard deviations from 3 independent experiments. (B) KIT-D816V cells treated with the indicated concentrations of PKC412 and imatinib were analyzed for cell cycle distribution. All values represent means and SDs from 3 independent experiments. (C) Representative histograms from flow cytometric analysis of PI-stained nuclei of Ba/F3 cells expressing KIT-D816V.
While KIT exon 8 mutations were frequently described in CBFB-MYH11-positive AML,3,5 we provide evidence that KIT- D816 mutations represent important cooperative mutations in AML1-ETO-positive AML. Despite an overall low frequency of KIT mutations in AML, they were found relatively frequently in AML1-ETO-positive AML. Importantly, these mutations are associated with a poor prognosis in AML1-ETO-positive AML but not in patients with a normal karyotype.
Despite the relatively high frequency of 10.5% [8 of 76 patients with t(8;21)] KIT-D816 mutations in t(8;21)-positive AML detected in our cohort, even higher frequencies of 40% (6 of 15 patients with core binding factor leukemias) and 17% [9 patients of 54 with t(8;21)] were described in previous studies.7,8 This may be based on different ethnic background or different selection criteria. Patients treated within the AMLCG study are relatively old (median age 60 years in the present cohort). It was already described that the frequency of cytogenetic subgroups differs between younger and elderly AML patients.8,35
In t(8;21)/KIT-positive AML the AML1-ETO depicts the socalled class II mutation that leads to a block in differentiation, whereas the KIT mutation represents the class I mutation that triggers excessive proliferation of the aberrant cell clone. An optimal cooperation of these 2 mutations could be postulated, as it could be shown that t(8;21) with KIT-D816 mutation had a significantly worse outcome compared with those without KIT-D816 mutations. The data presented in our study point to a tight association of KIT-D816 mutations and t(8;21), whereas other RTK mutations, FLT3-LM and FLT3-TKD, were more frequent in samples with t(15;17) and normal karyotype.
Unlike in AML1-ETO-positive AML an unfavorable impact of KIT-D816 on prognosis could not be shown in the normal-karyotype AML. A reason may be that normal-karyotype AML is a heterogenous group with different molecular mutations. A better subclassification of this group may be needed to work out a potential prognostic impact of KIT-D816 mutations in this cohort. The unfavorable impact of KIT-D816 mutations on the prognosis of patients with AML1-ETO was significant and might have direct therapeutic consequences. Patients with t(8;21)/AML1-ETO and inv(16)/CBFB-MYH11 are commonly treated with conventional chemotherapy. Allogeneic transplantation is reserved for the relapse only. The unfavorable prognosis associated with KIT-D816 mutations in AML1-ETO-positive AML shows that these patients might benefit from early hematopoietic cell transplantation (HCT). As indicated by the in vitro results presented in this study, these patients could also be considered for treatment with specific PTK inhibitors like PKC412.
An association of trisomy 4 in t(8;21)-positive AML with KIT-D816 mutations has been described previously.26,36 KIT is localized on chromosome 4 and thus trisomy 4 leads to an increased gene dosage of KIT. The Kasumi-1 cell line that is positive for AML1-ETO carries trisomy 4 and an amplification of KIT could be shown.37 The mechanism of an elevated mutation rate is still unclear but has previously been described also for MLL-PTD in trisomy 1138 and AML1 mutations in trisomy 21.39 The elevated mutation rate might be due to an up-regulation of the KIT gene at the genomic level, which has recently been described for seminomas.40
Recently, Schmidt et al41 reported mutations in the MET protooncogene in papillary renal carcinomas. This disease is characterized by trisomy of chromosome 7 containing the MET gene, trisomy of chromosomes 16 and 17, and the loss of Y chromosome in men. One of the affected positions, MET-D1246, was found to be homologous to KIT-D816 so that this aspartate residue in tyrosine kinases of different families might define a hot spot for somatic or germ line mutations, as shown for MET-D1246H and MET-D1246N, respectively. Thus, this nonhematopoietic malignancy has some striking parallels to AML positive for KIT-D816 mutation: the relevant gene is duplicated by a trisomy of the respective chromosome, one point mutation occurring is homologous to KIT-D816, and the disease is associated with the loss of the Y chromosome.
Another possible mechanism of cooperation between AML1-ETO and KIT could rely on a potential direct interaction of AML1 with the KIT promoter. Several studies have shown that AML1-ETO acts as a dominant-negative regulator of AML1 target genes.9,42 To our knowledge, no direct interactions between AML1 or AML1-ETO as transcription factors and the KIT gene have been reported yet, but it should be noted that the expression of the ETV6-AML1 fusion protein in t(12;21)-positive acute lymphoblastic leukemia led to an increase in primitive c-kit-positive multipotent progenitors in a murine bone marrow transplantation model.43 In addition, Wang et al8 have identified 11 types of KIT mutations including 6 previously undescribed ones among 26 (48.1%) of 54 cases with t(8;21). They could provide evidence that KIT seems to be the second but crucial genetic hit in AML1-ETO-positive AML. These authors hypothesized that up-regulation of KIT protein may be an alternative mechanism to activation of KIT by mutation and this may explain the higher KIT expression in patients with t(8;21).
In the present study, we were able to show, using in vitro systems, that the transforming KIT-D816V mutation was susceptible to inhibition by PKC412 in contrast to imatinib and SU5614.
Receptor tyrosine kinases like KIT are able to activate complex intracellular signal transduction pathways upon ligand binding and dimerization by catalyzing the transfer of the γ-phosphoryl group of adenosine triphosphate (ATP) to tyrosine residues of adaptor and target proteins.1,44 Selective PTK inhibitors like imatinib and PKC412 that compete with ATP have been successfully used in the treatment of patients with acute and chronic myeloid leukemias.31
We have shown that activating mutations in the activation loop of KIT (D816V) were strongly associated with AML1-ETO-positive AML and a poor clinical outcome. As described previously, the KIT-D816V mutant induced IL-3-independent growth in Ba/F3 cells and was resistant to the growth inhibitory activity of imatinib and SU5614. In contrast, the PTK inhibitor PKC412 that has been described to inhibit protein kinase C (PKC), KDR, KIT, FLT3, PDGFRA, and PDGFRB30,45-47 was identified as a potent inhibitor of KIT-WT and KIT-D816V. Moreover, PKC412, but not imatinib or SU5614, induced efficient dephosphorylation of autophosphorylated KIT-D816V receptor.
These profound differences in the sensitivity of the KIT-D816V mutant to different PTK inhibitors probably rely on the binding mode of these compounds. Structural studies have shown that imatinib binds the kinase domain of KIT as well as ABL and PDGFRA in the inactive (ie, catalytically quiescent) state.47-49 A point mutation within the activation loop like Asp816Val in KIT might confer constitutive activation and resistance to imatinib by destabilizing the equilibrium between the active and inactive state of the kinase.48 In contrast, PKC412 is thought to bind within the ATP binding pocket of the active conformation of PDGFRA,47 which is probably also the case for the KIT receptor.
These data provide a structural basis for the sensitivity of the KIT-D816V mutant to PKC412 and support our hypothesis that this compound might have therapeutic activity in patients with KIT-D816V-positive AML. PKC412 has recently been evaluated as a single agent in a phase 1 clinical trial for clinical activity in 32 patients with a variety of solid tumors50 and it has clinical activity in AML patients carrying activating FLT3 mutations.51 A combination of conventional chemotherapy with PTK inhibitors might be an attractive therapeutic option for patients with poor-prognosis KIT-D816V-positive/AML1-ETO-positive AML mutations.
Prepublished online as Blood First Edition Paper, October 27, 2005; DOI 10.1182/blood-2005-04-1466.
Supported in part by grants from the Nationales Genomforschungsnetz (NGFN) (Bundesministerium für Bildung und Forschung [BMBF] NGFN-Förderkennzeichen 01GSS0105-Teilprojekt 2) and the Deutsche Forschungsgemeinschaft (DFG Sp 556/1-3).
S.S. and K.S. are the principal investigators; S.S., T.M.K., W.K., and C.S. analyzed data; S.S., C.S., K.S., W.H., and T.H. contributed to the design of the study; and S.S. and T.M.K. conducted the work and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We greatly acknowledge the excellent technical work of Gudrun Mellert, Claudia Tschulik, Madlen Fuchs, Nina Leopold, and Theresa Förster. We would like to thank Novartis for kindly providing the compounds imatinib mesylate and PKC412 for the in vitro studies.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal