Circulating antiplasmin-cleaving enzyme (APCE) has a role in fibrinolysis and appears structurally similar to fibroblast activation protein (FAP), a cell-surface proteinase that promotes invasiveness of certain epithelial cancers. To explore this potential relationship, we performed comparative structure/function analyses of the 2 enzymes. APCE from human plasma and recombinant FAP (rFAP) exhibited identical pH optima of 7.5, extinction coefficients (
Introduction
Human α2-antiplasmin (α2AP) becomes rapidly cross-linked to fibrin by activated clotting factor XIII (FXIIIa) and protects fibrin by rapidly inhibiting plasmin within the fibrin clot.1,2 Fibrin containing reduced α2AP activity is susceptible to fibrinolysis, whereas fibrin containing a normal amount of α2AP is strikingly resistant to lysis by plasmin.2-6 Human α2AP was thought to have an N-terminal Asn and a 39-residue signal peptide until later work demonstrated that α2AP was secreted from liver cells as a 464-residue protein with an additional 12 residues at the N-terminus (Met-α2AP), making the signal peptide 27 residues.7-10 The N-terminal 12-residue peptide of Met-α2AP was reported to be cleaved in the circulation by an unknown proteinase.10 In normal human plasma, the ratio of Met-α2AP to Asn-α2AP was estimated as about 30%:70%.9,10 We showed that during clot formation Asn-α2AP becomes cross-linked to fibrin by FXIIIa about 13 times faster than Met-α2AP, and as a consequence, clot lysis rates slow in direct proportion to the ratio of Asn-α2AP/Met-α2AP in human plasma.5
We purified the plasma proteinase that converts Met-α2AP to Asn-α2AP and termed it antiplasmin-cleaving enzyme (APCE).5 Its N-terminal amino acid sequence corresponded to residues 24-38 of fibroblast activation protein (FAP), also known as seprase, a type II integral membrane glycoprotein, which is predicted to have its first 6 N-terminal residues within the fibroblast cytoplasm, followed by a 20-residue transmembrane domain, and then a 734-residue extracellular C-terminal catalytic domain.5,11,12 FAP is a proline-specific enzyme that exhibits gelatinase activity as well as dipeptidyl peptidase activity.13 FAP and dipeptidyl peptidase IV (DPP IV), which is another post-prolyl peptidase, are members of the S9b peptidase family in which the order of Ser/Asp/His in the active site is different from that in the chymotrypsin-like serine proteinase family.14 FAP is reported to be expressed by fibroblasts of reactive stroma of epithelial-derived cancers, but not by counterpart normal tissues.13,15,16 Subsequent studies showed that FAP is also expressed by several types of neoplastic parenchymal cancer cells and also by endothelial cells of newly formed vessels within certain neoplasms.17-21 In a murine breast cancer model, cells transfected to express FAP form more rapidly growing tumors when compared to control transfectants.22 Although these findings suggest that FAP may be a potential target for diagnostic and therapeutic approaches in certain cancers,23 its biologic function remains unknown. To date no certain physiologic substrate for FAP has been identified.
Schmidt et al24 indicated that no detectable soluble FAP, or a derivative of such, could be found in human blood; however, our finding of APCE in human plasma5 is inconsistent with this. Interestingly, Collins et al25 have found FAP-like activity in bovine serum, which is in accord with our report of APCE in human plasma. APCE may be a soluble, circulating derivative of FAP, resulting from cleavage of the Cys23-Ile24 bond in the transmembrane or extracellular domain of FAP, and if so, its presence in plasma could have an impact on the use of therapeutic agents designed to target FAP at the tissue level.23,24 We now report direct evidence that the 2 proteins are essentially identical. We obtained sufficient quantities of APCE from human plasma and soluble human rFAP from a yeast expression system to allow determinations and comparisons of previously unreported molecular structure/function details for the 2 proteinases.
Materials and methods
Materials
Human plasma Met-α2AP was isolated from a mixture of Met-α2AP and Asn-α2AP using Met-α2AP–specific antibody as previously described5 ; a percent extinction coefficient of α2AP at 280 nm of 6.70 and a molecular weight (MW) of 70 000 were used to determine molar concentration.26 Hybridoma cells secreting the F19 monoclonal antibody to FAP were purchased from American Type Culture Collection (Manassas, VA) and grown in serum-free media; the F19 antibody was purified from hybridoma cell culture media using MEP-Hypercel chromatography (Pall, East Hills, NY). An expression sequence tagged (EST) cDNA clone (GenBank accession no. AV698972) was obtained from Dr Ze-Guan Han, through the Chinese National Human Genome Center at Shanghai, China. On determining its sequence, using M13-forward and M13-reverse primers in conjunction with 4 specific primers, the EST clone was shown to contain full-length FAP cDNA (GenBank accession no. NM_004460). Other reagents included Pichia pastoris expression vector pPICZαA and P pastoris X-33 (Invitrogen, San Diego, CA), TPCK-treated trypsin (Sigma, St Louis, MO), and Z-Gly-Pro-AMC and Ala-Pro-AFC (Bachem, Torrance, CA). 7-Amido-4-methylcoumarin (AMC) and 7-amino-4-trifluoromethylcoumarin (AFC) were obtained from MP Biochemicals (Aurora, OH). A fluorescence resonance energy transfer (FRET) peptide, Arg-Lys(DABCYL)-Thr-Ser-Gly-Pro-Asn-Gln-Glu-Gln-Glu(EDANS)-Arg was synthesized in-house as previously described.5 Asn-Gln-Glu-Gln-Glu(EDANS)-Arg was prepared as follows: (1) the Pro-Asn bond of the FRET peptide was cleaved by APCE, and (2) the fluorescent fragment, Asn-Gln-Glu-Gln-Glu(EDANS)-Arg, was purified by high-performance liquid chromatography (HPLC) and confirmed by mass and amino acid sequence analyses. Institutional review board (IRB) approval was obtained from University of Oklahoma Health Sciences Center for these studies (IRB no. 12240).
Isolation of APCE
APCE was partially purified by ammonium sulfate precipitation and hydrophobic interaction chromatography as previously described.5 The activity peak was applied to an immunoaffinity column made with F19 monoclonal antibody and eluted with 5 M LiCl. Based on Z-Gly-Pro-AMC cleaving activity, the active fractions were pooled and brought to 6% ammonium sulfate, applied to a phenyl-5PW column (Tosohaas, Montgomeryville, PA) equilibrated in 6% ammonium sulfate, and eluted with 12.5 mM sodium phosphate, pH 7.5. Before storing at -80°C, glycerol was added to purified APCE to give a final concentration of 20%.
Construction of rFAP expression vector and transformation of P pastoris
Human FAP cDNA with XhoI and NotI sites at the 5′ and 3′ ends, respectively, was amplified as described previously,2 using 2 oligonucleotide primers (5′-ATCTCTCGAGAAAAGAATTGTCTTACGCCCTTCAAG for the XhoI site and 5′-CAGCCGCGGCCGCTTAGTCTGACAAAGAGAAAC for the NotI site) and the template full-length FAP cDNA. Using the introduced restriction sites, the amplified DNA was ligated into the P pastoris vector pPICZαA for secretional expression. The expression vector was digested with SacI and then transformed into P pastoris X-33 by the EasyComp transformation procedure (Invitrogen instruction manual). Transformants were selected on YPDS agar plates containing Zeocin 100 μg/mL.
Expression and purification of recombinant FAP
Zeocin resistants (100 colonies) were screened by determining rFAP activity in expression culture medium. The 5-mL cultures were grown overnight in buffered glycerol complex medium (BMGY medium) at 29°C and cells were harvested by centrifugation at 2000g. Cell pellets were resuspended with 5 mL BMMY medium, which is BMGY with 5 mL methanol per liter in place of glycerol, and then induction was continued with fresh methanol (5 mL/L) added every 24 hours for 3 days. Z-Gly-Pro-AMC cleaving activity in the culture medium was monitored.
The yeast cells displaying highest activity were grown to A600 of 2 to 3 in 200 mL BMGY medium in a 2-L baffled flask at 29°C. The cells were harvested and resuspended in 200 mL BMMY medium and cultured at 29°C for 5 days and then centrifuged at 12 800g for 40 minutes at 4°C to pellet the yeast cells. The supernatant was filtered through 0.1-μm Millipore membranes to remove solid residuals and purified by the same procedure as described for APCE.
Size exclusion chromatography
Native molecular mass for rFAP or APCE was determined at room temperature on a Superdex-200 PC 3.2/30 column (Amersham Bioscience, Piscataway, NJ), using 50 mM sodium phosphate-150 mM NaCl, pH 7.4, at a flow rate of 45 μL/min. Commercially obtained protein standards (Bio-Rad, Hercules, CA) were used for calibration.
Extinction coefficient
APCE or rFAP concentration was determined by amino acid analysis and absorbance at 280 nm measured in 12.5 mM sodium phosphate buffer, pH 7.5. Percent extinction coefficient was expressed as (ϵmolar)10 = (ϵpercent) × MW, where ϵmolar (A280 nm/M) and ϵpercent (
Enzyme assays
APCE or rFAP was incubated in 25 mM sodium phosphate buffer, pH 7.5, containing 1.0 mM EDTA and 4% methanol in a total volume of 200 μL for 20 minutes at 22°C, using as substrates Z-Gly-Pro-AMC (25-400 μM), Ala-Pro-AFC (50-800 μM), or a FRET peptide, Arg-Lys(DABCYL)-Thr-Ser-Gly-Pro-Asn-Gln-Glu-Gln-Glu(EDANS)-Arg (5-80 μM) that was designed to contain the APCE-sensitive Pro12-Asn13 bond within the Thr9-Gln16 sequence of Met-α2AP.5 Fluorescence was monitored with time at excitation and emission wavelengths of 360 and 460 nm for Z-Gly-Pro-AMC and the FRET peptide, and 360 and 530 nm for Ala-Pro-AFC, using a BIO-TEK FL600 fluorescence plate reader (Winooski, VT). To obtain standard curves, dilutions of AMC, AFC, and Asn-Gln-Glu-Gln-Glu(EDANS)-Arg were prepared in the same assay buffer and corresponding fluorescence was measured. All enzyme kinetic parameters were computed by fitting data to the Michaelis-Menten equation,27 using the commercially available program EZ-FIT (Perrella Scientific, Amherst, NH).
Zymography to measure gelatinase activity of APCE or rFAP was performed using gelatin zymograms, and renaturing and developing solutions (Invitrogen). Sample loading, electrophoresis, and incubation of each gel in the renaturing and developing buffers were carried out according to the accompanying protocol. After overnight incubation in the developing buffer at 37°C, each gel was stained with Coomassie brilliant blue G-250 to see the area of gelatin digestion as a clear band and rFAP or APCE protein as a darker band in the background.
LC/MS analysis of Met-α2AP digested by rFAP or APCE
Selected concentrations of Met-α2AP were incubated with 0.85 μg rFAP or APCE in 100 μL of 25 mM sodium phosphate-1.0 mM EDTA buffer, pH 7.5. After incubating for 5 hours, an internal standard peptide (FRQLTSKPNQEQV, 0.113 μg, 144 pmol) was dissolved in the reaction mixture and cold acetone (4 × sample volume) was added to precipitate proteins. The sample was incubated at -80°C for 60 minutes and centrifuged for 10 minutes at 16 000g at 4°C to remove precipitated proteins. The supernatant was removed and dried by vacuum centrifugation and dissolved in 5% acetic acid.
Liquid chromatography-mass spectrometry (LC/MS) analysis of samples was performed on a Magic 2002 HPLC system (Michrom BioResources, Auburn, CA) equipped with a Jupiter Proteo reverse-phase column (1.0 mm × 150 mm; Phenomenex, Torrance, CA). The column was equilibrated with 5% acetonitrile/water containing 0.1% TFA. On sample injection, the solvent composition was increased to 10% acetonitrile and a linear gradient was applied to 40% acetonitrile over 60 minutes. Peptides were detected at 215-nm wavelength. The HPLC apparatus was connected to an electrospray mass spectrometer (QSTAR; Applied Biosystems, Foster City, CA) equipped with an ion spray source operated in the positive ion mode. Scanning was performed over an m/z range of 350 to 1800 amu. Both the internal standard peptide and peptide products of digestion were located by reconstructed ion current analysis of data for each peptide over a 1.5 amu window for both the singly and doubly charged forms of each peptide, based on the peptide's predicted monoisotopic molecular mass. Quantitation was performed by summing all detected isotopes of each peptide.
Results
Expression and purification of rFAP
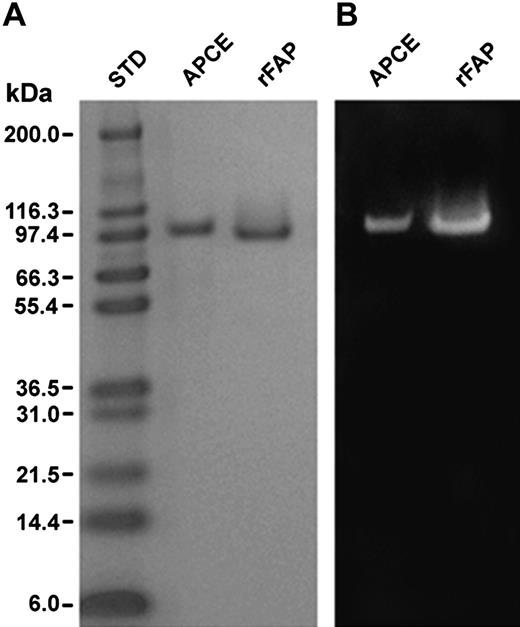
We have suggested that APCE may be the N-terminally cleaved form of FAP, partially because the N-terminal 15 amino acids of APCE are identical to residues 24-38 of FAP.5 Therefore, the 23 residues were removed by amplifying the coding region of full-length FAP cDNA between amino acid residues 24 and 760. Using the amplified DNA and P pastoris vector pPICZαA, the expression vector was constructed for secretion of rFAP into media. Figure 1A shows that purified rFAP or APCE gives a single band of 93 kDa or 97 kDa, respectively, on reduced sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Identical patterns for both enzymes were also observed in the absence of reducing agent. Figure 1B shows that the F19 monoclonal antibody to FAP13 cross-reacts with both APCE and rFAP.
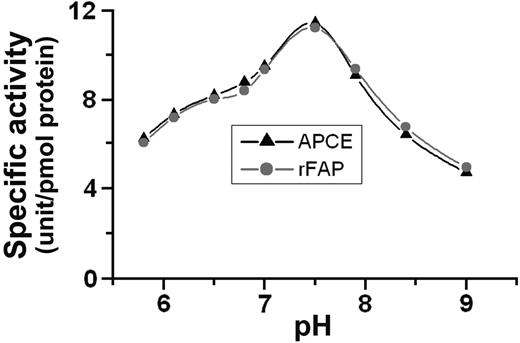
Our purification process for rFAP produced about 42 μg of pure protein from 1 L culture media to give a 17% yield. Protein concentration of rFAP or APCE was calculated using percent extinction coefficients of 20.5 or 20.2, respectively, at 280 nm as determined by us. Figure 2 shows the effects of pH on the specific activity of rFAP and APCE. At this time we have no explanation for the wide range of enzyme activity over the pH spectrum examined these studies. For each enzyme, the pH optimum was 7.5 for the cleavage of the Pro-Asn bond in the FRET peptide substrate, Arg-Lys(DABCYL)-Thr-Ser-Gly-Pro-Asn-Gln-Glu-Gln-Glu(EDANS)-Arg. Both enzymes also showed the same pH optima with the substrate Z-Gly-Pro-AMC (data not shown).
SDS-PAGE and Western blot analysis of purified APCE and rFAP. (A) Coomassie-stained reduced SDS-PAGE analysis on a 4% to 12% gradient gel. (B) Western blot of a duplicate gel to that in panel A, using a monoclonal antibody (F19) to native FAP. Although the loading in each lane (1.4 μg protein) is equivalent, the F19 antibody appears to bind rFAP to a greater extent, possibly relating to differences in glycosylation.
SDS-PAGE and Western blot analysis of purified APCE and rFAP. (A) Coomassie-stained reduced SDS-PAGE analysis on a 4% to 12% gradient gel. (B) Western blot of a duplicate gel to that in panel A, using a monoclonal antibody (F19) to native FAP. Although the loading in each lane (1.4 μg protein) is equivalent, the F19 antibody appears to bind rFAP to a greater extent, possibly relating to differences in glycosylation.
pH curve. FRET peptide, Arg-Lys(DABCYL)-Thr-Ser-Gly-Pro-Asn-Gln-Glu-Gln-Glu(EDANS)-Arg was incubated with purified APCE or rFAP at the indicated pH. Two buffer systems were used: 0.1 M sodium phosphate for the pH range 5.8-7.5 and 0.1 M Tris-HCl for the pH range 7.9-9.0; each value represents the average of duplicate incubations. One unit of enzyme activity is defined as picomoles of Asn-Gln-Glu-Gln-Glu(EDANS)-Arg released per minute at 22°C.
pH curve. FRET peptide, Arg-Lys(DABCYL)-Thr-Ser-Gly-Pro-Asn-Gln-Glu-Gln-Glu(EDANS)-Arg was incubated with purified APCE or rFAP at the indicated pH. Two buffer systems were used: 0.1 M sodium phosphate for the pH range 5.8-7.5 and 0.1 M Tris-HCl for the pH range 7.9-9.0; each value represents the average of duplicate incubations. One unit of enzyme activity is defined as picomoles of Asn-Gln-Glu-Gln-Glu(EDANS)-Arg released per minute at 22°C.
Sequence analysis of rFAP and APCE
The N-terminal amino acid sequences of rFAP and APCE were determined by using an automated protein sequencer. As shown in Figure 3, the N-terminal sequence of purified rFAP (EENTMRALTL, FAP residues 35-44) is different from the N-terminal sequence of APCE (IVLRPSPVHNSEENT, FAP residues 24-38). Although the N-terminal amino acid of rFAP was expected to be Ile24, in fact, it was Glu35. Various constructs were designed with a His(6)- or Glu-Ala-His (6)-tag attached N-terminally, or not at all, and every instance resulted in the expression of a stable, active enzyme, but with the same dominant N-terminal sequence. A dominant amino acid sequence of 10 amino acid residues (EENTMRALTL, residues 35-44) and a minor sequence containing residues 29-37 (SRVHNXEEN) were obtained in about an 80:20 ratio, indicating the homogeneity of purified rFAP and suggesting that 11 N-terminal residues had been deleted or cleaved during culture or enzyme preparation. As can be seen in Figure 1A, molecular size determination of rFAP (93 kDa) and APCE (97 kDa) by SDS-PAGE support rFAP as being smaller, probably due to glycosylation differences between human and yeast-expressed proteins and the deletion of 11 N-terminal residues. To compare the internal sequences of rFAP and APCE, each protein was subjected to SDS-PAGE, reduced, alkylated, and digested with trypsin.28 Each trypsin digest was analyzed by LC/MS-mass spectrometry (LC/MS/MS) to obtain molecular weights and MS/MS fragment ion spectra to use with the MASCOT MS/MS ion search engine to query the NCBI comprehensive nonidentical protein database (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=MIS). As shown in Figure 3, amino acid sequences of peptides obtained from trypsin digests of rFAP and APCE are identical to the sequence derived from FAP cDNA sequence.
Homodimer formation required for APCE or rFAP activity
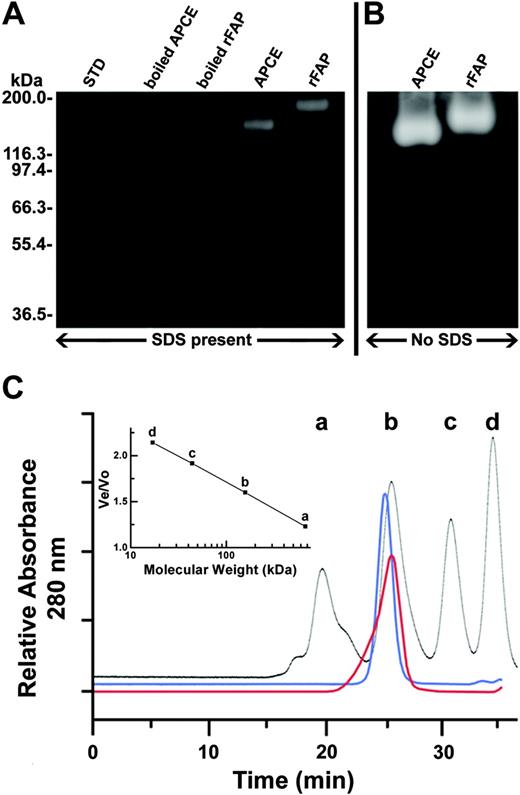
FAP appears to be composed of two 97-kDa identical subunits in a homodimeric structure, suggested as necessary for both dipeptidyl peptidase and gelatinase activities.11,13 Definitive data, however, showing that dimerization is required for activity are lacking. As shown in Figure 4, we examined this directly using zymography and size exclusion chromatography. When boiled in SDS, APCE and rFAP showed 97- and 93-kDa bands, respectively, and given complete denaturation, no gelatinase activity was observed (Figure 4A). However, nonboiled samples, from which SDS was removed by incubating the zymogram-gelatin gel in a renaturing buffer after electrophoresis (Figure 4A), clearly showed both 170-kDa and 180-kDa gelatinase activities for APCE and rFAP, respectively. Also noted were remaining inactive 97-kDa and 93-kDa monomer bands for both enzymes, which despite renaturation efforts, lacked gelatinase activity. The molecular size of APCE was consistent with prior reports of 175 kDa for enzymatically active FAP homodimer and 95 kDa for the inactive monomer.11,13,29 Without SDS in the samples, the gelatinase activity band of rFAP migrated slightly slower than that of APCE, and both band intensities increased about 20-fold compared with nonboiled SDS-containing sample bands; monomeric bands were not apparent (Figure 4B). Although by SDS-gel electrophoreses the monomer band of APCE migrates more slowly than that of rFAP (Figure 4A), the dimeric form of APCE migrates faster than that of rFAP in the non-SDS gel (Figure 4B). We speculate that this may be due to the negative surface charge of APCE being higher than for rFAP due to differences in glycosylation. Similar size discrepancy has been reported in native and recombinant DPP IV proteins, which are in the same S9b peptidase family as FAP.30 Using size exclusion chromatography, we also assessed whether APCE or rFAP remains as an enzymatically active dimer in phosphate-buffered saline, pH 7.4. As shown in Figure 4C, APCE is present as a 175-kDa protein peak, whereas rFAP gives a 153-kDa peak. No other peaks are present, particularly in the monomeric 95-kDa range. The rFAP peak showed a broader mass distribution than APCE that is probably due to heterogeneous glycosylation by the yeast-expression system.31 The broader peak observed for rFAP on size exclusion chromatography is also consistent with the broader band for rFAP by SDS-PAGE when compared to APCE (Figure 1). Both the 175-kDa and 153-kDa peak fractions have gelatinase and Ala-Pro-AFC–cleaving activities. These data from zymography and size exclusion chromatography suggest that homodimers are required for enzymatic activity and monomers lack such function.
Sequence alignment. FAP sequence (black) from GenBank (accession no. NM_004460) aligned with APCE (blue) and rFAP (red) N-terminal sequences and tryptic peptides characterized by LC/MS/MS.
Sequence alignment. FAP sequence (black) from GenBank (accession no. NM_004460) aligned with APCE (blue) and rFAP (red) N-terminal sequences and tryptic peptides characterized by LC/MS/MS.
Comparison of APCE with rFAP for molecular size and gelatinase activity. A protein load of 1.4 μg was applied to each electrophoretic lane. Each was stained with Coomassie brilliant blue G-250. Clear zones are due to gelatinase activity. (A) Electrophoresis in the presence of SDS on polyacrylamide gel containing copolymerized gelatin. (B) Electrophoresis in the absence of SDS, but on the identical type of gel. Comparable gelatinolysis is observed for the 2 enzymes; however, the mobilities of the single broad zones of lysis differ slightly and cannot be related to molecular weight standards because electrophoresis was done without SDS. (C) Analytical size exclusion chromatography of APCE (blue) and rFAP (red). Standard (black): a, thyroglobulin (670 kDa); b, γ-globulin (158 kDa); c, ovalbumin (44 kDa); and d, myoglobin (17 kDa). The inset shows a standard plot of the logarithmic molecular weight against the elution volume (Ve)/void volume (Vo).
Comparison of APCE with rFAP for molecular size and gelatinase activity. A protein load of 1.4 μg was applied to each electrophoretic lane. Each was stained with Coomassie brilliant blue G-250. Clear zones are due to gelatinase activity. (A) Electrophoresis in the presence of SDS on polyacrylamide gel containing copolymerized gelatin. (B) Electrophoresis in the absence of SDS, but on the identical type of gel. Comparable gelatinolysis is observed for the 2 enzymes; however, the mobilities of the single broad zones of lysis differ slightly and cannot be related to molecular weight standards because electrophoresis was done without SDS. (C) Analytical size exclusion chromatography of APCE (blue) and rFAP (red). Standard (black): a, thyroglobulin (670 kDa); b, γ-globulin (158 kDa); c, ovalbumin (44 kDa); and d, myoglobin (17 kDa). The inset shows a standard plot of the logarithmic molecular weight against the elution volume (Ve)/void volume (Vo).
Substrate specificity of rFAP and APCE
To determine if rFAP has the same specificity as APCE, enzymatic activities of rFAP and APCE were compared using selected synthetic peptides as substrates (Table 1). These analyses clearly demonstrated that rFAP possesses the same, or closely similar, kinetic parameters as APCE. For Z-Gly-Pro-AMC substrate, human rFAP and APCE have Km values of 0.101 mM and 0.124 mM. These values are about 2-fold smaller when compared to a Km of 0.270 mM for bovine FAP toward Z-Gly-Pro-AMC.25 Using Z-Gly-Pro-AMC, we determined kcat values for human rFAP and APCE to be 30.5 minute-1 and 32.2 minute-1; kcat for bovine FAP has not been reported. For Ala-Pro-AFC substrate, human rFAP and APCE have Km and kcat values of 0.323 mM and 64.7 minute-1 and 0.272 mM and 59.7 minute-1, respectively, in contrast to 0.20 mM and 120 minutes-1, which Sun et al32 reported for full-length human rFAP. The modest discrepancies between our data and theirs are likely due to different buffer pHs used in the 2 sets of experiments. Also, compared to their full-length rFAP, our N-terminal for rFAP was shorter by 34 amino acids, thus lacking the cytosolic and transmembrane domains, which could have diminished solubility and affected kinetic parameters. The kinetic efficiencies (kcat/Km values) of rFAP and APCE for cleavage of Z-Gly-Pro-AMC, Ala-Pro-AFC, and FRET peptide are essentially the same toward each substrate. However, the kcat/Km value for cleavage of FRET peptide, remembering that it is modeled on the sequence surrounding the APCE-cleavage site (Pro12-Asn13) of Met-α2AP, is about 8-fold higher than for Z-Gly-Pro-AMC and about 12-fold higher than for Ala-Pro-AFC.
Kinetic parameters for rFAP and APCE
. | rFAP . | . | . | APCE . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Substrate . | Km, mM . | kcat, min-1 . | kcat/Km, min-1/mM . | Km, mM . | kcat, min-1 . | kcat/Km, min-1/mM . | ||||
| Z-Gly-Pro-AMC | 0.101 ± 0.018 | 30.5 ± 3.2 | 306 ± 38 | 0.124 ± 0.024 | 32.2 ± 7.1 | 260 ± 13 | ||||
| Ala-Pro-AFC | 0.323 ± 0.052 | 64.7 ± 4.6 | 202 ± 18 | 0.272 ± 0.014 | 59.7 ± 8.1 | 218 ± 19 | ||||
| FRET peptide | 0.029 ± 0.004 | 72.2 ± 12.5 | 2491 ± 159 | 0.026 ± 0.005 | 63.4 ± 10.1 | 2445 ± 109 | ||||
. | rFAP . | . | . | APCE . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Substrate . | Km, mM . | kcat, min-1 . | kcat/Km, min-1/mM . | Km, mM . | kcat, min-1 . | kcat/Km, min-1/mM . | ||||
| Z-Gly-Pro-AMC | 0.101 ± 0.018 | 30.5 ± 3.2 | 306 ± 38 | 0.124 ± 0.024 | 32.2 ± 7.1 | 260 ± 13 | ||||
| Ala-Pro-AFC | 0.323 ± 0.052 | 64.7 ± 4.6 | 202 ± 18 | 0.272 ± 0.014 | 59.7 ± 8.1 | 218 ± 19 | ||||
| FRET peptide | 0.029 ± 0.004 | 72.2 ± 12.5 | 2491 ± 159 | 0.026 ± 0.005 | 63.4 ± 10.1 | 2445 ± 109 | ||||
Each value is the mean ± SD of 3 experiments.
FRET peptide is Arg-Lys(DABCYL)-Thr-Ser-Gly-Pro-Asn-Gln-Glu-Gln-Glu(EDANS)-Arg.
Met-α2AP cleavage by rFAP
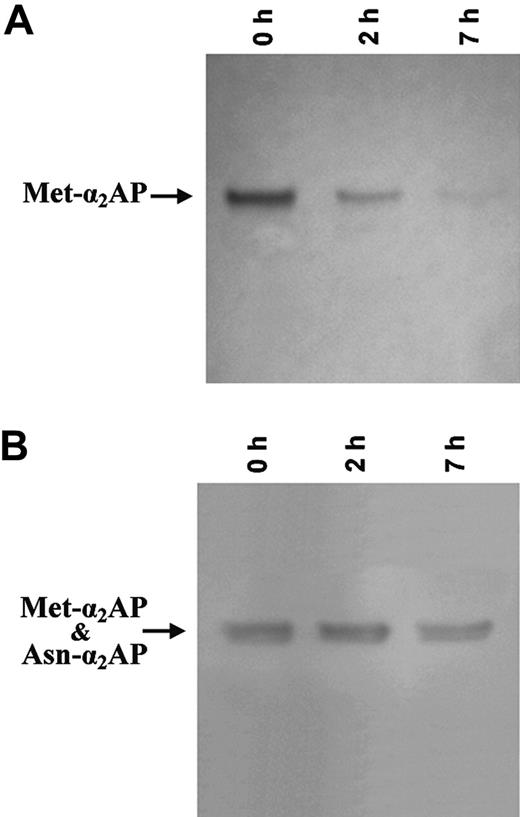
Because our kinetic studies and FRET peptide data suggested that rFAP, just as APCE,5 may specifically cleave Met-α2AP at Pro12-Asn13, rFAP was incubated with Met-α2AP and the mixture examined at selected times by Western blot analysis. As demonstrated in Figure 5A, the intensity of the Met-α2AP band decreased with time when immunoblotted with a specific antibody to the 12-residue N-terminal peptide of Met-α2AP.5 Figure 5B, however, shows that a blot immunostained with an antibody that detects both N-terminal forms of α2AP did not change through the entire time course, and bands suggesting fragmentation were never generated. N-terminal analyses of the 0-hour sample showed only the Met-α2AP sequence, MEPLGRQLTSGP, whereas the 7-hour sample had only the Asn-α2AP N-terminal sequence, NQEQVSPLTLLK. These data indicate that rFAP, like APCE, specifically cleaves the Pro12-Asn13 bond of Met-α2AP.
Immunoblots of Met-α2AP cleaved by rFAP to Asn-α2 AP. Met-α2AP was incubated with purified rFAP (0.2 μg) for selected times as shown at the top of each lane. At each time the incubation mixture was analyzed using 2 different antibodies. (A) rFAP diminished Met-α2AP band intensity with time as determined by immunoblotting with an antibody specific for the N-terminal peptide of Met-α2AP. (B) The α2AP band intensity was not diminished when blotted by an antibody to the C-terminal region common to both Met-α2AP and Asn-α2AP.
Immunoblots of Met-α2AP cleaved by rFAP to Asn-α2 AP. Met-α2AP was incubated with purified rFAP (0.2 μg) for selected times as shown at the top of each lane. At each time the incubation mixture was analyzed using 2 different antibodies. (A) rFAP diminished Met-α2AP band intensity with time as determined by immunoblotting with an antibody specific for the N-terminal peptide of Met-α2AP. (B) The α2AP band intensity was not diminished when blotted by an antibody to the C-terminal region common to both Met-α2AP and Asn-α2AP.
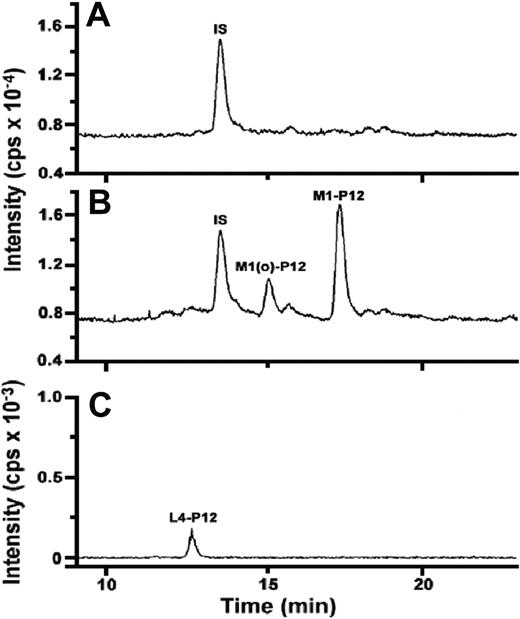
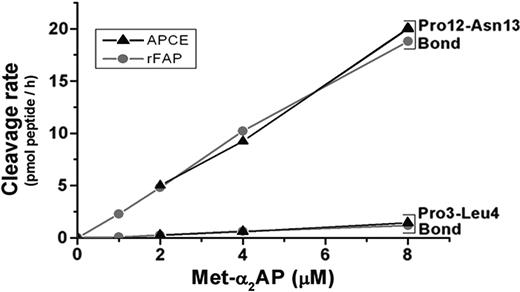
Given cleavage of the Pro12-Asn13 bond in Met-α2AP, we questioned whether the Pro3-Leu4 bond might also be cleaved during incubation with rFAP. This prompted the use of LC/MS to analyze peptides generated during incubation of rFAP with Met-α2AP. Figure 6A (internal standard peptide [IS] plus Met-α2AP incubated without rFAP) and Figure 6B (IS plus Met-α2AP incubated with rFAP) each depict LC/MS total ion current analysis of the digest between retention times of 9 and 23 minutes. In Figure 6A, IS shows the expected peak. In Figure 6B, when Met-α2AP was incubated with rFAP, the control IS and the N-terminal 12 amino acid peptide (M1-P12), MEPLGRQLTSGP, and its oxidized form, M1(o)-P12, were observed. As shown in Figure 6C, a 9-residue L4-P12 peptide (LGRQLTSGP) that is clearly a cleavage product of the N-terminal 12 amino acids was located by reconstructed ion current analysis over a 1.5-amu window for the doubly charged form of the peptide based on its predicted monoisotopic molecular mass. The amounts of M1-P12, M1(o)-P12, and L4-P12 peptides were determined by comparison of each peak area with the IS peak area, 144 pmol of IS having been added to each sample. As shown in Figure 7, cleavage rates of the Pro3-Leu4 bond and the Pro12-Asn13 bond in Met-α2AP by rFAP or APCE were determined by the amount of generated L4-P12 peptide, and the sum of L4-P12, M1-P12, and M1(o)-P12 peptides, assuming that the Pro12-Asn13 bond is cleaved before Pro3-Leu4. Cleavage rates of Pro3-Leu4 and Pro12-Asn13 increased linearly with substrate concentrations up to 8 μM. Under competitive conditions, the ratio of hydrolytic rates of each cleavage site can be described as a relative kcat/Km value.33 Based on cleavage rates of Pro3-Leu4 and Pro12-Asn13 in the presence of 2, 4, or 8 μM Met-α2AP, the relative kcat/Km values for Pro3-Leu4 and for Pro12-Asn13 were determined for each substrate concentration and averaged for each of the bond cleavages to yield: for rFAP, 1 and 16.6 ± 1.2 (mean ± standard deviation [SD]), whereas for APCE, the relative kcat/Km for Pro3-Leu4 and for Pro12-Asn13 were 1 and 16.3 ± 3.2 (mean ± SD).
LC/MS analysis of peptides released from the rFAP digest of Met-α2AP. (A) Total ion current (m/z = 350-1800 amu) of a control sample containing Met-α2AP but lacking rFAP, showing only an internal standard peak (IS). (B) Total ion current of Met-α2AP incubated with rFAP: IS, oxidized N-terminal 12 amino acid M1(o)-P12 peptide and the 12 amino acid M1-P12 peptide are observed. (C) Reconstructed ion currents (464.4-465.9 amu) of the rFAP-treated sample for the doubly charged ions represented the L4-P12 peptide, indicating the presence of an additional cleavage site within the N-terminal 12 residue peptide.
LC/MS analysis of peptides released from the rFAP digest of Met-α2AP. (A) Total ion current (m/z = 350-1800 amu) of a control sample containing Met-α2AP but lacking rFAP, showing only an internal standard peak (IS). (B) Total ion current of Met-α2AP incubated with rFAP: IS, oxidized N-terminal 12 amino acid M1(o)-P12 peptide and the 12 amino acid M1-P12 peptide are observed. (C) Reconstructed ion currents (464.4-465.9 amu) of the rFAP-treated sample for the doubly charged ions represented the L4-P12 peptide, indicating the presence of an additional cleavage site within the N-terminal 12 residue peptide.
Discussion
The major goals of this study were to determine if APCE is a soluble form of FAP and to systematically characterize the molecular and kinetic properties of both enzymes. Despite the identity of limited amino acid sequences we previously reported for APCE and FAP,5 concern remained about differences between the 2; namely, FAP has a 23-residue N-terminal extension with Met at the N-terminus, whereas APCE has Ile as its N-terminus (corresponding to Ile24 in FAP), cleaves Met-α2AP and circulates in blood, unlike what has been reported for FAP.24 Our initial approach was to develop methods for obtaining sufficient amounts of apparently homogenous APCE from human plasma for characterization. Purification yields of APCE from human plasma were improved 4-fold, using a modification of our previous method.5 To date, efforts at structure/function analyses have been limited by low yields of FAP from baculovirus or mammalian cell expression systems and, in addition, by uncertain purity. We chose the P pastoris expression system and found that rFAP was secreted into culture media from which we could obtain about 42 μg pure rFAP per liter. Pure rFAP, possessing only amino acid sequence deduced from FAP cDNA, was found to be a soluble, stable, and active enzyme. Our yield of purified protein was about 5-fold greater than that reported for insect cells with recombinant baculovirus expressing the 760-residue full-length protein.32 Chinese hamster ovary cells expressing full-length FAP provided only trace amounts of recombinant protein.34
Based on comparison of APCE and rFAP amino acid sequences to the published sequence for FAP cDNA, we found that APCE and rFAP each contained only the extracellular C-terminal catalytic domain and no N-terminal cytoplasmic or transmembrane domains; although rFAP is N-terminally shorter by 11 amino acids, it has the same specific activity as APCE. In-gel tryptic digestion and then LC/MS/MS analyses were used to characterize peptides from APCE and rFAP. The analyses identified 30 peptides from APCE and 21 from rFAP, equivalent to 48% and 30%, respectively, of the amino acid sequence predicted from FAP cDNA (Figure 3). APCE and rFAP were both purified using the F19 monoclonal antibody specific for FAP,35 and both enzymes were recognized by the same antibody in Western blot analyses (Figure 1B). Hence, data comparing amino acid sequences and antibody cross-reactivities support APCE being a soluble derivative of membrane-bound FAP.
We examined whether a homodimeric structure for APCE or rFAP is required for gelatinase and dipeptidyl peptidase activities, using gelatin zymography and size exclusion chromatography. The homodimeric form of rFAP or APCE cleaves gelatin, Ala-Pro-AFC and Z-Gly-Pro-AMC, whereas monomers of either had no such activities. Although previous data suggested that homodimer formation was needed for enzymatic function,11,13,29 this could only be inferred from zymography that could not provide molecular weight data and no size exclusion chromatography was done; hence, homodimer structure remained uncertain. The recently published crystal structure of FAP showed that the enzymatically active, soluble form of recombinant FAP expressed by a baculovirus system is a homodimer, but whether monomers have enzymatic activity was not reported.36 Each subunit comprises 2 distinct domains, an α/β-hydrolase domain (residues 27-53 and 493-760) and a β-propeller domain (54-492).36 Based on its crystal structure, a small region of the propeller (residues 232-258) and 3 regions of the hydrolase domain (residues 651-657, 706-731, and 737-757) can be predicted to participate in the dimerization of FAP. One might speculate that based on this information, homodimers should be separable into monomers by site-directed mutagenesis or peptides that disrupt dimerization so that it could be determined if monomers possess enzymatic activity, but as yet, this has not been done with FAP. Data in the present study, however, suggest that monomers lack enzymatic activity, reinforcing the speculation that if a peptide inhibitor were designed to compete with dimer interface sequences, and thereby prevent dimerization, then it might be possible to abate FAP activity in pathologic conditions such as certain neoplasms.23,37,38
rFAP or APCE-catalyzed cleavage of Met-α2AP as a function of Met-α2AP concentration. Cleavage rates are given in picomoles of peptides released per hour for 1 pmol enzyme.
rFAP or APCE-catalyzed cleavage of Met-α2AP as a function of Met-α2AP concentration. Cleavage rates are given in picomoles of peptides released per hour for 1 pmol enzyme.
Although our data support that APCE is soluble FAP and that its homodimeric structure appears to be required for activity expression, we also explored whether the enzymes shared substrate specificities. The synthetic substrates, Ala-Pro-AFC and Z-Gly-Pro-AMC, and the FRET peptide, synthesized to contain P4-P4′ residues of Met-α2AP (Table 1), were used for kinetic assessments. We determined that each enzyme had a pH 7.5 optimum at which all kinetic analyses were then performed. Enzyme concentrations and kcat values were calculated using the subunit molecular weights and extinction coefficients as we determined for APCE and rFAP. The kinetic parameters for each enzyme were identical within acceptable error, which again supports rFAP and APCE being essentially the same enzyme. FAP has been reported to prefer Ala-Pro-AFC over Lys-Pro-AFC, and Gly-Pro-AFC.13 Z-Gly-Pro-AMC, essentially representing a tripeptide, has a 2 to 3 times higher affinity for both rFAP and APCE than does Ala-Pro-AFC (Table 1), which suggests that both enzymes are actually endoproteinases with the ability to cleave bonds very near the N-terminus of the substrate. The FRET peptide has the highest affinity among the 3 substrates for rFAP and APCE, and the kinetic efficiency (kcat/Km) of rFAP and APCE toward this substrate is the highest, indicating that other residues within the P4-P4′ region must contribute to the substrate specificity of both enzymes.39
rFAP, just as APCE, also cleaves the Pro12-Asn13 bond of Met-α2AP. Initially for calculating kinetic parameters, we tried to quantitate a large fragment (residues 13-464) from the cleavage of Met-α2AP by densitometric scanning of time-dependent disappearance of Met-α2AP bands on immunoblots. But due to the large inherent error we encountered with this method, we elected to quantitate the N-terminal cleavage product (residues 1-12; MEPLGRQLTSGP) of the Pro12-Asn13 bond of Met-α2AP by LC/MS. We found small amounts of cleavage products corresponding to L4-P12 peptide and an oxidized version of the M1-P12 peptide along with the expected peptide M1-P12, all of which complicated data analyses for determining reliable individual Km and kcat values for rFAP or APCE toward the Pro12-Asn13 bond of Met-α2AP. Under competitive situations, however, the substrate preference of an enzyme can be expressed as relative kcat/Km values,33 which yields a value ratio of the Pro3-Leu4 bond and the Pro12-Asn13 bond as about 1:16. The big difference in the relative kcat/Km values can be explained by Gly and Asn (P2 and P1′) being more preferred by APCE (soluble FAP) than are Glu and Leu (P2 and P1′), the latter characterizing the Pro3-Leu4 bond region.39
Soluble FAP (APCE) circulating in human plasma appears to have a role in making α2AP more efficient for protection of extravascular fibrin that forms as a host defense to staunch hemorrhage.5 Because the source of the plasma form of this enzyme was uncertain, it seemed important to know if cell membrane-bound FAP, wherever expressed in vivo under normal conditions, might be shed to somehow enter the circulation. We reasoned that if APCE is indeed a soluble form of FAP, the 2 should be virtually identical. Endothelial cells might produce this enzyme, but whereas cultured endothelial cells have been reported to express FAP mRNA, translation of FAP protein was not documented.17 FAP is not expressed by normal human liver, but in the cirrhotic liver a strong correlation exists between FAP expression and severity of liver fibrosis.34,40 Our results support soluble FAP as a derivative of membrane-bound FAP, with a role in human fibrinolysis, but otherwise, the origin and additional functions of membrane-bound or soluble FAP under normal conditions remain enigmatic.
Prepublished online as Blood First Edition Paper, October 13, 2005; DOI 10.1182/blood-2005-08-3452.
Supported by the William K. Warren Medical Research Center and National Institutes of Health grant HL072995.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal