Abstract
The flow-responsive transcription factor KLF2 is acquiring a leading role in the regulation of endothelial cell gene expression. A genome-wide microarray expression profiling is described employing lentivirus-mediated, 7-day overexpression of human KLF2 at levels observed under prolonged flow. KLF2 is not involved in lineage typing, as 42 endothelial-specific markers were unaffected. Rather, KLF2 generates a gene transcription profile (> 1000 genes) affecting key functional pathways such as cell migration, vasomotor function, inflammation, and hemostasis and induces a morphology change typical for shear exposure including stress fiber formation. Protein levels for thrombomodulin, endothelial nitric oxide synthase, and plasminogen activator inhibitor type-1 are altered to atheroprotective levels, even in the presence of the inflammatory cytokine TNF-α. KLF2 attenuates cell migration by affecting multiple genes including VEGFR2 and the potent antimigratory SEMA3F. The distribution of Weibel-Palade bodies in cultured cell populations is normalized at the single-cell level without interfering with their regulated, RalA-dependent release. In contrast, thrombin-induced release of Weibel-Palade bodies is significantly attenuated, consistent with the proposed role of VWF release at low–shear stress regions of the vasculature in atherosclerosis. These results establish that KLF2 acts as a central transcriptional switch point between the quiescent and activated states of the adult endothelial cell.
Introduction
The vascular endothelium plays a leading role in maintaining vascular homeostasis by controlling various processes in the vascular wall, on the endothelial cell surface, and in the blood stream. Although the differential phenotype of endothelial cells at different positions of the vascular tree has become an intriguing field of study over the past decades, all endothelial cells are derived from the same origin.1 During embryogenesis, in a process known as vasculogenesis, the vascular tract develops from the vascular plexus, which in turn is derived from the blood islands that constitute both endothelium-forming and blood-forming cells. Blood islands develop from the hemangioblast, the common precursor for the endothelial cell and the hematopoietic cell.2 Throughout the further development and maturation of the vascular network, the endothelial-forming cells differentiate into a fully functional endothelial cell and finally acquire their endothelial-specific properties that are involved in the regulation of hemostasis, endothelial barrier function, inflammatory response, vasomotor function, leukocyte extravasation, and angiogenesis.3 Therefore, in contrast to the lineage-specific gene expression that uniquely defines the endothelial precursor during the various stages of vasculogenesis, the expression of a discrete group of mostly endothelial-specific genes is vital to the functionally differentiated mature endothelium. It has been postulated that shear stress, the tangential force exerted by the flowing blood on the vascular endothelial lining, is of prime importance in determining endothelial phenotype.4,5
Over recent years, the zinc finger transcription factor lung Krüppel-like factor (LKLF/KLF2) has emerged as a prime and pivotal candidate for directly relaying biomechanical shear forces into a gene transcription profile that might determine endothelial phenotype in response to flow.6-10 We previously demonstrated that KLF2 mRNA and protein levels are substantially and uniquely induced by prolonged steady or pulsatile laminar flow both in vitro and in vivo.6,10 Furthermore, we have demonstrated that KLF2 is specifically expressed in endothelial cells of the adult vasculature, but its expression drops to low levels near sites of flow disturbance, coinciding with regions that show pathologic changes of the vascular wall.10 The remarkably stable transcriptional induction of KLF2 specifically by flow therefore suggests a role for KLF2 as an intermediary transcriptional regulator of endothelial flow–specific gene expression. Indeed, we showed KLF2 to be essential in the shear-induced upregulation of endothelial nitric oxide synthase (eNOS), one of the prime flow-responsive endothelial genes, by combined siRNA and flow experiments.9 In addition, KLF2 has recently been identified as a pivotal transcription factor in regulating endothelial vasomotor function, inflammatory response, hemostasis, and angiogenesis.7-10 Still, despite this seemingly central role in various aspects of endothelial physiology, early studies using knock-out mice have demonstrated that the absence of KLF2 does not impede primary vasculogenesis.11 Rather, the embryonic lethal phenotype is caused by the fact that KLF2 is essential for the stabilization of the immature vascular tract by recruited smooth muscle cells. Given the tentative pivotal role of flow on endothelial physiology, a dissection of KLF2-induced gene expression profiles from general flow-induced processes would be a first step in identifying the specific role of KLF2 in the endothelial gene expression repertoire and differentiation.
We investigated the steady-state changes in endothelial gene expression that are caused by a prolonged and stable overexpression of KLF2. By using genome-wide microarray-based gene expression analysis, we established the full repertoire of both directly and indirectly KLF2-regulated endothelial cell gene expression. The primary bioinformatics analysis is complemented by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) and protein and cell biologic analysis, most specifically illustrated by KLF2 effects on von Willebrand Factor (VWF), a key endothelial marker protein. These data reveal that KLF2 systematically controls the expression of many genes that are involved in the functionally mature differentiated endothelium but is not involved in determining endothelial lineage typing.
Materials and methods
Cell culturing and lentiviral KLF2 overexpression
Human umbilical vein endothelial cell (HUVEC) isolation, culturing, and lentiviral overexpression of KLF2 was previously described.6,10,12 Recombinant human vascular endothelial growth factor (VEGF) 165 (rhVEGF165) and rhTNF-α (R&D Systems, Minneapolis, MN) were used at concentrations of 40 and 10 ng/mL, respectively.
RNA isolation, microarray expression profiling, and pathway analysis
Total RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA) and enriched for poly-A+ using the Oligotex mRNA Minikit (Qiagen, Hilden, Germany). Aminoallyl (AA)–labeled first-strand cDNA probes were synthesized from 1.5 μg poly-A+ RNA using SuperScript II (Invitrogen) with a molar ratio of aminoallyl-dUTP (Sigma, St Louis, MO) to dUTP of 4:1. Labeled cDNA was purified using the QIAquick PCR Purification Kit (Qiagen), and Cy3 or Cy5 mono-reactive dyes (Amersham Biosciences, Piscataway, NJ) were coupled according to the manufacturer's instructions. Purified Cy3- and Cy5-labeled cDNAs were hybridized to the microarrays for 20 hours at 40°C in Microarray Hybridization Solution (Amersham) and 35% formamide (Sigma). Microarrays were glass based containing 60-mer oligonucleotide sequences (Sigma/Compugen Library), which represent 18 650 human genes (Micro Array Department, Swammerdam Institute of Life Sciences, Amsterdam, The Netherlands). Images were acquired using the Agilent-II scanner (Agilent Technologies, Palo Alto, CA) and processed by ArrayVision 8.0 software (Imaging Research, St Catharines, ON, Canada). Background-subtracted intensities were Loess normalized (Limma package, Bioconductor Software, http://www.bioconductor.org) and imported into the Rosetta Resolver database and analysis software (Rosetta Biosoftware, Seattle, WA). Statistical analysis was performed using false discovery rate (FDR)–corrected P values, involving a recalculation of the P values using the Benjamini-Hochberg correction for multiple testing.13 Genes with a Locuslink14 ID (16 234) were imported into pathway analysis software (MAPPfinder version 2.0; Gladstone Institutes, San Francisco, CA)15 and PubMed-mining software (Ingenuity Pathway Analysis version 2; Ingenuity Systems, Mountain View, CA) for further analysis.
Semiquantitative real-time RT-PCR
Real-time RT-PCR was performed on total RNA isolated using the Absolutely RNA RT-PCR Miniprep Kit (Stratagene, La Jolla, CA) as described.10 All primers were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi).16 After correction for the house-keeping controls hypoxanthine phosphoribosyltransferase 1 and large ribosomal phosphoprotein P0, the mRNA levels were expressed as ratios compared with the mock-transduced control cultures.
Western blotting and immunofluorescence
Western blotting and immunofluoresence were performed as described.10,17 VEGF receptor 2 (VEGFR2/KDR), phospho-VEGFR2, thrombomodulin (TM), plasminogen activator inhibitor type 1 (PAI-1), eNOS, and RalA antibodies were from R&D Systems, Abcam (Cambridge, United Kingdom), DakoCytomation (Glostrup, Denmark), Biopool (Umea, Sweden), and BD Transduction Laboratories (San Diego, CA), respectively. Specific peptide antisera against human KLF2 were previously described.10
For immunofluorescence, KLF2- and mock-transfected HUVECs were grown on gelatin-coated glass coverslips, fixed with paraformaldehyde, and embedded in 10% (wt/vol) Mowiol (Calbiochem, San Diego, CA). Secondary antibodies were TRITC labeled, and phalloidin-FITC/TRITC was used as a stain for F-actin. Photomicrographs were acquired using a Zeiss Axioplan 2 microscope equipped with 10 × /0.3 numeric aperture (NA), 20 × /0.5 NA, 63 × /1.4 NA, and 100 ×/1.5 NA Plan Neofluar objectives (Zeiss, Oberkochen, Germany) and a Coolsnap HQ digital camera (Roper Scientific, Ottobrunn, Germany).
VWF was detected using a polyclonal anti-VWF antibody (DakoCytomation) and a Texas Red–labeled secondary goat antirabbit antibody. Cells were embedded and viewed by confocal microscopy using a Zeiss LSM510. Images were generated by making optical sections (Z-stacks with 400-nm intervals) and 3-dimensional analysis using depth-coding software (application for Zeiss LSM510 Version 2.3) as described.18 Z-stacks were taken after stimulation and Weibel-Palade bodies (WPBs) were counted.
Protein-C activation assay
Assays were performed as described.19 Confluent monolayers of HUVECs were washed 3 times and assay buffer containing 12 nM thrombin and 5 ng/mL protein C was added. After 20 minutes, an aliquot was removed and protein-C activation was quenched by adding hirudin (Sigma) to 40 nM. The chromogenic substrate S-2366 was added and the rate of change in optical absorbance at 405 nm was monitored with the EL808 Microplate Reader (Bio-TEK, Winooski, VT).
Migration assay
Wounding assays were performed as described.20 Briefly, confluent HUVEC monolayers, which had been transduced with lentivirus expressing the KLF2 mRNA (Lenti-KLF2)10 or a mock lentivirus (Lenti-mock), were grown on fibronectin. Cells were removed by scraping across the surface of a 6-well culture plate using a modified plastic cell scraper. Standard phase-contrast microphotographs were acquired using an Olympus CKX31 microscope equipped with a 4 × /0.13 NA objective (Olympus, Tokyo, Japan) and a Sony DXC-950P digital camera (Sony, Tokyo, Japan), and were analyzed in Adobe Photoshop 5.0 (Adobe Systems, San Jose, CA). The numbers of pixels that the wounds had decreased in width after 24, 48, or 72 hours were converted to distance (μm) by calibrating the setup with a Buerker cell counting chamber (Fein-Optik, Bad Blankenburg, Germany).
Assaying von Willebrand factor release
The thrombin- or forskolin-induced secretion of VWF from KLF2-transduced or mock-transduced HUVECs was assessed by quantification of secreted VWF antigen and by counting the decrease in the number of WPBs in fixed cells, essentially as described.21 In brief, endothelial cells grown for 24 hours in 6-well plates were washed 2 times with serum-free medium (SFM) containing 1% human albumin. Subsequently, the cells were preincubated with SFM for 1 hour, which was substituted with SFM containing either forskolin or epinephrine in combination with 100 μM isobutylmethylxanthine (IBMX; Sigma-Aldrich Chemie, Steinheim, Germany), thrombin alone, or no stimulating agent.
Statistical analysis
Data are given as mean with error bars representing the standard error of the mean (SEM) for the indicated number of experiments. The unpaired Student t test was used to calculate the statistical significance of the expression ratios versus control cultures. P values less than .05 were considered statistically significant.
Results
Identification of the KLF2 transcriptional targets
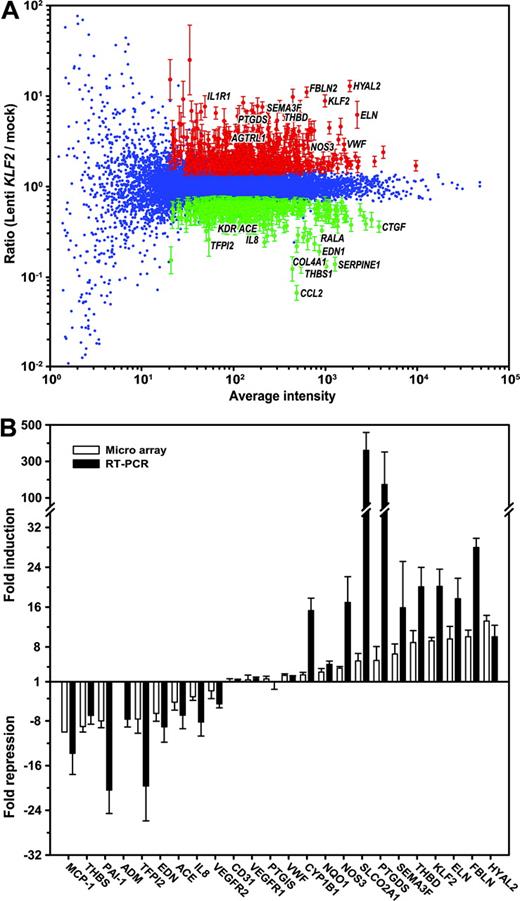
We previously described a lentivirus-driven overexpression approach, where human KLF2 is expressed using the relatively moderate phosphoglycerate kinase (PGK) promoter, thereby increasing KLF2 expression to levels similar to those observed in HUVECs that are exposed to prolonged pulsatile flow.10 Following transduction of 5 independent HUVEC cultures with the KLF2 lentivirus or mock lentivirus, cells were cultured for a period of 7 days to allow the establishment of a steady-state gene expression pattern. The stable overexpression of the KLF2 mRNA was confirmed by real-time RT-PCR, showing a 20 ± 3-fold (n = 5; P < .006) induction compared with mock transduction (Figure 1B). In order to determine the complete repertoire of KLF2 downstream genes in endothelial cells, gene expression analysis was performed on these 5 independent overexpression experiments using oligonucleotide-based microarrays containing specific 60-mer oligonucleotides for 18 600 human genes. For each experiment, fluorescently labeled cDNA probes, derived from the mock and KLF2 transductions, were hybridized to the arrays with replicate dye-swap hybridizations. Thus, a total of 10 experimental and technical replicate expression data sets comparing mock and KLF2 transductions were obtained, and significant differential gene expression was statistically evaluated using the Rosetta Resolver Gene Expression Data Analysis System. Figure 1A shows the composite plot of the complete 10-array data set. After data filtering (ie, absolute ratio > 1.5, 10-base log of average intensity > 1.3, and FDR-corrected P < .001), a total of 1039 (600 induced, 439 repressed) genes remained that were statistically significant differentially expressed (Tables S1-S2, available on the Blood website; see the Supplemental Tables link at the top of the online article). Real-time RT-PCR was used to verify the KLF2-mediated transcriptional regulation of a selection of genes (Figure 1B). The results show that all 24 genes tested were regulated to similar or higher extents when probed by independent RT-PCR analysis. Thus, the high number of replicates together with a thorough downstream statistical analysis have reliably identified the KLF2-responsive gene repertoire.
Lineage-specific endothelial gene expression is not controlled by KLF2
The genomic profiling indicates a pleiotropic effect of KLF2 on endothelial gene expression, raising the possibility that KLF2 is involved in determining endothelial lineage. The endothelium expresses a number of lineage marker genes that are already expressed during the early stages of vascular patterning and primary vascular development.1,2,22,23 The gene expression data presented here demonstrate that prolonged KLF2 overexpression only marginally affects the expression of these marker genes that are important during early vasculogenesis (Table 1; Figure 1B), with the exception of VEGFR2. Real-time RT-PCR analysis indeed validated the significant regulation of VEGFR1 (FLT1) and VEGFR2 (KDR) by a fold change of 1.7 ± 0.3 and –5.0 ± 0.7, respectively (Figure 1B). Thus, KLF2 is not a key regulator of the expression of the genes that are known to define the endothelial lineage and that are involved in vascular patterning, vasculogenesis, and angiogenesis during embryonic development.
Effect of KLF2 on the expression of endothelial lineage markers
Gene symbol . | Gene description* . | Fold change . | P . |
|---|---|---|---|
| ANGPT1 | Angiopoietin 1, transcript variant 1 | 1.05 | .93 |
| ANGPT2 | Angiopoietin 2 | -1.14 | .65 |
| BMP2 | Bone morphogenetic protein 2 | -1.42 | < .001 |
| BMPER | BMP-binding endothelial regulator precursor protein | 1.02 | .75 |
| BMPRIA | Bone morphogenetic protein receptor, type IA | 1.46 | < .001 |
| CD34 | CD34 antigen | 1.06 | .60 |
| CDH5 | VE-cadherin (vascular endothelium) | -1.08 | .56 |
| CLDN5 | Claudin 5 | -1.08 | .37 |
| EDG1-7 | Endothelial differentiation G-protein—coupled receptors 1-7 | -1.3-1.5 | .008-.9 |
| EFNA1-5 | Ephrin-A1, A2, A3, A4, A5 | -1.7-1.1 | < .001-.9 |
| EFNB1-3 | Ephrin-B1, B2, B3 | -1.3-1.5 | .002-.1 |
| ENG | Endoglin (Osler-Rendu-Weber syndrome 1) | 1.30 | .001 |
| EPHB4 | Ephrin receptor B4 | 1.52 | < .001 |
| FLT1 | Vascular endothelial growth factor receptor-1 (VEGFR1/Flt) | 1.40 | < .001 |
| FZD4 | Frizzled homolog 4 (Drosophila) | -1.25 | .012 |
| GJA4 | Gap junction protein alpha 4 (connexin 37) | -1.46 | < .001 |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | -1.03 | .74 |
| HEY2 | Hairy/enhancer-of-split related with YRPW motif 2 | -1.01 | .90 |
| JAG2 | Jagged 2, transcript variant 1 | -1.90 | < .001 |
| KDR | Vascular endothelial growth factor receptor-2 (VEGFR2/KDR/Flk-1) | -2.61 | < .001 |
| NOTCH1 | Homolog of Drosophila Notch (TAN1) | 1.09 | .39 |
| NOTCH2 | Homolog of Drosophila Notch | 1.14 | .044 |
| NOTCH3 | Homolog of Drosophila Notch | -1.32 | .002 |
| NOTCH4 | Homolog of Drosophila Notch (INT3) | -1.45 | < .001 |
| NRP1 | Neuropilin 1 | -1.44 | .029 |
| NUMB | Numb homolog (Drosophila) | -1.07 | .36 |
| PECAM1 | Platelet/endothelial cell adhesion molecule (CD31) | 1.11 | .22 |
| PROM1 | Prominin 1 | 1.13 | .16 |
| SEMA3C | Semaphorin 3C | 1.10 | .16 |
| SIAH1 | Seven in absentia homolog 1 (Drosophila) | -1.15 | .065 |
| TIE | Tyrosine kinase with immunoglobulin and EGF homology domains | -1.00 | .96 |
Gene symbol . | Gene description* . | Fold change . | P . |
|---|---|---|---|
| ANGPT1 | Angiopoietin 1, transcript variant 1 | 1.05 | .93 |
| ANGPT2 | Angiopoietin 2 | -1.14 | .65 |
| BMP2 | Bone morphogenetic protein 2 | -1.42 | < .001 |
| BMPER | BMP-binding endothelial regulator precursor protein | 1.02 | .75 |
| BMPRIA | Bone morphogenetic protein receptor, type IA | 1.46 | < .001 |
| CD34 | CD34 antigen | 1.06 | .60 |
| CDH5 | VE-cadherin (vascular endothelium) | -1.08 | .56 |
| CLDN5 | Claudin 5 | -1.08 | .37 |
| EDG1-7 | Endothelial differentiation G-protein—coupled receptors 1-7 | -1.3-1.5 | .008-.9 |
| EFNA1-5 | Ephrin-A1, A2, A3, A4, A5 | -1.7-1.1 | < .001-.9 |
| EFNB1-3 | Ephrin-B1, B2, B3 | -1.3-1.5 | .002-.1 |
| ENG | Endoglin (Osler-Rendu-Weber syndrome 1) | 1.30 | .001 |
| EPHB4 | Ephrin receptor B4 | 1.52 | < .001 |
| FLT1 | Vascular endothelial growth factor receptor-1 (VEGFR1/Flt) | 1.40 | < .001 |
| FZD4 | Frizzled homolog 4 (Drosophila) | -1.25 | .012 |
| GJA4 | Gap junction protein alpha 4 (connexin 37) | -1.46 | < .001 |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | -1.03 | .74 |
| HEY2 | Hairy/enhancer-of-split related with YRPW motif 2 | -1.01 | .90 |
| JAG2 | Jagged 2, transcript variant 1 | -1.90 | < .001 |
| KDR | Vascular endothelial growth factor receptor-2 (VEGFR2/KDR/Flk-1) | -2.61 | < .001 |
| NOTCH1 | Homolog of Drosophila Notch (TAN1) | 1.09 | .39 |
| NOTCH2 | Homolog of Drosophila Notch | 1.14 | .044 |
| NOTCH3 | Homolog of Drosophila Notch | -1.32 | .002 |
| NOTCH4 | Homolog of Drosophila Notch (INT3) | -1.45 | < .001 |
| NRP1 | Neuropilin 1 | -1.44 | .029 |
| NUMB | Numb homolog (Drosophila) | -1.07 | .36 |
| PECAM1 | Platelet/endothelial cell adhesion molecule (CD31) | 1.11 | .22 |
| PROM1 | Prominin 1 | 1.13 | .16 |
| SEMA3C | Semaphorin 3C | 1.10 | .16 |
| SIAH1 | Seven in absentia homolog 1 (Drosophila) | -1.15 | .065 |
| TIE | Tyrosine kinase with immunoglobulin and EGF homology domains | -1.00 | .96 |
Gene expression profiling of Lenti-KLF2– and Lenti-mock–transduced HUVECs. Microarray dye-swap hybridization analysis was performed comparing the gene expression profiles of 5 independent HUVEC isolates that were analyzed at 7 days after transduction with the Lenti-KLF2 virus and Lenti-mock virus. The composite plot of the 10 hybridizations is shown in panel A. The Loessnormalized weighted average hybridization signal intensity on the horizontal axis is plotted against the 10-base log of the ratio of Lenti-KLF2 over Lenti-mock. The genes with a statistically significant fold change (P < .01; according to the Rossetta Resolver error models and statistical evaluation) are shown in red (induced by KLF2) and green (repressed by KLF2) with error bars derived from the Rosetta Resolver error model. (B) Real-time semiquantitative RT-PCR verification of the microarray data on a selection of the genes presented in Tables 1 and 2. The RT-PCR was performed on the same cDNA preparations that were used for the microarray hybridization (n = 5). The data are represented as the mean ± SEM fold induction or fold repression compared with the Lenti-mock controls (▪). Also, the fold change in expression derived from the microarray data (□) is included for comparison.
Gene expression profiling of Lenti-KLF2– and Lenti-mock–transduced HUVECs. Microarray dye-swap hybridization analysis was performed comparing the gene expression profiles of 5 independent HUVEC isolates that were analyzed at 7 days after transduction with the Lenti-KLF2 virus and Lenti-mock virus. The composite plot of the 10 hybridizations is shown in panel A. The Loessnormalized weighted average hybridization signal intensity on the horizontal axis is plotted against the 10-base log of the ratio of Lenti-KLF2 over Lenti-mock. The genes with a statistically significant fold change (P < .01; according to the Rossetta Resolver error models and statistical evaluation) are shown in red (induced by KLF2) and green (repressed by KLF2) with error bars derived from the Rosetta Resolver error model. (B) Real-time semiquantitative RT-PCR verification of the microarray data on a selection of the genes presented in Tables 1 and 2. The RT-PCR was performed on the same cDNA preparations that were used for the microarray hybridization (n = 5). The data are represented as the mean ± SEM fold induction or fold repression compared with the Lenti-mock controls (▪). Also, the fold change in expression derived from the microarray data (□) is included for comparison.
KLF2 controls endothelial functional differentiation
The large number of KLF2 downstream genes that were identified necessitates sorting them into distinct processes that are of particular interest to endothelial cell biology. Therefore, a combination of Gene Ontology24 ordering, GenMAPP pathway analysis, and Ingenuity PubMed text mining was performed to provide a global functional view on the KLF2 effect. This approach ranks genes into the different processes in which they are involved, consistent with the complex matrix of genes and functions in biology. Thus, a number of categories were identified showing a high level of regulation by KLF2. Remarkably, they describe typical vascular biologic processes in which the fully differentiated endothelium of the healthy, mature vessel wall plays a key role (Table 2). The direct or indirect involvement of KLF2 in the expression of a number of the key genes for these distinct functional classes was validated by real-time PCR (Figure 1B) according to their primary function in hemostasis (SERPINE1, TFPI2, VWF, THBD), inflammation (MCP1, IL8), vascular tone regulation (ADM, EDN1, ACE, NQO1, NOS3, PTGIS, SLCO2A1, PTGDS), angiogenesis/migration (VEGFR-1 and VEGFR-2, NOS3, SEMA3F), and composition of the endothelial basement membrane/vascular wall matrix (THBS, ELN, FBLN, HYAL2).
KLF2-responsive genes assigned to the principal categories of endothelial function
Functional category and gene symbols . | Fold change* . |
|---|---|
| Cell migration | |
| ADAM15 | 4.1 |
| CCL2 | -15.4 |
| CCR8 | 2.0 |
| CXCR4 | -1.9 |
| EDN1 | -5.3 |
| HPSE | 3.5 |
| ID1 | 2.6 |
| ID3 | -3.1 |
| IGF2 | 4.1 |
| IL8 | -2.9 |
| KDR | -2.6 |
| NOS3 | 2.7 |
| PTHLH | 7.5 |
| SEMA3F | 6.6 |
| SERPINE1 | -7.4 |
| THBS1 | -7.8 |
| Angiogenesis, vascularization, and development of the artery | |
| BMP4 | -2.7 |
| CCL2 | -15.4 |
| CTGF | -2.8 |
| ECM1 | 2.8 |
| FGF5 | -2.5 |
| HPSE | 3.5 |
| ID1 | 2.6 |
| ID3 | -3.1 |
| IL8 | -2.9 |
| ITGB5 | 3.2 |
| KDR | -2.6 |
| NOS3 | 2.7 |
| PPAP2B | 2.6 |
| PTHLH | 7.5 |
| PTPRJ | 3.3 |
| SEMA3F | 6.6 |
| SERPINE1 | -7.4 |
| THBS1 | -7.8 |
| TIMP1 | 2.7 |
| Vasomotor and blood pressure regulation | |
| ACE | -3.0 |
| ADM | -7.7 |
| AGTRL1 | 3.4 |
| APLN | -2.4 |
| EDN1 | -5.3 |
| NOS3 | 2.7 |
| PTHLH | 7.5 |
| Immune and lymphatic system development and function | |
| BMP4 | -2.7 |
| CIQR1 | -4.4 |
| CCL2 | -15.4 |
| CCND3 | 1.9 |
| CRP | -2.2 |
| CXCR4 | -1.9 |
| DLL1 | 2.2 |
| DOCK2 | -1.9 |
| EDN1 | -5.3 |
| FGR | 2.0 |
| ICOSL | -2.7 |
| ID1 | 2.6 |
| ID3 | -3.1 |
| IGF2 | 4.1 |
| ILIRL1 | 2.6 |
| IL8 | -2.9 |
| KCNN4 | 6.4 |
| KITLG | 3.5 |
| LBP | -2.2 |
| LGALS9 | 2.6 |
| LTC4S | 5.2 |
| MAPK9 | 3.0 |
| MVP | 2.0 |
| NDRG1 | 5.9 |
| NOS3 | 2.7 |
| PROS1 | 2.0 |
| PTPRJ | 3.3 |
| SFTPD | 5.7 |
| STAT1 | -2.1 |
| THBD | 6.1 |
| THBS1 | -7.8 |
| TRAF6 | -2.2 |
| Chemotaxis and infiltration of leukocytes, granulocytes, and lymphocytes | |
| CCL2 | -15.4 |
| CCR8 | 2.0 |
| CRP | -2.2 |
| CXCR4 | -1.9 |
| DAF | 2.0 |
| DOCK2 | -1.9 |
| ELN | 6.1 |
| ILIRL1 | 2.6 |
| IL8 | -2.9 |
| KITLG | 3.5 |
| LGALS9 | 2.6 |
| NFKBIA | 1.9 |
| NOS3 | 2.7 |
| RGS3 | -2.4 |
| SFTPD | 5.7 |
| THBS1 | -7.8 |
| Hemostasis and hematologic disorder | |
| BCL3 | 1.9 |
| CCR8 | 2.0 |
| CXCR4 | -1.9 |
| EPAS1 | 2.1 |
| IGF2 | 4.1 |
| ILIRL1 | 2.6 |
| IL8 | -2.9 |
| KITLG | 3.5 |
| LEPR | -1.9 |
| MT2A | -2.2 |
| NFKBIA | 1.9 |
| PROS1 | 2.0 |
| PTHLH | 7.5 |
| SERPINE1 | -7.4 |
| TFP1 | -1.9 |
| THBD | 6.1 |
| VWF | 2.5 |
Functional category and gene symbols . | Fold change* . |
|---|---|
| Cell migration | |
| ADAM15 | 4.1 |
| CCL2 | -15.4 |
| CCR8 | 2.0 |
| CXCR4 | -1.9 |
| EDN1 | -5.3 |
| HPSE | 3.5 |
| ID1 | 2.6 |
| ID3 | -3.1 |
| IGF2 | 4.1 |
| IL8 | -2.9 |
| KDR | -2.6 |
| NOS3 | 2.7 |
| PTHLH | 7.5 |
| SEMA3F | 6.6 |
| SERPINE1 | -7.4 |
| THBS1 | -7.8 |
| Angiogenesis, vascularization, and development of the artery | |
| BMP4 | -2.7 |
| CCL2 | -15.4 |
| CTGF | -2.8 |
| ECM1 | 2.8 |
| FGF5 | -2.5 |
| HPSE | 3.5 |
| ID1 | 2.6 |
| ID3 | -3.1 |
| IL8 | -2.9 |
| ITGB5 | 3.2 |
| KDR | -2.6 |
| NOS3 | 2.7 |
| PPAP2B | 2.6 |
| PTHLH | 7.5 |
| PTPRJ | 3.3 |
| SEMA3F | 6.6 |
| SERPINE1 | -7.4 |
| THBS1 | -7.8 |
| TIMP1 | 2.7 |
| Vasomotor and blood pressure regulation | |
| ACE | -3.0 |
| ADM | -7.7 |
| AGTRL1 | 3.4 |
| APLN | -2.4 |
| EDN1 | -5.3 |
| NOS3 | 2.7 |
| PTHLH | 7.5 |
| Immune and lymphatic system development and function | |
| BMP4 | -2.7 |
| CIQR1 | -4.4 |
| CCL2 | -15.4 |
| CCND3 | 1.9 |
| CRP | -2.2 |
| CXCR4 | -1.9 |
| DLL1 | 2.2 |
| DOCK2 | -1.9 |
| EDN1 | -5.3 |
| FGR | 2.0 |
| ICOSL | -2.7 |
| ID1 | 2.6 |
| ID3 | -3.1 |
| IGF2 | 4.1 |
| ILIRL1 | 2.6 |
| IL8 | -2.9 |
| KCNN4 | 6.4 |
| KITLG | 3.5 |
| LBP | -2.2 |
| LGALS9 | 2.6 |
| LTC4S | 5.2 |
| MAPK9 | 3.0 |
| MVP | 2.0 |
| NDRG1 | 5.9 |
| NOS3 | 2.7 |
| PROS1 | 2.0 |
| PTPRJ | 3.3 |
| SFTPD | 5.7 |
| STAT1 | -2.1 |
| THBD | 6.1 |
| THBS1 | -7.8 |
| TRAF6 | -2.2 |
| Chemotaxis and infiltration of leukocytes, granulocytes, and lymphocytes | |
| CCL2 | -15.4 |
| CCR8 | 2.0 |
| CRP | -2.2 |
| CXCR4 | -1.9 |
| DAF | 2.0 |
| DOCK2 | -1.9 |
| ELN | 6.1 |
| ILIRL1 | 2.6 |
| IL8 | -2.9 |
| KITLG | 3.5 |
| LGALS9 | 2.6 |
| NFKBIA | 1.9 |
| NOS3 | 2.7 |
| RGS3 | -2.4 |
| SFTPD | 5.7 |
| THBS1 | -7.8 |
| Hemostasis and hematologic disorder | |
| BCL3 | 1.9 |
| CCR8 | 2.0 |
| CXCR4 | -1.9 |
| EPAS1 | 2.1 |
| IGF2 | 4.1 |
| ILIRL1 | 2.6 |
| IL8 | -2.9 |
| KITLG | 3.5 |
| LEPR | -1.9 |
| MT2A | -2.2 |
| NFKBIA | 1.9 |
| PROS1 | 2.0 |
| PTHLH | 7.5 |
| SERPINE1 | -7.4 |
| TFP1 | -1.9 |
| THBD | 6.1 |
| VWF | 2.5 |
Fold change in the average micro array hybridization signal intensities (n=10) of Lenti-KLF2- over Lenti-mock-transduced HUVECs (P < .01).
KLF2 alters endothelial morphology and migration
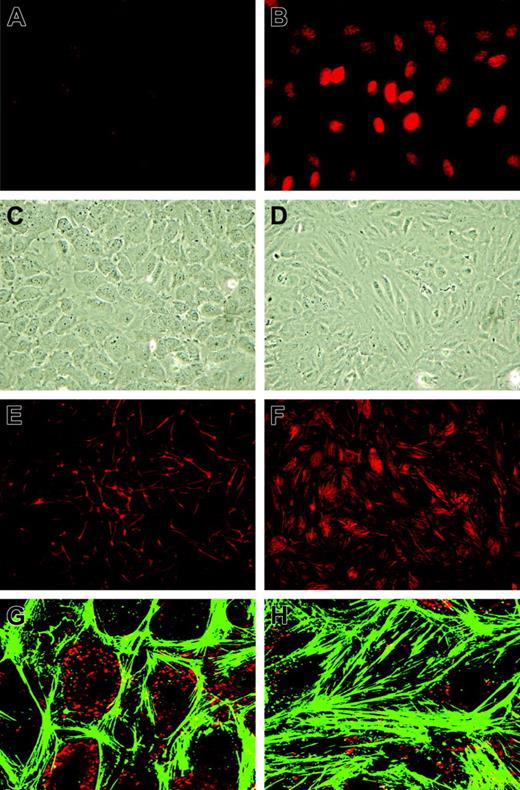
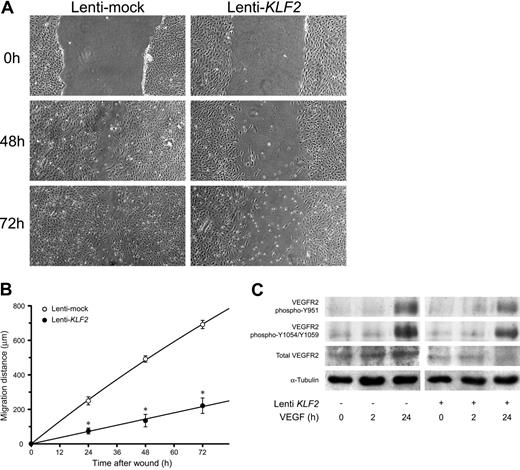
As many genes and processes are affected by sustained KLF2 expression, we first investigated its effect on classical cultured HUVEC phenotype. First, correct targeting of the KLF2 protein to the endothelial nucleus was evidenced by using specific peptide antisera10 in immunofluorescence (Figure 2A-B). Phase-contrast microscopy showed a stretched appearance of HUVECs overexpressing KLF2 for 5 days compared with the typical cobblestone shape of the confluent mock-transduced cultures (Figure 2C-D). Lenti-KLF2 cultures that were allowed to grow to complete confluence had a cell density of 596 ± 10 cells/mm2 compared with 382 ± 13 cells/mm2 for Lenti-mock cultures. Staining for F-actin using fluorescently labeled phalloidin revealed a major reorganization of the cytoskeleton after prolonged KLF2 overexpression, characterized by the formation of typical stress fibers (Figure 2E-H). As a substantial number of KLF2-sensitive genes have been implicated in cell motility by pathway analysis (Table 2), endothelial cell migration was studied using a standard wounding assay of HUVECs growing on a fibronectin matrix. Untransduced and mock-virus–transduced HUVEC cultures both exhibited a constant migration rate of approximately 10 μm/h (Figure 3A-B), closing the 1.5-mm wound within 72 hours. In marked contrast, wounded KLF2-overexpressing monolayers exhibited a greatly reduced migration rate of 2.8 μm/h and failed to close within 72 hours. As cell motility is an important aspect of angiogenesis, we next investigated the potential involvement of the decreased angiogenic response caused by the downregulation of VEGFR2 that was observed at the mRNA level (Figure 1B; Table 1). In agreement with the decrease in gene expression, the total VEGFR2 protein levels are shown to be decreased by KLF2 overexpression and were not significantly affected by stimulation with VEGF for up to 24 hours (Figure 3C). Unexpectedly, KLF2 overexpression did not have a large effect on the VEGF-induced phosphorylation of the VEGFR2 protein at residues Y951 and Y1059 that are essential for VEGF-induced proliferation and migration, respectively (Figure 3C). Thus, the observed KLF2-induced decrease in total VEGFR2 protein only marginally affects its VEGF-mediated phosphorylation that is responsible for cell migration. The robust 16-fold induction of semaphorin-3F (SEMA3F) by KLF2 (Figure 1B) can be essential with respect to cell migration,25 but further functional studies are presently precluded by a lack of specific antibodies.
KLF2 controls stable expression of endothelial marker genes
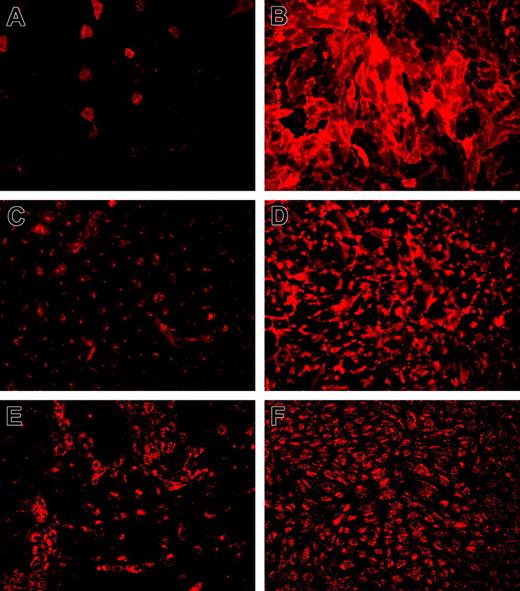
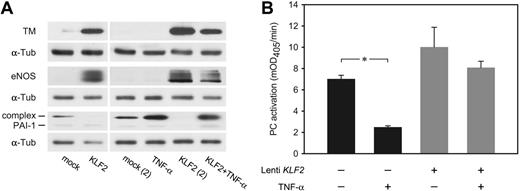
Among the genes shown to be regulated by KLF2 (Table 2) are several of the best-known endothelial signature genes, including TM (THBD), eNOS (NOS3), and VWF (VWF). Immunofluorescence confocal imaging analysis showed sustained expression of the endothelial marker protein VWF, whereas both TM and eNOS were expressed at highly increased levels (Figure 4). It is quite noteworthy that both TM and eNOS are specifically expressed at the plasma membrane in KLF2-expressing cells. The magnitude of the KLF2-mediated transcriptional regulation of TM and eNOS was confirmed at the protein level on KLF2 lentivirus-transduced HUVEC cultures (Figure 5A). Expression of these genes, which are also involved in hemostasis, is generally restricted to the differentiated endothelial cell and sensitive to stimuli such as inflammatory cytokines and growth factors.26,27 In contrast, KLF2-suppressed PAI-1 (SERPINE1) is only produced by activated or inflamed endothelium.28 As we previously showed that endogenous KLF2 expression is highly sensitive to inflammatory cytokines like TNF-α,6 we analyzed its effect under the sustained overexpression of KLF2. Protein levels of the uncomplexed form of PAI-1 were low in lysates of both mock and KLF2-overexpressing cells (Figure 5A). However, a prominent band at approximately 60 kDa was induced by TNF-α but absent after prolonged KLF2 overexpression. The reactivity of this band with the PAI-1 antibody suggests that the majority of PAI-1 is complexed to urokinase-type plasminogen activator as based on the molecular mass of the band.29
Morphologic changes of HUVECs induced by prolonged KLF2 overexpression. HUVECs were transduced with Lenti-mock (A,C,E,G) or Lenti-KLF2 (B,D,F,H) and analyzed 7 days after transduction. (A-B) Immunofluorescence using an anti–human KLF2 antibody. (C-D) Phase-contrast imaging. (E-F) Phalloidin-TRITC staining for F-actin stress fibers. (G-H) Close-ups of phalloidin-FITC staining (green) with VWF immunofluorescence (red) used for orientation. Images were acquired using a 20 × /0.5 NA objective (A-F) or a 63 × /1.4 NA oil objective (G-H).
Morphologic changes of HUVECs induced by prolonged KLF2 overexpression. HUVECs were transduced with Lenti-mock (A,C,E,G) or Lenti-KLF2 (B,D,F,H) and analyzed 7 days after transduction. (A-B) Immunofluorescence using an anti–human KLF2 antibody. (C-D) Phase-contrast imaging. (E-F) Phalloidin-TRITC staining for F-actin stress fibers. (G-H) Close-ups of phalloidin-FITC staining (green) with VWF immunofluorescence (red) used for orientation. Images were acquired using a 20 × /0.5 NA objective (A-F) or a 63 × /1.4 NA oil objective (G-H).
Interestingly, we found that sustained exogenous KLF2 expression leads to induced levels of TM and eNOS even after a 6-hour treatment with TNF-α at only slightly lower levels than in the absence of TNF-α (Figure 5A). Functional analysis of protein-C activation indeed showed that KLF2 is able to largely prevent the downregulation of TM-dependent protein-C activation normally caused by TNF-α (Figure 5B). These results reveal that the modulation of TM, eNOS, and PAI-1 expression by inflammatory cytokines alone is vastly compensated by the prolonged overexpression of KLF2.
KLF2 controls consistent von Willebrand factor expression without interfering with its regulated release
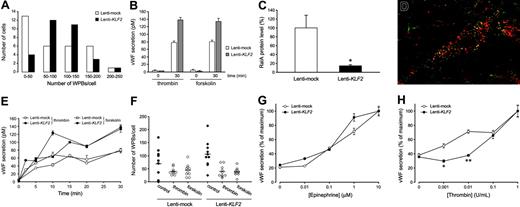
Immunohistochemical staining for the presence of WPBs, the endothelial-specific VWF storage organelles, is most often used to establish a specific and quiescent endothelial phenotype. We noted a 2-fold induction of the VWF mRNA by KLF2 in array and real-time RT-PCR analysis, and immunofluorescence confocal imaging suggested an increased expression of VWF in our cultures at the protein level (Figure 4E-F). In addition, it is quite noteworthy that WPB-stored VWF, which is normally heterogeneously expressed in cell culture, is now homogeneously distributed in the endothelial cell population after prolonged KLF2 expression, reminiscent of its consistent in vivo endothelial expression.6,10,30 In agreement with this macroscopic analysis, the quantification of the number of WPBs in an equal number of individual mock or KLF2 cells revealed that KLF2 invokes a more Gaussian distribution of the WPB number per cell over the entire population rather than enhancing the maximal number of WPBs per individual cell (Figure 6A). A direct involvement of the basal KLF2 levels of static cells in regulating the VWF expression levels is unlikely, given the lack of effect of our previously reported KLF2 siRNA10 on the number and the heterogeneous distribution pattern of WPBs (data not shown). Interestingly, we did notice a slight decrease in the basal secretion level of VWF in Lenti-KLF2 cells, indicating a more efficient storage in WPBs (Figure 6B).
In order to systematically study any potential changes in VWF storage and release in/from the WPBs, various aspects of induced VWF secretion were studied next. First, the amount of VWF released in the culture medium after treatment with thrombin or forskolin, both potent inducers of VWF release from these organelles, was determined (Figure 6B). In agreement with the KLF2-induced increase in VWF mRNA and protein storage, the maximal amount of VWF protein that can be released from these cultures is almost 2-fold increased by KLF2. Second, the mechanism of stimulated WPB exocytosis involves activation of the small GTP-binding protein RalA (v-ral simian leukemia viral oncogene homolog A).21 The microarray analysis presented here shows a 3.1-fold (P < .001) repression of the RalA mRNA after prolonged KLF2 overexpression (Table 2), which is confirmed by Western blotting showing a 7-fold reduction of RalA protein levels (Figure 6C). However, colocalization of RalA with VWF in a subset of WPBs is still observed in dual immunofluorescence irrespective of the lower RalA levels in KLF2-overexpressing cells (Figure 6D). These findings raise the question whether the acute VWF secretion by the endothelium is significantly affected by KLF2. Time course experiments did not reveal differences in the stimulated WPB release kinetics (Figure 6E), and an equal decrease in the number of WPBs after thrombin or forskolin treatment was observed (Figure 6F). These findings led us to conclude that, despite the decreased RalA levels, the mechanism of VWF secretion from the WPBs is basically unaffected by KLF2, in agreement with the consistent colocalization of RalA with WPBs in KLF2-overexpressing cells. Finally, the sensitivity of the stimulated VWF release response after treatment with increasing doses of the physiologically relevant agents thrombin and epinephrine was assessed. No significant difference between the dose response of epinephrine-induced VWF secretion was observed (Figure 6G). In marked contrast, thrombin failed to induce VWF secretion at concentrations below 0.1 U/mL (Figure 6H), showing a KLF2-mediated desensitized thrombin response. In conclusion, increased stimulated VWF secretion was observed in KLF2-overexpressing cells as a result of a more efficient VWF storage and WPB distribution over the cell population rather than that the basic mechanism of VWF secretion is affected by KLF2.
KLF2 reduces HUVEC migration without affecting VEGFR2-dependent VEGF signaling. The effect of a 7-day lentivirus-mediated overexpression of KLF2 on HUVEC migration was determined using a standard wounding assay. Untransduced (data not shown), Lenti-mock (○), and Lenti-KLF2 (•) transduced HUVEC cultures were wounded with a modified cell scraper, and photomicrographs were taken directly and after culturing the same cultures for an additional 24, 48, and 72 hours under standard conditions (A). Images were acquired using a 4 ×/0.13 NA objective. Migration distances were determined by measuring the width of the wounds at all time intervals (n = 3; B). (C) The effect of KLF2 on downstream VEGFR2 signaling was assessed by Western blotting using antibodies for total VEGFR2 and 2 specific phosphorylation states (Y951 and Y1054/1059). Following the 7 days after Lenti-KLF2 and Lentimock transduction, HUVEC cultures were treated with VEGF for 0, 2, and 24 hours before total protein lysates were made. Equal amounts of total proteins were loaded onto the gel and α-tubulin was used as an additional control for equal loading. *P < .01.
KLF2 reduces HUVEC migration without affecting VEGFR2-dependent VEGF signaling. The effect of a 7-day lentivirus-mediated overexpression of KLF2 on HUVEC migration was determined using a standard wounding assay. Untransduced (data not shown), Lenti-mock (○), and Lenti-KLF2 (•) transduced HUVEC cultures were wounded with a modified cell scraper, and photomicrographs were taken directly and after culturing the same cultures for an additional 24, 48, and 72 hours under standard conditions (A). Images were acquired using a 4 ×/0.13 NA objective. Migration distances were determined by measuring the width of the wounds at all time intervals (n = 3; B). (C) The effect of KLF2 on downstream VEGFR2 signaling was assessed by Western blotting using antibodies for total VEGFR2 and 2 specific phosphorylation states (Y951 and Y1054/1059). Following the 7 days after Lenti-KLF2 and Lentimock transduction, HUVEC cultures were treated with VEGF for 0, 2, and 24 hours before total protein lysates were made. Equal amounts of total proteins were loaded onto the gel and α-tubulin was used as an additional control for equal loading. *P < .01.
Immunofluorescence analysis of typical endothelial marker genes that are under transcriptional control of KLF2. Immunofluorescence for TM (A-B), eNOS (C-D), and VWF (E-F) was performed on HUVEC cultures 7 days after transduction with mock (A,C,E) and KLF2 (B,D,F) lentivirus. Detergent permeabilization was omitted for TM staining, establishing a fully surface-exposed antibody-accessible localization. Expression of induced eNOS is predominantly located at the plasma membrane near intercellular junctions as also observed in sheared cultures compared with more Golgi-localized protein in mock-transduced cells. Images were acquired using a 10 ×/0.3 NA objective.
Immunofluorescence analysis of typical endothelial marker genes that are under transcriptional control of KLF2. Immunofluorescence for TM (A-B), eNOS (C-D), and VWF (E-F) was performed on HUVEC cultures 7 days after transduction with mock (A,C,E) and KLF2 (B,D,F) lentivirus. Detergent permeabilization was omitted for TM staining, establishing a fully surface-exposed antibody-accessible localization. Expression of induced eNOS is predominantly located at the plasma membrane near intercellular junctions as also observed in sheared cultures compared with more Golgi-localized protein in mock-transduced cells. Images were acquired using a 10 ×/0.3 NA objective.
Discussion
KLF2-null mice die during embryogenesis due to a defective vasculature,11 but the exact stage of endothelial differentiation that is critically affected by the absence of KLF2 remains to be identified. We have recently shown that KLF2 is responsible for the shear stress–modulated expression of several genes involved in the regulation of vascular tone, including eNOS, which is one of the prime functions of the arterial endothelium.10 In the current study, we describe the steady-state changes in endothelial gene expression after a prolonged stable overexpression of human KLF2 from the moderate PGK promoter, obtaining similar levels as achieved by prolonged flow exposure. The lentivirus-driven expression of KLF2 has several advantages over other expression systems, notably the ability of stable integration and prolonged overexpression, reaching near to 100% transduction efficiencies, without inducing substantial aspecific (inflammatory) side effects.31 Indeed, we did not detect any appreciable changes in genome-wide gene expression or cell morphology when comparing mock-transduced cells with normal HUVECs. Thus, a steady-state gene expression pattern develops that solely reflects the cumulative effect of KLF2 expression in the absence of other flow-induced signal transduction pathways. Discrimination between direct and indirect transcriptional targets of KLF2 is not feasible with this approach. Still, as, for example, eNOS expression is highly increased, nitric oxide is a potential mediator of indirect KLF2 effects and therefore forms a fundamental part of the integrative effects of KLF2 on endothelial cell function.32,33 We believe that the long-term overexpression of KLF2 thus resembles more the in vivo and developmental situation. In fact, we observed that several of the most notable effects on endothelial cell biology as shown in Figures 2 and 4 only started to develop after 3 to 4 days of continued exposure to elevated KLF2, with cells starting to resemble sheared endothelium in vitro and in vivo. The previously reported single-gene effects of mostly murine KLF26-10 in human endothelial cells have now been extended to over 1000 genes for human KLF2 and we established that previously identified direct targets like eNOS and TM7,8 remain elevated for 1 to 2 weeks at the RNA and protein level. Furthermore, we now show that 42 endothelial signature genes including VEGF-receptor1/2, Tie1/2, EphB2/B3, PDGFb receptor, their ligands, and VE-cadherin are indeed not significantly affected by KLF2. Null mice for these genes all have defects in the organization/sprouting of endothelial cells during vasculogenesis and are therefore lethal at an earlier stage in embryogenesis than KLF2–/– mice. Therefore, KLF2 is unlikely to be involved in primary endothelial patterning during early vasculogenesis.34,35
KLF2 overcomes the inflammatory TNF-α effect on the expression of hemostatic genes. HUVECs transduced with mock and KLF2 lentivirus for 7 days were either unstimulated or stimulated with TNF-α for 6 hours. (A) TM, eNOS, and PAI-1 protein levels were determined by Western blotting using α-tubulin as equal loading control. Two independent experiments are shown for the KLF2 overexpression with and without TNF-α. (B) Using a chromogenic assay, the effect of the induction of TM expression by KLF2 on the formation of active protein C (PC) was studied in the absence or presence of TNF-α for 6 hours. The amount of active protein C formed after 20 minutes was 1.4-fold increased after KLF2 overexpression compared with mock controls. TNF-α had no significant effect on protein-C activation with KLF2 overexpression, in contrast to a 3.5-fold reduction in mock-transduced or untreated cells. *P = .037. mOD indicates milli–optical density.
KLF2 overcomes the inflammatory TNF-α effect on the expression of hemostatic genes. HUVECs transduced with mock and KLF2 lentivirus for 7 days were either unstimulated or stimulated with TNF-α for 6 hours. (A) TM, eNOS, and PAI-1 protein levels were determined by Western blotting using α-tubulin as equal loading control. Two independent experiments are shown for the KLF2 overexpression with and without TNF-α. (B) Using a chromogenic assay, the effect of the induction of TM expression by KLF2 on the formation of active protein C (PC) was studied in the absence or presence of TNF-α for 6 hours. The amount of active protein C formed after 20 minutes was 1.4-fold increased after KLF2 overexpression compared with mock controls. TNF-α had no significant effect on protein-C activation with KLF2 overexpression, in contrast to a 3.5-fold reduction in mock-transduced or untreated cells. *P = .037. mOD indicates milli–optical density.
KLF2 increases VWF secretion in HUVEC cultures by inducing a homogenous distribution of WPBs over the cell population. (A) Histogram of the number of WPBs that were counted in randomly chosen cells in mock (□) and KLF2 (▪) lentivirus-transduced HUVECs. Prolonged KLF2 expression induces a more homogeneous Gaussian distribution of the WPB number per cell over the entire cell population. (B) The thrombin- and forskolin-stimulated secretion of VWF in the medium was assayed after 0 and 30 minutes in mock (□) and KLF2 (▦) lentivirus-transduced HUVECs. (C) The amount of RalA 7 days after transduction of HUVECs with mock and KLF2 lentivirus quantified from Western blots (n = 4). (D) Dual immunofluorescence for VWF (red) and RalA (green) in HUVECs 7 days after transduction with KLF2 lentivirus. Colocalization of RalA with VWF is shown in yellow. Image was acquired using a 100 ×/1.5 NA oil objective. (E) Time course of the regulated VWF secretion after thrombin (circles) or forskolin (squares) stimulation of Lenti-mock (open symbols) and Lenti-KLF2 (filled symbols) HUVEC cultures. (F) The number of WBPs in VWF-expressing cells of mock-(circles) and KLF2-transduced (diamonds) HUVEC cultures was determined with (open symbols) or without (filled symbols) prior stimulation with thrombin or forskolin for 30 minutes. (G-H) Stimulated VWF secretion was measured by enzyme-linked immunosorbent assay (ELISA) following a 30-minute exposure of Lenti-mock (○) and Lenti-KLF2 (•) transduced HUVECs to increasing concentrations of epinephrine (G) and thrombin (H). *P < .01 and **P < .001.
KLF2 increases VWF secretion in HUVEC cultures by inducing a homogenous distribution of WPBs over the cell population. (A) Histogram of the number of WPBs that were counted in randomly chosen cells in mock (□) and KLF2 (▪) lentivirus-transduced HUVECs. Prolonged KLF2 expression induces a more homogeneous Gaussian distribution of the WPB number per cell over the entire cell population. (B) The thrombin- and forskolin-stimulated secretion of VWF in the medium was assayed after 0 and 30 minutes in mock (□) and KLF2 (▦) lentivirus-transduced HUVECs. (C) The amount of RalA 7 days after transduction of HUVECs with mock and KLF2 lentivirus quantified from Western blots (n = 4). (D) Dual immunofluorescence for VWF (red) and RalA (green) in HUVECs 7 days after transduction with KLF2 lentivirus. Colocalization of RalA with VWF is shown in yellow. Image was acquired using a 100 ×/1.5 NA oil objective. (E) Time course of the regulated VWF secretion after thrombin (circles) or forskolin (squares) stimulation of Lenti-mock (open symbols) and Lenti-KLF2 (filled symbols) HUVEC cultures. (F) The number of WBPs in VWF-expressing cells of mock-(circles) and KLF2-transduced (diamonds) HUVEC cultures was determined with (open symbols) or without (filled symbols) prior stimulation with thrombin or forskolin for 30 minutes. (G-H) Stimulated VWF secretion was measured by enzyme-linked immunosorbent assay (ELISA) following a 30-minute exposure of Lenti-mock (○) and Lenti-KLF2 (•) transduced HUVECs to increasing concentrations of epinephrine (G) and thrombin (H). *P < .01 and **P < .001.
The present study has identified a KLF2-induced gene expression profile that triggers an enhanced healthy and quiescent endothelial phenotype. The concerted regulation of multiple instead of single genes within various functional categories makes a strong observation and directly implies their potential physiologic significance. Some recent studies have functionally substantiated our findings with regard to angiogenesis, inflammation, hemostasis, and vascular tone.7-10 In the current study, the KLF2-mediated modulation of multiple genes that are involved in cell migration (Table 2) is shown to result in inhibition of wound healing (Figure 3), thereby suggesting a novel way in which KLF2 can inhibit the angiogenic response in addition to recent in vivo findings.9 Furthermore, an important role for KLF2 in modulating the expression of the basal levels of many anti-inflammatory and proinflammatory genes is revealed (Table 2), including MCP-1 and TM. In agreement with published data,7 the response of the endothelium to inflammatory stimuli is repressed, as shown by the reduced efficacy of TNF-α to modulate the expression of TM and PAI-1 in KLF2-overexpressing HUVECs (Figure 5). Finally, we have now extended the subset of well-known flow-responsive genes that are involved in controlling vascular tone and owe their shear stress sensitivity to their direct transcriptional control by KLF2, as we have recently demonstrated by combining flow and KLF2-RNA interference.10 The most notable exception to this set is prostacyclin synthase (PGIS), which is unchanged by KLF2 itself. The reported increased PGI2 bioavailability in response to flow might very well be accomplished by the highly increased expression of the KLF2-induced prostaglandin transporter SLCO2A1, which itself was previously shown to be highly responsive to shear stress.36
Collectively, the presented data lead to the novel insight that KLF2 gene regulation is almost entirely dedicated to the control of many of the genes that define the key functions of the normal functioning endothelium of the completely developed healthy arterial tree. The large number of downstream transcriptional targets of KLF2 (Table 2) precludes a simple direct transcription factor-to-function relation. For example, PAI-1 is not only critical in controlling hemostasis/fibrinolysis but also intricately involved in cell migration,37 and TM is not only involved in anticoagulation but also contributes to embryonic development38 and has anti-inflammatory properties.39 The reduction of cell migration can therefore be related to a collection of genes, including a potential contribution of VEGFR2, which was reported to be responsible for reduced in vivo angiogenesis.9 Phosphorylation of VEGFR2 residues Y951 and Y1059 is respectively associated with VEGF-induced cell migration and proliferation.40 However, KLF2 did not significantly change the VEGF-induced phosphorylation at these sites (Figure 3C), making the involvement of the KLF2-mediated reduction in VEGFR2 expression in the inhibition of migration unlikely. In contrast to the lack of Y951 phosphorylation in pericyte-associated mature endothelial cells in vivo,41 we did not observe Y951 silencing in our cell culture experiments, likely due to the absence of pericytes and shorter VEGF stimulation time intervals. Since angiogenesis is not affected in KLF2-null mice and VEGFR2–/– mice die at a much earlier time in development than KLF2–/– mice,11,42 VEGFR2 repression alone does not seem to offer a satisfactory explanation for the observed abrogation of endothelial cell migration. As shown in Table 2, many more genes involved in migration are affected by KLF2 including SEMA3F, a secreted member of the class 3 semaphorins. A recent report established the major inhibitory effect of SEMA3F on the migration of HUVECs on fibronectin in a similar migration assay as used in the current study.25 In addition, the expression of important basement membrane and/or vascular matrix proteins (fibulin-2, elastin, and collagen type IV, α1) is significantly affected by KLF2 (Table 2), providing the possibility that a change in composition of the endothelial matrix profoundly alters cell migration. Furthermore, observed changes in expression of matrix metalloproteinases and their inhibitors (TIMP3) could contribute as well.30
The modestly higher VWF mRNA levels in the KLF2-overexpressing cultures (Figure 1B) result in an increased average protein level of VWF per individual cell as a consequence of the increase in the number of cells that are filled with WPBs (Figures 4E-F and 6A). This pattern of expression is much more reminiscent of in vivo vascular endothelium, where consistent VWF expression is a hallmark of an intact endothelial cell monolayer.6,10,43 Studies using VWF–/–/LDLR–/– double knock-out mice have demonstrated that the absence of VWF in regions of disturbed flow largely prevents formation of atherosclerotic lesions at these predisposed sites.44 In this respect, the absence of KLF2 at low-shear sites10 might attenuate the storage of VWF in WPBs or lead to increased basal release of proatherogenic VWF at these sites. In contrast to epinephrine and forskolin, the secretion of VWF induced by physiologic levels of thrombin is significantly attenuated by KLF2, providing an additional antithrombotic effect of KLF2 that is consistent with the role of VWF release at low–shear stress regions of the vasculature in atherosclerosis.44 The restriction of this effect to thrombin can be well explained by the considerable increase of functional surface TM levels observed in KLF2-overexpressing cells (Figures 4A-B and 5). The high affinity of thrombin for TM thus results in effective competition between TM and the thrombin receptor,45 the latter being essential for the thrombin-induced but not epinephrine-induced WPB exocytosis. Both the lack of effect of the siRNA-mediated knock-down of KLF2 on the WPBs in normal HUVECs (data not shown) and the published moderately suppressive effect of KLF2 on isolated VWF promoter constructs8 suggest that the change in cellular WPB distribution that is induced by KLF2 (Figures 4E-F and 6) is likely due to other intracellular or paracrine signaling events. This latter concept is in agreement with the KLF2 knock-out mouse phenotype where the absence of KLF2 in the endothelium of KLF2–/– mice causes a poorly formed tunica media, resulting in blood vessel rupture.11 As smooth muscle cells lack detectable KLF2,6 this effect should also be attributed to endothelial paracrine factors. However, due to the unexpectedly high number of KLF2-responsive genes, which include multiple endothelial marker genes and genes that encode for many secreted proteins, prostaglandin, and proteoglycan molecules, the definition of an obvious molecular mechanism that accounts for the destabilization of the vessel wall in KLF2–/– mice is precluded at present.
In summary, our gene expression analysis, extended by protein, cell biologic, and functional data, has established that KLF2 controls molecular processes that are important for the endothelial cell in the context of the fully developed, mature vessel. In conjunction with recent findings on the function of KLF2 in the endothelium and the KLF2-null phenotype,7-11 this underscores the concept that the flow response of KLF2 in the mature arterial vessel wall is of major importance to vascular homeostasis. Thus, the presence of a functional KLF2 gene is crucial for obtaining a mature healthy endothelial phenotype, involving various specific functions that can be modulated by (patho)physiologic stimuli as a normal part of endothelial physiology. An imbalance of these particular functions that are transcriptionally controlled by flow-responsive but cytokine-sensitive KLF2 would lead to endothelial dysfunction, the key initiator of vascular pathologies like atherogenesis.
Prepublished online as Blood First Edition Paper, February 2, 2006; DOI 10.1182/blood-2005-08-3465.
Supported by the Netherlands Heart Foundation, The Hague (project grants NHS2000.144 and NHS2000.097 and Molecular Cardiology Program grant M93.007); NWO-Genomics, The Hague (grant 050-10-014); and the European Union (European Vascular Genomics Network grant LSHM-CT-2003-503254).
R.J.D. and R.A.B. contributed equally to this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal