The congenital dyserythropoietic anemias (CDAs) are a heterogeneous group of genetic disorders of erythropoiesis characterized by anemia secondary to ineffective erythropoiesis and striking morphologic abnormalities of erythroblasts (reviewed in Wickramasinghe and Wood1 ). The autosomal recessive inheritance of CDA type I has been well documented and linkage analysis in Israeli Arab families has localized the gene to chromosome 15q15.2 Analysis of genes in this region revealed mutations in a gene designated CDAN1 that were associated with CDA type I disease in both an Israeli Arab family and in sporadic cases, mostly of European origin.3-5 In the present study, we confirmed several of these mutations of the CDAN1 gene in affected patients, report a new mutation in this gene, and provide convincing evidence that not all affected patients show linkage of CDA I to 15q15, thereby demonstrating genetic heterogeneity in this condition.
In 3 unrelated consanguineous families, 13 affected CDA I individuals (diagnosed by demonstrating the Swiss-cheese appearance of erythroblast heterochromatin on electron microscopy of bone marrow from at least 1 affected member) showed mild to moderate anemia and a high or high-normal mean corpuscular volume (MCV) (Table 1). Patients from 2 unrelated consanguineous families (families A and B, both of Lebanese descent) demonstrated homozygosity for microsatellite markers in the CDA I locus across 45 members from 2 generations, and mutational analysis revealed homozygosity for the same missense mutation (3238C > T) that characterized the Israeli Arab family. It therefore seems likely that a founder effect is responsible for the disease in this population.
Essential clinical and laboratory data in the 3 families (A-C) and 6 sporadic patients with CDA type I
Patient/sex . | Year of birth . | Hb, g/L . | MCV, fL . | Retics, % or 109/L . | Bilirubin, mM . | sTK, U/L . | sTfR, U/L . | Ferritin, ng/mL . | Genotype . |
|---|---|---|---|---|---|---|---|---|---|
| A1/F | 1944 | 90 | 109 | — | 46 | — | — | 1341 | R1041W homozygote |
| A2/M | 1956 | 112 | 98 | 5.0% | 32 | 99 | 9.1 | 812 | R1041W homozygote |
| A3*/M | 1967 | 140 | 95 | — | — | 17 | 12.8 | — | R1041W homozygote |
| A4/M | 1979 | 124 | 101 | — | 15 | 179 | 13.7 | 650 | R1041W homozygote |
| A5/F | 1987 | 107 | 93 | 2.3% | 39 | 111 | 14 | 359 | R1041W homozygote |
| A6/F | 1992 | 121 | 97 | — | — | 130 | 12.4 | — | R1041W homozygote |
| A7/M | 1946 | 116 | 100 | — | — | 92 | 12.2 | — | R1041W homozygote |
| A8/F | 1950 | 97 | 97 | — | — | 161 | 11.6 | — | R1041W homozygote |
| A9/M | 1986 | 100 | 97 | — | — | 86 | 12.5 | — | R1041W homozygote |
| B3/F | 1960 | 79 | 103 | 67 | 42 | 187 | 59.7 | 1184 | R1041W homozygote |
| B7/M | 1974 | 88 | 105 | 54 | 110 | 222 | 57.1 | 1592 | R1041W homozygote |
| C3/F | 1980 | 103 | 97 | 106 | — | 78 | 25.2 | 232 | WT/WT |
| C5/F | 1973 | 77 | 95 | 61 | 63 | 88 | 43.5 | 260 | WT/WT |
| E/M | 1929 | 112 | 104 | 27 | 77 | 44 | 35.2 | 766 | D1042/WT |
| F/M | 1953 | 120 | 102 | 73 | 38 | 117 | 42.6 | 735 | P671L/WT |
| G/F | 1950 | 82 | 104 | 15 | 27 | — | 11.4 | 1755 | N598S/R724W |
| H/M | 1950 | 99 | 99 | 146 | 34 | 144 | 60.9 | 1190 | DeIV372/WT |
| I/M | 1977 | 110 | 92 | 93 | 32 | 167 | 59.6 | 382 | WT/WT |
| J/F | 1927 | 105 | 111 | 43 | 25 | — | 33.1 | 766 | P671L/WT |
Patient/sex . | Year of birth . | Hb, g/L . | MCV, fL . | Retics, % or 109/L . | Bilirubin, mM . | sTK, U/L . | sTfR, U/L . | Ferritin, ng/mL . | Genotype . |
|---|---|---|---|---|---|---|---|---|---|
| A1/F | 1944 | 90 | 109 | — | 46 | — | — | 1341 | R1041W homozygote |
| A2/M | 1956 | 112 | 98 | 5.0% | 32 | 99 | 9.1 | 812 | R1041W homozygote |
| A3*/M | 1967 | 140 | 95 | — | — | 17 | 12.8 | — | R1041W homozygote |
| A4/M | 1979 | 124 | 101 | — | 15 | 179 | 13.7 | 650 | R1041W homozygote |
| A5/F | 1987 | 107 | 93 | 2.3% | 39 | 111 | 14 | 359 | R1041W homozygote |
| A6/F | 1992 | 121 | 97 | — | — | 130 | 12.4 | — | R1041W homozygote |
| A7/M | 1946 | 116 | 100 | — | — | 92 | 12.2 | — | R1041W homozygote |
| A8/F | 1950 | 97 | 97 | — | — | 161 | 11.6 | — | R1041W homozygote |
| A9/M | 1986 | 100 | 97 | — | — | 86 | 12.5 | — | R1041W homozygote |
| B3/F | 1960 | 79 | 103 | 67 | 42 | 187 | 59.7 | 1184 | R1041W homozygote |
| B7/M | 1974 | 88 | 105 | 54 | 110 | 222 | 57.1 | 1592 | R1041W homozygote |
| C3/F | 1980 | 103 | 97 | 106 | — | 78 | 25.2 | 232 | WT/WT |
| C5/F | 1973 | 77 | 95 | 61 | 63 | 88 | 43.5 | 260 | WT/WT |
| E/M | 1929 | 112 | 104 | 27 | 77 | 44 | 35.2 | 766 | D1042/WT |
| F/M | 1953 | 120 | 102 | 73 | 38 | 117 | 42.6 | 735 | P671L/WT |
| G/F | 1950 | 82 | 104 | 15 | 27 | — | 11.4 | 1755 | N598S/R724W |
| H/M | 1950 | 99 | 99 | 146 | 34 | 144 | 60.9 | 1190 | DeIV372/WT |
| I/M | 1977 | 110 | 92 | 93 | 32 | 167 | 59.6 | 382 | WT/WT |
| J/F | 1927 | 105 | 111 | 43 | 25 | — | 33.1 | 766 | P671L/WT |
M indicates male; F, female; Hb, hemoglobin; sTK, serum thymidine kinase; sTfR, serum transferrin receptor; Retics, reticulocytes; and —, not done. Reference ranges: retics, 0.5%-3.8% (20-130 × 109/L); bilirubin, < 17 mM; sTK, 4.2-6.6 U/L; sTfR, 2.3-8.4 U/L; and ferritin, 22-402 ng/mL.
patient on interferon therapy.
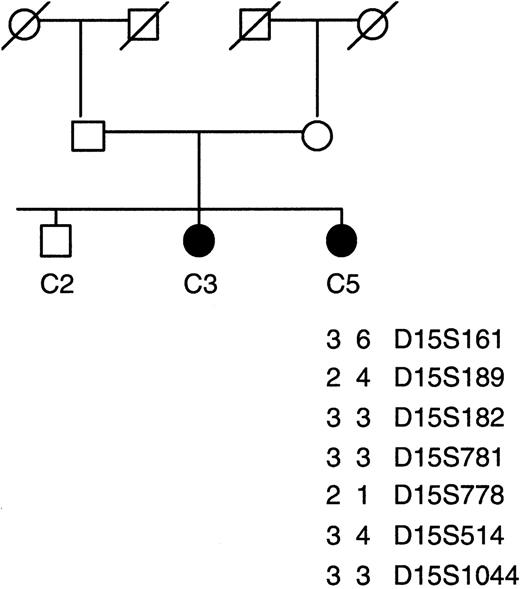
Haplotype analysis using microsatellite markers in consanguineous family C with familial CDA1. A total of 19 markers spanning the 8-Mb region of the CDAN1 locus were used for linkage analysis; only informative markers are shown. The index patient C5 lacks homozygosity over this region.
Haplotype analysis using microsatellite markers in consanguineous family C with familial CDA1. A total of 19 markers spanning the 8-Mb region of the CDAN1 locus were used for linkage analysis; only informative markers are shown. The index patient C5 lacks homozygosity over this region.
In affected individuals of the other consanguineous CDA I family (C; Pakistani), with hematologic results typical of CDA I and a substantial proportion of the erythroblasts showing the “Swiss-cheese” abnormality on electron microscopy, there was no homozygosity for the microsatellite markers across the extended CDAN1 locus and sequence analysis detected no mutations in the CDAN1 gene (Figure 1). Clearly the disease is not linked to chromosome 15q15 in this family, strongly suggesting that there is at least one alternative genetic locus for CDA I.
We also investigated 6 sporadic cases with CDA I for CDAN1 gene mutations by bidirectional genomic sequencing. Six mutant alleles were found in these cases, 5 previously reported (3242A > T, 2129C > T [2 patients], 1910A > G, and 2287C > T) together with a previously unreported 3 base pairs in frame deletion. resulting in deletion of valine 372. In 4 of the 6 sporadic cases, only 1 mutation in the CDAN1 gene could be demonstrated, a situation also seen in half of the sporadic cases described previously,3-5 and in 1 case, with multiple dysmorphic features, no mutations were detected. Mutations in regions of the CDAN1 gene that were not sequenced (promoter, introns) or in regulatory sequences may explain these findings. Given the genetic heterogeneity of this condition, however, it is plausible that a mutation in a second gene could interact with a CDAN1 mutation to result in the CDA I phenotype as a result of digenic inheritance.
A more comprehensive understanding of CDA I will require a better knowledge of the function of CDAN1 protein, its cellular distribution and intracellular localization, and the existence of any putative binding partners. The genetic heterogeneity demonstrated here indicates that this should be a fruitful field for the study of erythropoiesis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal