Abstract
Previous work has demonstrated that WT1 (–Ex5/–KTS) potentiates granulocyte colony-stimulating factor (G-CSF)–mediated granulocytic differentiation. This WT1 isoform suppresses cyclin E, which may contribute to the prodifferentiation effect by slowing proliferation, but WT1 target genes that affect survival might also be involved. We screened a cDNA array and identified the bCL2 family member A1/BFL1 as a new WT1 target gene in 32D cl3 murine myeloblast cells. Induction of WT1 (–Ex5/–KTS) expression is accompanied by up-regulation of A1 on the cDNA array, and this up-regulation was confirmed by semiquantitative reverse transcription–polymerase chain reaction (RT-PCR). Moreover, both promoter-reporter assays and chromatin immunoprecipitation assays suggest that this isoform of WT1 activates the promoter directly. Constitutive expression of A1 in 32D cl3 cells induces spontaneous granulocytic differentiation, with both morphologic and cell-surface antigen changes, as well as resistance both to chemotherapy and to withdrawal of interleukin-3 (IL-3). Finally, we note an association between WT1 expression and A1 expression in primary acute myeloid leukemia samples. Taken together, these results demonstrate that A1 is a new WT1 target gene involved in both granulocytic differentiation and resistance to cell death, and suggests that these genes might play an important role in the biology of high-risk leukemias.
Introduction
WT1 is a zinc finger transcription factor that was originally identified as a tumor suppressor gene in children with hereditary syndromes predisposing to Wilms tumor.1,2 The wild-type gene is expressed in the genitourinary system, and germ-line deletion of WT1 in mice results in failure of genitourinary development with widespread apoptosis.3 WT1 is also expressed in hematopoietic stem/progenitor cells, and there is strong evidence that WT1 plays both developmental and antiapoptotic roles in the myeloid lineage. In normal bone marrow, WT1 expression is limited to CD34+/CD38– cells.4,5 As these cells begin to express CD38, WT1 expression is lost. Different isoforms of WT1 have been shown to either inhibit6 or potentiate7 myelopoiesis in cell culture models. WT1 overexpression has been reported in acute myeloid leukemia (AML), and is correlated with poor survival,8 probably due to chemotherapy resistance. Because chemotherapy induces apoptosis in susceptible cells, this finding provides additional support for an antiapoptotic function of WT1.
WT1 mRNA is subject to alternative splicing at 2 independent sites, resulting in the expression of 4 major protein isoforms. One splicing event includes or excludes exon 5 from the mature mRNA, and the other inserts a 3–amino acid sequence (lysine-threonine-serine [KTS]) between exons 9 and 10, which encode the third and fourth zinc fingers.9 The resulting protein products are called WT1 (+Ex5/+KTS), WT1 (+Ex5/–KTS), WT1 (–Ex5/+KTS), and WT1 (–Ex5/–KTS). WT1 isoforms have distinct effects on target promoters,10-12 and the presence or absence of the KTS insert can affect subcellular localization of the WT1 protein13 as well as interactions with RNA splicing apparatus.14 Disruption of the normal ratio of isoforms in humans affects genitourinary development,15,16 demonstrating the importance of proper WT1 RNA splicing for normal development. Thus, for a full understanding of the role of WT1 in normal and malignant myelopoiesis, the functions of individual WT1 isoforms must be addressed.
The 32D cl3 murine myeloblast cell line is a useful model of myeloid differentiation. Inoue et al demonstrated that high-level expression of WT1 (+Ex5/+KTS) inhibits granulocyte colony-stimulating factor (G-CSF)–mediated differentiation of these cells.6 In contrast, we found that expression of WT1 (–Ex5/–KTS) accelerated G-CSF–mediated differentiation.7 We have subsequently attempted to identify target genes that might mediate the effects of WT1, and have reported that WT1 suppresses the expression of cyclin E in this cell line.17 The present work identifies the antiapoptotic gene A1 (designated BFL1 in humans) as a new WT1 target gene. A1 is a bCL2 family member that has been implicated in myeloid differentiation and in resistance to apoptosis in hematopoietic cells.18 Induction of WT1 (–Ex5/–KTS) in 32D cl3 cells up-regulates A1 expression. Constitutive expression of A1 in these cells results in spontaneous differentiation and resistance to the apoptotic effects of cytokine withdrawal and chemotherapeutic agents. These findings suggest that WT1 (–Ex5/–KTS) mediates differentiation and resistance to chemotherapy, at least in part through induction of A1 expression.
Materials and methods
Cell lines and plasmids
CV1, NIH 3T3, and 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum. U937 cells were grown in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum. 32D cl3 cells were grown in Iscove MDM (IMDM) supplemented with 10% heat-inactivated fetal calf serum and 1 ng/mL interleukin-3 (IL-3; R&D Systems, Minneapolis, MN). Stably transfected cell lines were maintained in this medium supplemented with 1 mg/mL G418 (Invitrogen, Gaithersburg, MD). Cells were attached to glass slides using a Shandon CytoSpin apparatus (Thermo Electron Corp, Pittsburgh, PA), and stained with Wright/Giemsa stain. Photomicrographs were produced using a Nikon E-800 upright microscope (Melville, NY) with a 40 × Plan Apo lens with a numeric aperture of 0.95. The camera was a Nikon DXM 1200F digital camera. The acquisition software was ACT-1, provided by Nikon.
The plasmid pcDNA3-HA-Bfl1, which includes the A1 cDNA and a hemagglutinin (HA) tag downstream of the cytomegalovirus (CMV) immediate-early promoter, was a generous gift from Dr G. Chinnadurai (St Louis University School of Medicine, MO). A plasmid which contains 1375 bp of the murine A1 promoter cloned upstream of the firefly luciferase gene was a generous gift from Dr C. Gelinas (University of Medicine and Dentistry of New Jersey [UMDNJ]–Robert Wood Johnson Medical School, New Brunswick, NJ). The cDNA for WT1 (–Ex5/–KTS) was also cloned into a retroviral vector (MIGR-1; a gift from Linzhao Chen, Johns Hopkins University), with expression driven by the murine stem-cell virus long-terminal repeat (LTR) and the gene for green fluorescent protein (GFP) cloned downstream of an internal ribosome entry site. Deletion mutagenesis of the A1 promoter was performed using the Erase-a-Base Kit (Promega, Madison, WI) according to the manufacturer's protocol.
Transfection
32D cl3 cells were transfected using TransIT LT1 (Panvera, Madison, WI) and Lipofectamine 2000 (Invitrogen), following the manufacturer's protocols. Cells (107) were transfected with 15 μg pcDNA3-HA-Bfl1 that had been linearized by digestion with PvuI. After 48 hours, the cells were cloned by limiting dilution in 96-well plates and selected for resistance to G418. Transient transfections were performed using TransIT LT1 according to the manufacturer's protocol. For retrovirus experiments, transduction competent virus was generated by cotransfecting 293T cells with the MIGR-1 vector and an amphotrophic virus and harvesting supernatant. U937 cells were transduced with retrovirus in RetroNectin (TaKaRa, Shiga, Japan)–coated plates using Polybreen (Sigma-Aldrich, St Louis, MO) to enhance efficiency.
Western blotting
Cells were harvested in 1 × LDS sample buffer (Invitrogen), and electrophoresis and blotting were performed on reduced samples, using the Invitrogen XCell II SureLock blot system. Proteins were separated on 4% to 12% bis-tris gels using MOPS buffer and blotted onto PVDF membranes. Antibodies against WT1 (C-19; Santa Cruz Biotechnology, Santa Cruz, CA), hemagglutinin (Sigma-Aldrich) and horseradish peroxidase–conjugated secondary antibodies (Amersham, Piscataway, NJ) were used, and antibody binding was revealed by enhanced chemiluminescence (ECL Western blotting analysis system; Amersham).
RT-PCR
Total RNA was extracted in TriZol (Invitrogen), and cDNA was reverse transcribed using random primers and Superscript II (Invitrogen), according to the manufacturer's protocol. The sequences of primers used to detect expression of A1 were as follows: 5′-GAAGATGGCATCATTAACTG-3′ and 5′-TCCATTCTGCCGTATCCATT-3′. Polymerase chain reaction (PCR) was performed with a melting temperature of 94°C, followed by annealing at 50°C for 30 seconds, and then extension at 72°C for 45 seconds for a total of 25 cycles. PCR products were resolved by electropheresis on a 2% agarose gel. Conditions and primers for WT1, actin, and 36B4 reverse transcription (RT)–PCR have been previously described.7,19
Gene array
RNA was isolated from cells and reverse transcribed as described. Radiolabeled cDNA was prepared using [α-32P]-dCTP and hybridized to the Mouse Apoptosis Expression Array (catalog no. GA004; R&D Systems) according to the manufacturer's protocol.
Luciferase assay
Luciferase assays were performed using the Dual Luciferase Reporter Assay System (Promega), and results were normalized by cotransfection with Renilla luciferase expressed from the simian virus 40 (SV40) promoter.
Flow cytometry
Cells were incubated either with phycoerythin (PE)–conjugated anti-GR1, PE-conjugated anti-CD69, or fluorescein isothiocyanate (FITC)–conjugated anti-CD74, or with their respective isotype controls (all from Becton Dickinson, San Jose, CA) and fixed with 1% paraformaldehyde in phosphate-buffered saline (PBS). Flow cytometric analysis was performed using a FACSCalibur (Becton Dickinson) and the CellQuest Pro software (Becton Dickinson). Numerous positive events (10 000) were analyzed in each sample.
Chromatin immunoprecipitation assay
Histones were crosslinked to DNA by adding formaldehyde to a concentration of 1%. Crosslinking was stopped by adding glycine to a final concentration of 0.125 M. Cells were lysed, and the DNA was sheared by sonication in a Gene Pulser II (BioRad, Hercules, CA) using 5 10-second pulses at 40% power. After microcentrifugation, the supernatant was precleared with a slurry of salmon sperm DNA and protein A agarose. Nonspecifically bound material was removed by microcentrifugation, and the supernatant was incubated with WT1 antibody (C-19) overnight. Immune complexes were collected with salmon sperm/protein A agarose slurry and washed extensively. Crosslinks were reversed by adding 5 M NaCl to the eluates, and the DNA was recovered by extraction in phenol/chloroform/isoamyl alcohol followed by ethanol precipitation. The presence of A1 promoter in the immune precipitates was confirmed by PCR. The sequences of primers for the A1 promoter following the chromatin immunoprecipitation (ChIP) assay were as follows: 5′-GGGATGAATAATGTTCCATAA-3′ and 5′-AAGCTGTTGAGGCAATGT-3′. PCR was performed with a melting temperature of 94°C for 15 seconds, followed by 47°C for 30 seconds, and then 72°C for 45 seconds for 40 cycles.
Chemosensitivity
Cells (105 per well) were plated in 96-well plates and treated for 24 hours with either doxorubicin or cytarabine (both from Sigma-Aldrich). An MTT assay (using a kit from Chemicon International, Temecula, CA) was performed to quantitate viable cell number. Annexin V expression was measured using a kit (Annexin-V FLUOS Staining Kit; Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol.
Results
Gene expression profiling demonstrates A1 is a WT1 target gene
We have previously demonstrated that WT1 (–Ex5/–KTS) potentiates G-CSF–mediated granulocytic differentiation of the 32D cl3 murine myeloblast cell line.7 Because this is the isoform of WT1 that has been shown most conclusively to function as a transcription factor, we undertook a gene expression profiling approach to identifying WT1 target genes that might influence granulocytic differentiation. 32D cl3 cell lines stably expressing WT1 (–Ex5/–KTS) under the control of the inducible metallothionein promoter have been described in detail.7 We used radiolabeled cDNA from the 32DWT1 cell line before and after induction of WT1 expression with ZnCl2 to probe a commercially prepared mouse gene array, chosen because of the presence of genes related to both apoptosis and hematopoiesis. As a control, we used cDNA from the 32DV4 cell line (which was transfected with the empty expression vector used to generate the 32DWT1 cell line) before and after treatment with ZnCl2. Of the 232 evaluable genes on the array, 74 (32%) were either up- or down-regulated in the ZnCl2-treated 32DWT1 cells but not in 32DV4 cells (Table 1). Metallothionein was up-regulated in both WT1-expressing cells and in control cells, providing internal validation of the system. Among the down-regulated genes were a number of death effectors, such as ICAD, ALG4, and CIDE-A. Down-regulation of these genes is consistent with the observation that overexpression of WT1 is associated with resistance to apoptosis. Among the genes up-regulated in WT1-expressing cells but not in the control cells were several previously identified WT1 target genes: RBBP4,20 insulin-like growth factor-II,21 thrombospondin,22 and ODC.23 Among the other genes whose expression levels were affected by induction of WT1 was the bCL2 family member A1 (Table 1). A complete evaluation of the gene expression profile associated with induction of WT1 (–Ex5/–KTS) in these cells will be reported separately.
Gene expression changes in 32DWT1 cells treated with ZnCl2
Gene . | UniGene no. . | Fold change with ZnCl2 . |
|---|---|---|
| Caveolin 2 | Mm.31915 | 0.15 |
| Dffa | Mm.41433 | 0.26 |
| Pdcd11 | Mm.41166 | 0.32 |
| CIDE-A | Mm.449 | 0.36 |
| RBBP4 | Mm.12145 | 2.67 |
| IGF-II | Mm.3862 | 2.96 |
| PTEN | Mm.220979 | 3.33 |
| Cyclin A2 | Mm.4189 | 4.2 |
| 14-3-3 η | Mm.332314 | 4.22 |
| Sfrp5 | Mm.103731 | 4.55 |
| PCNA | Mm.7141 | 5.02 |
| CDK4 | Mm.6839 | 6.23 |
| Granzyme B | Mm.14874 | 10.72 |
| NAIP | Mm.89961 | 11.2 |
| ODC | Mm.34102 | 16.95 |
| CBP | Mm.132238 | 17.53 |
| MT-2 | Mm.147226 | 39 |
| Granzyme A | Mm.15510 | — |
| Sfrp2 | Mm.19155 | — |
| BID | Mm.235081 | — |
| A1 | Mm.196770 | — |
| CDK6 | Mm.31672 | — |
| Thrombospondin | Mm.4159 | — |
| APEX | Mm.203 | — |
Gene . | UniGene no. . | Fold change with ZnCl2 . |
|---|---|---|
| Caveolin 2 | Mm.31915 | 0.15 |
| Dffa | Mm.41433 | 0.26 |
| Pdcd11 | Mm.41166 | 0.32 |
| CIDE-A | Mm.449 | 0.36 |
| RBBP4 | Mm.12145 | 2.67 |
| IGF-II | Mm.3862 | 2.96 |
| PTEN | Mm.220979 | 3.33 |
| Cyclin A2 | Mm.4189 | 4.2 |
| 14-3-3 η | Mm.332314 | 4.22 |
| Sfrp5 | Mm.103731 | 4.55 |
| PCNA | Mm.7141 | 5.02 |
| CDK4 | Mm.6839 | 6.23 |
| Granzyme B | Mm.14874 | 10.72 |
| NAIP | Mm.89961 | 11.2 |
| ODC | Mm.34102 | 16.95 |
| CBP | Mm.132238 | 17.53 |
| MT-2 | Mm.147226 | 39 |
| Granzyme A | Mm.15510 | — |
| Sfrp2 | Mm.19155 | — |
| BID | Mm.235081 | — |
| A1 | Mm.196770 | — |
| CDK6 | Mm.31672 | — |
| Thrombospondin | Mm.4159 | — |
| APEX | Mm.203 | — |
Listed are genes noted to be significantly up- or down-regulated upon WT1 induction in 32DWT1 cells, but not (with the exception of metallothionein) in 32DV4 cells treated with ZnCl2. Metallothionein was up-regulated in control cells, which is the expected response to treatment with heavy metal.
— indicates a gene that was expressed only after treatment with ZnCl2.
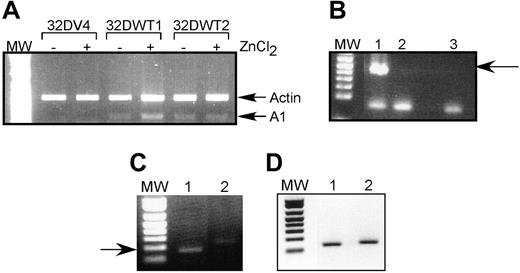
We chose to pursue the possibility that A1 is a direct WT1 target gene because (1) there is no detectable A1 expression in control cells but significant expression in WT1-expressing cells; (2) A1 is up-regulated by G-CSF, and our previous work demonstrated that this isoform of WT1 potentiates the effects of G-CSF; and (3) A1 has been directly implicated in granulocytic differentiation. We used semiquantitative RT-PCR to confirm that A1 mRNA is up-regulated by WT1 (–Ex5/–KTS). Control 32DV4 cells did not express detectable amounts of A1 mRNA, and ZnCl2 had no effect on A1 expression in these cells. In contrast, induction of WT1 (–Ex5/–KTS) in 32DWT1 cells by the addition of ZnCl2 also induced the expression of A1 (Figure 1A). 32DWT2 cells, which constitutively express WT1 because of leaky activity of the metallothionein promoter, also express A1 independent of the addition of ZnCl2. Quantification of these data by scanning densitometry revealed that induction of WT1 expression in the 32DWT1 cells led to a 3-fold increase in A1 expression, and up-regulation of WT1 above the basal level of expression in the 32DWT2 cells caused a 60% increase in A1 expression. These findings confirm the cDNA array results, demonstrating a correlation between WT1 (–Ex5/–KTS) expression and A1 expression.
We also investigated the effect of WT1 on A1 expression in U937 cells, a human leukemia cell line that does not express endogenous WT1. U937 cells were transduced with a retroviral vector directing the expression of WT1 (–Ex5/–KTS) or with the corresponding empty vector. Successfully transduced, GFP-positive cells were collected and assayed for expression of A1 and WT1 by semiquantitative RT-PCR. As was seen in 32D cl3 cells, expression of exogenous WT1 (–Ex5/–KTS) (Figure 1B) results in increased expression of the endogenous A1 gene (Figure 1C). The samples express similar amounts of the ribosomal RNA 36B4 (Figure 1D).
Association between WT1 expression and A1 expression in primary AML samples
We next evaluated the expression of WT1 and A1 in primary AML samples. In a panel of 19 samples collected from patients with poor-prognosis AML (patients with high initial white blood cell counts; complex cytogenetics; monosomies 5, 7, or 9; or refractory disease), we detected WT1 expression in 15 (78.9%) samples, and we detected A1 expression in 14 (73.7%) samples. Coexpression of WT1 and A1 was seen in 12 samples (Table 2), and the relative rate of A1 expression was 1.6-fold higher in WT1-positive samples than in WT1-negative samples. These data support (likelihood ratio 2.7) the alternative hypothesis that WT1 up-regulates A1 expression not only in cell lines, but also in the subset of AML patients with the worst prognosis.
A1 mRNA is up-regulated by WT1 (–Ex5/–KTS). (A) Semiquantitative RT-PCR was performed as described on RNA isolated from control 32DV4 cells and from 32DWT1 and 32DWT2 cells treated overnight with (+) and without (–) 50 μM ZnCl2. Arrows indicate the Actin (control) and A1 PCR products. MW indicates the position of the molecular-weight markers. (B) RT-PCR was performed as described on RNA isolated from U937 cells transduced with the empty MIGR-1 retroviral vector (lane 2) or the same vector with the WT1 (–Ex5/–KTS) cDNA (lane 1). A water control is in lane 3. The arrow indicates the position of the WT1 band. (C) RT-PCR was performed on the same samples using A1 primers. The arrow indicates the position of the A1 band. (D) RT-PCR was performed on the same samples using 36B4 primers.
A1 mRNA is up-regulated by WT1 (–Ex5/–KTS). (A) Semiquantitative RT-PCR was performed as described on RNA isolated from control 32DV4 cells and from 32DWT1 and 32DWT2 cells treated overnight with (+) and without (–) 50 μM ZnCl2. Arrows indicate the Actin (control) and A1 PCR products. MW indicates the position of the molecular-weight markers. (B) RT-PCR was performed as described on RNA isolated from U937 cells transduced with the empty MIGR-1 retroviral vector (lane 2) or the same vector with the WT1 (–Ex5/–KTS) cDNA (lane 1). A water control is in lane 3. The arrow indicates the position of the WT1 band. (C) RT-PCR was performed on the same samples using A1 primers. The arrow indicates the position of the A1 band. (D) RT-PCR was performed on the same samples using 36B4 primers.
WT1 directly up-regulates A1
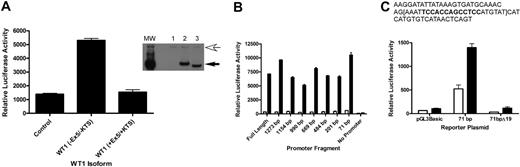
To address whether A1 is a direct target of WT1 (–Ex5/–KTS), we performed promoter-reporter assays using a reporter plasmid containing the firefly luciferase gene under the control of the murine A1 promoter. CV1 cells, a green monkey kidney cell line that does not express WT1 (as verified by RT-PCR; data not shown), were transiently transfected with the luciferase reporter plasmid and with a plasmid directing the constitutive expression of the cDNA for either WT1 (–Ex5/–KTS), WT1 (+Ex5/+KTS), or a control plasmid. Cotransfection with WT1 (–Ex5/–KTS), but not with WT1 (+Ex5/+KTS) or the control, resulted in a 5-fold increase in A1 promoter activity (Figure 2A), suggesting that WT1 (–Ex5/–KTS) directly regulates the activity of the A1 promoter, and that this is an isoform-specific effect. Western blotting confirmed that the WT1 isoforms are expressed at equal levels (Figure 2A inset), demonstrating that the difference in activity is not related to differences in isoform expression. Similar results were seen using NIH 3T3 cells (data not shown).
Inspection of the DNA sequence of the A1 promoter did not reveal a perfect consensus WT1 binding site, so a series of promoter mutants, with progressive deletions from the 5′ end of the promoter, was generated. Each of these deletion mutants was tested in the promoter-reporter assay in transiently transfected CV1 cells. Significant activity was seen in even the smallest deletion mutant analyzed (Figure 2B). This small fragment of the promoter, 71 bp in length, contains a sequence, TCCACCAGCCTCC, that is similar to a putative WT1 binding site ((TCC)n24 ). In an attempt to confirm that this sequence mediates responsiveness to WT1 (–Ex5/–KTS), 19 base pairs, including this sequence, were deleted from the 71-bp promoter fragment, and this construct, designated 71bpΔ19, was tested for WT1 responsiveness in transient transfection assays. This construct is not activated by WT1 (–Ex5/–KTS), but it has basal activity equivalent to the luciferase construct lacking a promoter (Figure 2C). These data are consistent with WT1 responsiveness requiring these 19 base pairs, but in the absence of significant basal activity it is also possible that deletion of these 19 base pairs destroys the function of the promoter fragment entirely. Further work is ongoing to clarify the role of this small sequence in mediating WT1 responsiveness and to identify the key residues for WT1 binding.
A1 is a direct WT1 target. (A) CV1 cells were transiently transfected with either a control plasmid or a plasmid directing the expression of WT1 (–Ex5/–KTS) or WT1 (+Ex5/+KTS) along with a reporter plasmid containing the firefly luciferase gene controlled by the murine A1 promoter. Luciferase assays were normalized by cotransfection with a Renilla luciferase gene. Error bars represent the standard error of triplicate assays. Inset shows a Western blot of lysate from transiently transfected cells. Lane 1 shows control plasmid; Lane 2, WT1 (+Ex5/+KTS); and Lane 3, WT1 (–Ex5/–KTS). The top arrow indicates a nonspecific band that confirms equal protein loads in each lane, and the bottom arrow indicates the position of WT1. MW is the molecular-weight markers. (B) Serial deletion mutants of the A1 promoter were generated as described, and these were cotransfected into CV1 cells along with either the control plasmid or the WT1 (–Ex5/–KTS) expression vector. Luciferase assays were performed as described, and were normalized by cotransfection with Renilla luciferase. Error bars represent the standard error of triplicate assays. □ indicates control; ▪, WT1 (–Ex5/–KTS). (C) The sequence of the 71-bp promoter fragment, with the putative WT1 binding site in bold type. The 19-bp deletion is contained within the brackets. The indicated plasmids (pGL3Basic, which lacks a promoter, the 71-bp promoter fragment, and the deletion mutant of the 71-bp fragment) were transfected into CV1 cells along with either a control plasmid or the WT1 (–Ex5/–KTS) expression vector and luciferase assays were performed. Assays were normalized by cotransfection with Renilla luciferase, and error bars represent the standard error of triplicate assays. □ indicates control; ▪, WT1 (–Ex5/–KTS).
A1 is a direct WT1 target. (A) CV1 cells were transiently transfected with either a control plasmid or a plasmid directing the expression of WT1 (–Ex5/–KTS) or WT1 (+Ex5/+KTS) along with a reporter plasmid containing the firefly luciferase gene controlled by the murine A1 promoter. Luciferase assays were normalized by cotransfection with a Renilla luciferase gene. Error bars represent the standard error of triplicate assays. Inset shows a Western blot of lysate from transiently transfected cells. Lane 1 shows control plasmid; Lane 2, WT1 (+Ex5/+KTS); and Lane 3, WT1 (–Ex5/–KTS). The top arrow indicates a nonspecific band that confirms equal protein loads in each lane, and the bottom arrow indicates the position of WT1. MW is the molecular-weight markers. (B) Serial deletion mutants of the A1 promoter were generated as described, and these were cotransfected into CV1 cells along with either the control plasmid or the WT1 (–Ex5/–KTS) expression vector. Luciferase assays were performed as described, and were normalized by cotransfection with Renilla luciferase. Error bars represent the standard error of triplicate assays. □ indicates control; ▪, WT1 (–Ex5/–KTS). (C) The sequence of the 71-bp promoter fragment, with the putative WT1 binding site in bold type. The 19-bp deletion is contained within the brackets. The indicated plasmids (pGL3Basic, which lacks a promoter, the 71-bp promoter fragment, and the deletion mutant of the 71-bp fragment) were transfected into CV1 cells along with either a control plasmid or the WT1 (–Ex5/–KTS) expression vector and luciferase assays were performed. Assays were normalized by cotransfection with Renilla luciferase, and error bars represent the standard error of triplicate assays. □ indicates control; ▪, WT1 (–Ex5/–KTS).
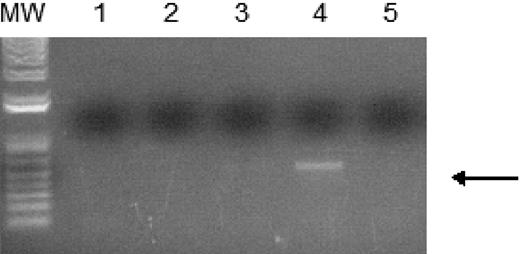
WT1 (–Ex5/–KTS) binds to the A1 promoter. Chromatin immunoprecipitation assays were performed as described using 32DV4 and 32DWT1 cells treated overnight with 50 μM ZnCl2. The arrow indicates the position of the PCR product representing the A1 promoter, present in the 32DWT1 sample but not in the 32DV4 sample. Lane 1 is the control with no template, lanes 2 and 5 are the no-antibody controls from 32DV4 and 32DWT1 cells, respectively, and lanes 3 and 4 are the samples from 32DV4 cells (Lane 3) and 32DWT1 cells (Lane 4). The identity of the PCR fragment was confirmed by sequencing.
WT1 (–Ex5/–KTS) binds to the A1 promoter. Chromatin immunoprecipitation assays were performed as described using 32DV4 and 32DWT1 cells treated overnight with 50 μM ZnCl2. The arrow indicates the position of the PCR product representing the A1 promoter, present in the 32DWT1 sample but not in the 32DV4 sample. Lane 1 is the control with no template, lanes 2 and 5 are the no-antibody controls from 32DV4 and 32DWT1 cells, respectively, and lanes 3 and 4 are the samples from 32DV4 cells (Lane 3) and 32DWT1 cells (Lane 4). The identity of the PCR fragment was confirmed by sequencing.
In order to confirm that WT1 (–Ex5/–KTS) directly binds to the A1 promoter, ChIP assays were performed. 32DWT1 and 32DV4 cells were treated overnight with ZnCl2, and histone/WT1 complexes were immunoprecipitated with an anti-WT1 antibody. A fragment of the A1 promoter was immunoprecipitated from the 32DWT1 cell lysate, but not from control cell lysate (Figure 3). This is consistent with a direct, physical interaction between WT1 (–Ex5/–KTS) and theA1 promoter, demonstrating that A1 is a direct target of the transcriptional regulatory activity of this WT1 isoform.
Expression of HA-tagged A1 in stable transfectants. 32D cl3 cells were transfected with pcDNA3-HA-Bfl1 as described, and G418-resistant clones were isolated. Western blotting with an antibody against the HA tag was performed on total cellular protein from 3 of these transfectants: 32DA1.1 (Lane 2), 32DA1.2 (Lane 3), and 32D A1.3 (Lane 4), as well as protein from untransfected 32D cl3 cells (Lane 1). The arrow indicates the presence of HA-tagged A1 in the transfected cells but not in the parental cells. Molecular-weight markers are shown in the lane marked MW.
Expression of HA-tagged A1 in stable transfectants. 32D cl3 cells were transfected with pcDNA3-HA-Bfl1 as described, and G418-resistant clones were isolated. Western blotting with an antibody against the HA tag was performed on total cellular protein from 3 of these transfectants: 32DA1.1 (Lane 2), 32DA1.2 (Lane 3), and 32D A1.3 (Lane 4), as well as protein from untransfected 32D cl3 cells (Lane 1). The arrow indicates the presence of HA-tagged A1 in the transfected cells but not in the parental cells. Molecular-weight markers are shown in the lane marked MW.
Generation of stably transfected cell lines
Lin et al reported that 32D cl3 cells stably transfected with A1 develop morphologic changes reminiscent of granulocytic differentiation upon withdrawal of IL-3.25 Because the WT1 (–Ex5/–KTS) isoform both up-regulates A1 expression and also potentiates G-CSF–mediated granulocytic differentiation, we investigated in more detail the effect of constitutive A1 expression on myeloid differentiation in this cell line. We transfected 32D cl3 cells with a plasmid (pcDNA3-HA-Bfl1) containing the cDNA for Bfl1, the human homolog of A1, with an HA epitope tag, under the control of the CMV immediate early promoter. Three G418 resistant cell lines were generated, designated 32DA1.1, 32DA1.2, and 32DA1.3. Whole-cell lysates of the cell lines were subjected to Western blot analysis to confirm A1 expression using an antiserum that detects the HA tag. Significant expression of the HA-tagged protein is seen in each cell line (Figure 4).
Effect of A1 on survival and proliferation
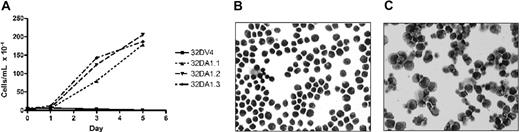
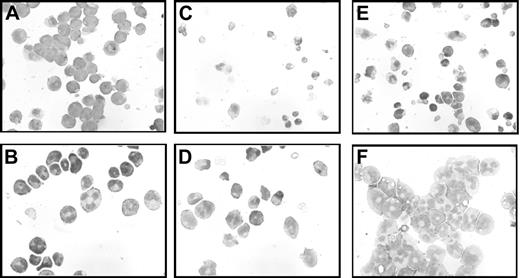
We previously demonstrated that induction of WT1 (–Ex5/–KTS) slowed the proliferation of 32D cl3 cells in G-CSF, but not in IL-3.7 We reasoned that if this isoform of WT1 acts, at least in part, through up-regulation of A1, constitutive expression of A1 might also affect proliferation. 32DA1.1, 32DA1.2, and 32DA1.3 cells, as well as control cells, were rinsed extensively in cytokine-free medium and plated in either this same medium or in medium supplemented with IL-3 or with G-CSF. The cells constitutively expressing A1 grew in either IL-3 or in G-CSF with kinetics indistinguishable from control cells (data not shown). Interestingly, cells constitutively expressing A1 survived and proliferated even in the absence of cytokine (Figure 5A). We examined the morphology of our A1-expressing cell lines grown with or without IL-3 for 8 days. While there were more cells that appeared to be dying in the IL-3–deprived cultures, the cells growing in IL-3 (Figure 5B) had a similar appearance to the surviving cells growing without cytokine (Figure 5C). These photomicrographs are of 32DA1.3 cells, but the other cell lines behaved similarly (data not shown). These data suggest that while A1 allows survival in the absence of IL-3, it does not mediate the growth inhibitory effects of WT1 (–Ex5/–KTS).
32DA1 cells survive without IL-3. Control 32DV4 cells, as well as 32DA1.1, 32DA1.2, and 32DA1.3 cells were washed extensively in cytokine-free medium and plated in cytokine-free medium. The number of living cells in each culture was quantified daily by counting cells that were able to exclude Trypan blue (A). After 8 days in culture with (B) or without (C) IL-3, cytospin preparations were evaluated. Photomicrographs of the 32DA1.3 line are shown.
32DA1 cells survive without IL-3. Control 32DV4 cells, as well as 32DA1.1, 32DA1.2, and 32DA1.3 cells were washed extensively in cytokine-free medium and plated in cytokine-free medium. The number of living cells in each culture was quantified daily by counting cells that were able to exclude Trypan blue (A). After 8 days in culture with (B) or without (C) IL-3, cytospin preparations were evaluated. Photomicrographs of the 32DA1.3 line are shown.
Effect of A1 on differentiation
We next investigated the effect of A1 on the differentiation of our stably transfected cell lines. Morphologically, the A1-expressing cell lines have some characteristics of differentiated cells, including condensed and circular nuclei, even when growing in IL-3 (Figure 6B). This is in contrast with the very immature appearance of control 32DV4 cells growing in IL-3 (Figure 6A). As noted, 32D cl3 cells constitutively expressing A1 and then grown for a prolonged period without IL-3 appear to be morphologically identical to cells grown in IL-3 (Figures 5B-C) and mostly have an immature morphology. We observed a period immediately after IL-3 deprivation during which the number of cells in these cultures remained constant, followed by a period of rapid growth, and we hypothesized that a subpopulation of the cells deprived of IL-3 might spontaneously differentiate, and that the delayed proliferation might reflect undifferentiated cells “outgrowing” the differentiated cells. To address this possibility, we examined the morphology of cells deprived of cytokine for only 48 hours. In these cultures, the predominant population of cells was morphologically quite differentiated, with multilobulated nuclei and pale cytoplasm (Figure 6D). These data are consistent with spontaneous differentiation in the absence of IL-3, and are a stark contrast to the rapid cell death seen in the control cells growing in the absence of IL-3 (Figure 6C).
Because we identified A1 as a WT1 target gene, and because WT1 (–Ex5/–KTS) potentiates G-CSF–mediated differentiation of 32D cl3 cells,7 we evaluated the effect of G-CSF on the differentiation of the A1 transfectant cell lines. Treating these cells with G-CSF resulted in the progressive appearance of morphologically mature granulocytes at a pace comparable with what was seen with WT1 (–Ex5/–KTS)–expressing cells (Figure 6F), and more rapidly than the control cells (Figure 6E). Thus, constitutive expression of A1 potentiates G-CSF–mediated differentiation of 32D cl3 cells, just as induction of WT1 (–Ex5/–KTS) does.
In addition to morphology, we also investigated the expression of lineage-specific cell-surface antigens in the A1-expresing cells. We evaluated the level of expression of Gr-1 (a neutrophil-specific antigen), CD69 (which is expressed on the surface of activated monocytes and neutrophils), and CD74 (the gamma chain of human leukocyte antigen [HLA]–DR, which is expressed on macrophages), by flow cytometry. Even growing in IL-3, cells constitutively expressing A1 expressed significantly higher levels of the granulocytic markers Gr-1 and CD69 than did the control cells (Table 3). We attempted to use expression of these markers to quantitate differences in response to the differentiating effects of G-CSF, but the basal expression of both markers in the A1-expressing cells was too high for significant differences to be detected. None of the cell lines tested expressed significant levels of CD74 in IL-3 or in G-CSF. These data further support the model that constitutive expression of A1 initiates granulocytic differentiation of 32D cl3 cells.
Cell-surface antigen expression in 32DA1 cells
Cell line . | Gr-1, % . | CD69, % . | CD74, % . |
|---|---|---|---|
| 32DV4 | 6.7 | 19.5 | 1.6 |
| 32DWT1 | 13.1 | 22.8 | 2.4 |
| 32DA1.1 | 91.7 | 99.2 | 1.8 |
| 32DA1.2 | 55.8 | 84.3 | 1.8 |
| 32DA1.3 | 67 | 85.6 | 5.2 |
Cell line . | Gr-1, % . | CD69, % . | CD74, % . |
|---|---|---|---|
| 32DV4 | 6.7 | 19.5 | 1.6 |
| 32DWT1 | 13.1 | 22.8 | 2.4 |
| 32DA1.1 | 91.7 | 99.2 | 1.8 |
| 32DA1.2 | 55.8 | 84.3 | 1.8 |
| 32DA1.3 | 67 | 85.6 | 5.2 |
Cells were grown in IL-3. Expression of the indicated cell surface antigen was quantified by FACS.
Morphology of 32DA1 cells in different growth conditions. 32DV4 (A,C,E) and 32DA1.3 cells (B,D,F) were grown in IL-3 (A-B), without cytokine (C-D), or in G-CSF (E-F). Photomicrographs of cells attached to glass slides using a Cytospin apparatus and then stained with Wright/Giemsa stain are shown.
Morphology of 32DA1 cells in different growth conditions. 32DV4 (A,C,E) and 32DA1.3 cells (B,D,F) were grown in IL-3 (A-B), without cytokine (C-D), or in G-CSF (E-F). Photomicrographs of cells attached to glass slides using a Cytospin apparatus and then stained with Wright/Giemsa stain are shown.
Effect of A1 expression on chemosensitivity
WT1 expression has been associated with a poor outcome in AML patients, which may reflect decreased sensitivity to chemotherapeutic agents. Up-regulation of A1 expression provides a plausible mechanism for this observation. Accordingly, we investigated whether WT1 (–Ex5/–KTS) and A1 affect the sensitivity of 32D cl3 cells to chemotherapeutic drugs commonly used to treat AML. Cell lines undergoing exponential growth in IL-3 were treated with various concentrations of doxorubicin or cytarabine, both agents incorporated in the majority of AML treatment protocols, and cell viability was measured after 24 hours using an MTT assay. 32DWT1 cells treated with ZnCl2 to induce expression of WT1 (–Ex5/–KTS) were significantly less sensitive to the cytotoxic effects of cytarabine (Figure 7A). Treating the control cells with ZnCl2 did not affect their sensitivity to the drug, and the control cells were more sensitive than the WT1-expressing cells. The cell lines constitutively expressing A1 showed decreased sensitivity to cytarabine (Figure 7A), consistent with the postulated antiapoptotic effect of A1. Similar effects were seen in cultures treated with doxorubicin (Figure 7B). For each drug, the decreased sensitivity is overcome at high drug doses, suggesting that this is a relative, rather than an absolute, resistance to the drugs. Relative, not absolute, chemoresistance is clinically reported in AML patients with high level WT1 expression.26 It is unlikely that this chemoresistance reflects the differentiation status of the cell lines. Even though the A1-expressing cells express cell surface markers of granulocytic differentiation, they proliferate at the same rate as the control cells. Because cytarabine and doxorubicin are both preferentially toxic to dividing cells, these cell lines should be equivalently susceptible to the effects of the drugs. Our data, therefore, support a model of WT1-related resistance to chemotherapy meditated through up-regulation of A1.
Because the MTT assay we used to assess chemosensitivity measures the number of living cells, differences between cell lines might reflect differences in proliferation induced by the chemotherapy drugs, rather than an A1-related resistance to apoptosis. In order to distinguish between these possibilities, we assessed the degree of apoptosis in cells treated with 1 μM cytarabine using an annexin-V assay. Up-regulation of WT1 (–Ex5/–KTS) in the 32DWT1 cells resulted in a 50% decrease in the proportion of apoptotic cells (Figure 7C). In contrast, addition of ZnCl2 to cultures of the control 32DV4 cells modestly increased the proportion of apoptotic cells. The 3 independently derived cell lines constitutively expressing A1 contained proportions of apoptotic cells comparable to the WT1-expressing cells (Figure 7C). Similar results were seen with doxorubicin-treated cultures (data not shown). Thus, the results from the MTT assay reflect resistance to apoptosis mediated by expression of A1, not differences in proliferation.
Discussion
Using a myeloid cell line stably expressing an inducible form of WT1 (–Ex5/–KTS), we have demonstrated that the bCL2 family member A1 (designated BFL1 in humans) is a WT1 target gene. A1 was initially identified from a gene array screen, and up-regulation was confirmed by semi-quantitative RT-PCR. Promoter-reporter assays demonstrate that WT1 (–Ex5/–KTS) can regulate the A1 promoter in transiently transfected cells, and a ChIP assay shows that this isoform of WT1 interacts with the A1 promoter in vivo. Deletion mutagenesis of the A1 promoter identified a 71-bp fragment capable of mediating responsiveness to WT1, and this fragment contains a sequence (TCCACCAGCCTCC) that is similar to a previously identified WT1 binding site ((TCC)n). Finally, in a set of poor-prognosis primary AML samples, the relative rate of A1 expression is 1.6 in samples with WT1 expression, supporting the hypothesis that WT1 promotes A1 expression with a likelihood ratio of 2.7. These findings clearly place A1/BFL1 onto the list of bona fide WT1 target genes and strengthen the association between WT1 and the regulation of apoptosis. Moreover, our findings that constitutive expression of A1 in 32D cl3 cells mediates spontaneous granulocytic differentiation provide a mechanism for our previous observation that WT1 (–Ex5/–KTS) potentiates the G-CSF-mediated differentiation of these cells.
Expression of A1 leads to resistance to chemotherapy. The indicated cell lines were incubated for 24 hours in the indicated concentration of drug (panel A, cytarabine [Ara-C]; panel B, doxorubicin), and surviving cells quantified using an MTT assay. Results were normalized to the value obtained with no chemotherapy. Error bars represent standard error of the mean of triplicate assays. (C) Annexin V staining was performed with the same cell lines treated with 1 μM Ara-C as described. Apoptotic cells were defined as those which were Annexin V positive and negative for propidium iodide.
Expression of A1 leads to resistance to chemotherapy. The indicated cell lines were incubated for 24 hours in the indicated concentration of drug (panel A, cytarabine [Ara-C]; panel B, doxorubicin), and surviving cells quantified using an MTT assay. Results were normalized to the value obtained with no chemotherapy. Error bars represent standard error of the mean of triplicate assays. (C) Annexin V staining was performed with the same cell lines treated with 1 μM Ara-C as described. Apoptotic cells were defined as those which were Annexin V positive and negative for propidium iodide.
The mechanism by which WT1 regulates apoptosis appears to vary by cell type. WT1 was first implicated in the control of apoptosis by Kreidberg et al who noticed widespread apoptosis in the region where kidneys should have developed in WT1 knockout mice.3 Subsequently, several groups have examined the effect of WT1 on bCL2 with conflicting results. Hewitt et al, for example, reported that WT1 represses the bCL2 promoter,27 and Heckman et al found that WT1 represses the wild type bCL2 allele in lymphoma cells containing a chromosomal translocation involving the other bCL2 locus.28 In contrast, Mayo et al reported that in Saos-2 and CV1 cells, WT1 upregulates bCL2, while bCL2 is strongly down-regulated by WT1 in HeLa cells.29 We have looked by several different techniques and have never seen an effect of WT1 on bCL2 expression in 32D cl3 cells (data not shown). Morrison et al recently reported that, in Saos-2 cells stably transfected with WT1 under the control of an inducible promoter, up-regulation of WT1 expression results in up-regulation of the proapoptotic BAK gene.30 As with bCL2, we are unable to detect up-regulation of BAK by WT1 in 32D cl3 cells (data not shown). These findings, combined with our data with A1, demonstrate that WT1 affects the expression of multiple bCL2 family members in a lineage-specific manner.
Terminal differentiation of myeloid cells requires the coordinated regulation of survival, slowed proliferation, and expression of lineage-specific genes. Our previous studies revealed that WT1 (–Ex5/–KTS) potentiates G-CSF–mediated differentiation but is insufficient to induce granulocytic differentiation in the absence of cytokine.7 We demonstrated that WT1 (–Ex5/–KTS) suppresses the cyclin E promoter,17 slowing proliferation, but had not yet identified a mechanism by which this WT1 isoform affects apoptosis and myeloid-specific gene expression. Prior work implicating A1 in the G-CSF receptor signaling pathway18 fills these gaps in our understanding of the effect of WT1 (–Ex5/–KTS) on myelopoiesis. We have presented data here demonstrating that constitutive expression of A1 causes morphologic and molecular differentiation of 32D cl3 cells in the absence of G-CSF (Figures 5 and 6 and Table 3) and renders the cells relatively resistant to apoptosis (Figure 7). The high level of expression of granulocytic markers in the A1 transfectants compared with the WT1-transfected cells parallels their morphologic differentiation states: 32DWT1 cells are morphologically immature and are quite similar to 32DV4 cells, both in morphology and in cell surface antigen expression, while 32DA1.1, 32DA1.2, and 32DA1.3 cells have a sizable population of morphologically mature cells even in the presence of IL-3 and accordingly express higher levels of cell surface markers of mature granulocytes. Our data support our contention that WT1 (–Ex5/–KTS) affects multiple processes required for terminal differentiation: slowing proliferation via suppression of cyclin E, resisting apoptosis and promoting the expression of lineage-specific genes through up-regulation of A1. However, because WT1 (–Ex5/–KTS) expression is insufficient to induce terminal differentiation of 32D cl3 cells, other signals are clearly necessary to complete this process. These findings fit well with observations that WT1 is expressed very early in the course of normal myeloid differentiation, in myeloblasts that coexpress CD34 and CD38.4,5 These cells are relatively long-lived, proliferate slowly, but remain capable of differentiation. The ability of WT1 (–Ex5/–KTS) to down-regulate cyclin E,17 thus inhibiting proliferation, while up-regulating A1, thus protecting from apoptosis, is consistent with an important role in hematopoietic stem cell biology.
How can the growing body of data associating WT1 expression with granulopoiesis be made to fit with a model of WT1 overexpression contributing to leukemogenesis, especially in poor prognosis AML? Work from our lab and from others has shown that WT1 isoforms have distinct effects on target promoters. For example, we showed that WT1 (–Ex5/–KTS) suppresses the cyclin E promoter, and WT1 (+Ex5/+KTS) inhibits this suppression.17 Because cells express multiple WT1 isoforms simultaneously, the net effect of WT1 expression is likely to be a combinatorial effect of all of the expressed forms. This holds even in the setting of AML with WT1 mutations. In many AML samples, coexpression of mutant and wild-type WT1 is seen.31 Overexpression of WT1 could result in phenotypic changes consistent with leukemic transformation if antiapoptotic genes are upregulated, but antiproliferative genes are not. Our data are consistent with this model: WT1 (–Ex5/–KTS) suppresses cyclin E, WT1 (+Ex5/+KTS) blocks this effect, and mutant forms of WT1 do not affect this promoter at all.17
WT1 (–Ex5/–KTS) upregulates the antiapoptotic gene A1, and our preliminary experiments demonstrate that at least some mutant forms of WT1 do as well (data not shown). Thus, the net effect of endogenous WT1 overexpression, even taking into account expression of mutant forms of WT1, might be to promote leukemogenesis by up-regulating antiapoptotic genes without dramatically affecting the expression of genes related to proliferation. Future work will address this by coexpressing multiple, rather than individual, WT1 isoforms.
Prepublished online as Blood First Edition Paper, February 16, 2006; DOI 10.1182/blood-2005-10-4025.
Supported by grants to D.M.L. from the American Cancer Society (IRG-58-005-41), the Lauri Strauss Leukemia Foundation, Bear Necessities Pediatric Cancer Foundation, Flight Attendant Medical Research Institute, and the Children's Cancer Foundation. D.M.L. was a Fellow of the Leukemia and Lymphoma Society while this research was being performed. L.A.S. was supported by a grant from the National Cancer Institute (T32CA60441).
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Alan Friedman, Curt Civin, and Robert Arceci for critical review of the manuscript, and Leslie Meszler and the Johns Hopkins University microscopy core for assistance with photomicrographs.


![Figure 7. Expression of A1 leads to resistance to chemotherapy. The indicated cell lines were incubated for 24 hours in the indicated concentration of drug (panel A, cytarabine [Ara-C]; panel B, doxorubicin), and surviving cells quantified using an MTT assay. Results were normalized to the value obtained with no chemotherapy. Error bars represent standard error of the mean of triplicate assays. (C) Annexin V staining was performed with the same cell lines treated with 1 μM Ara-C as described. Apoptotic cells were defined as those which were Annexin V positive and negative for propidium iodide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/12/10.1182_blood-2005-10-4025/4/m_zh80120696830007.jpeg?Expires=1769284626&Signature=h4tUBiJu9thmSd7p4VgW7YPSB8HzAAkbijXzYsBC3UpEfXf0jdHKPlJy7ETq0PtnDXN2PDiEDXFtTDEd-CpJebeFe4lFiqLVrY7AZg-88QdgNVTISqoOJQRjmTzLPriMaiAFLSuYbisrIvRDHX6k7GQshvR7zeL8EkuTngbp3BgkJGPta87WEeBHRTSmXEIX1yezx69msF~P6cUIYuSQoJwzDPgoNNSGGRX4eDVxbTsVUf1-vM54fUjmDjf92PFLnkXmyiF2lkKQQKoUlqUKqUq-jsOlDta~IRtPOeloaQFf80IuEnQwgj7tRGUlI8oNqBL9Ka5BZqJItFEAlwEuUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal