The PR (PRDI-BF1-RIZ) domain zinc finger protein 1 (PRDM1) is a transcription repressor with a pivotal role in plasma-cell differentiation. We identified clonal inactivating mutations in PRDM1 in the diffuse large B-cell lymphoma (DLBCL) cell line OCI-Ly3 and in 8 of 35 de novo clinical DLBCL samples. The mutational spectrum consists predominantly (7 cases) of single-nucleotide mutations affecting consensus splice donor sites, some of which are recurrent, that lead to splicing aberrations and premature translation termination. In 2 of these cases, point mutations appear to be caused by RNA editing with G-to-A and U-to-G conversions. Other mutations include frame-shift deletion and chromosomal inversion. Except for one mutant, which may act as a dominant-negative, all mutations are associated with either deletion or silencing of the paired PRDM1 allele. This study identifies PRDM1 inactivation as a recurrent genetic defect in DLBCL cells and establishes PRDM1 as a potential tumor suppressor gene in DLBCL. Moreover, it implies inhibition of terminal differentiation as a pathogenetic pathway in DLBCL, particularly for the activated B-cell–like DLBCL. It also demonstrates for the first time the potential role of RNA editing in lymphomagenesis.
Introduction
The PR (PRDI-BF1-RIZ) domain zinc finger protein 1 (PRDM1) was originally identified as positive regulatory domain I–binding factor 1 (PRDI-BF1), which functions as an inducible repressor of β-interferon gene transcription in response to viral infection.1 It belongs to the PRDM gene family of transcription repressors, characterized by the presence of DNA-binding Kruppel-type zinc fingers and the PR domain at the amino terminus. The PR domain is evolutionarily conserved, related to the SET domain, and implicated in chromatin-mediated gene expression.2 Its transcription repressor activity depends on the formation of a protein complex with co-repressors of the Groucho family,3 histone deacetylases,4 and histone H3 methyltransferase.5 Similar to other PRDM family members, PRDM2 (RIZ)6 and PRDM3 (MDS1-EVI1),7 PRDM1 exists as 2 isoforms: PRDM1α and PRDM1β. The latter is functionally impaired and differs from the originally described PRDM1α by lacking the amino-terminal 101 amino acids and having a disrupted PR domain.8
PRDM1 acts as a master regulator in the differentiation of mature B lymphocytes to plasma cells.9 The mouse ortholog of PRDM1, Blimp-1, was originally identified as a gene that is induced upon interleukin-2 (IL-2)– and IL-5–dependent terminal differentiation of mouse lymphoma cell line BCL-1.10 It is expressed in vivo in all plasma cells, including short-lived and long-lived plasma cells, and in a subset of germinal center (GC) B cells, which express plasmacytic markers.11 Blimp1 is required for the formation of mature plasma cells and pre–plasma memory cells,12 as well as maintenance of long-lived plasma cells.13 Ectopic expression of Blimp1 in mature B-cell lines10 and primary splenocytes14-16 can induce plasma-cell differentiation. Microarray profiling demonstrates that Blimp1 orchestrates plasma-cell differentiation by repressing genetic programs associated with activated B cells and/or GC B cells, including those controlling cell proliferation, and by activating genetic programs associated with plasma-cell functions, including apoptosis.17 Several key transcription factors, including c-MYC,18 CIITA,15,19 and PAX-5,14 have been shown to be transcription targets of Blimp1.
We hypothesize that genetic alterations in PRDM1 may play a role in the development of human lymphomas based on the following observations. First, other members of the PRDM gene family are implicated in tumorigenesis. For example, PRDM2 is frequently inactivated in different human cancers.20 Mice deficient for full-length PRDM2 (RIZ1) develop tumors, particularly diffuse large B-cell lymphomas (DLBCLs), in multiple tissues.21 Alterations in PRDM3 (MSD1-EVII) are implicated in the pathogenesis of acute leukemias.22 Second, BCL-6, which has been shown to be deregulated in DLBCLs,23 may contribute to the development of these lymphomas by inhibiting PRDM1-dependent terminal differentiation.24,25 It is conceivable that PRDM1 may be targeted directly by genetic alterations in DLBCLs as an alternative means of inactivation of the terminal differentiation pathway. Third, PRDM1 is localized on chromosome 6q21,26 a region frequently deleted in non-Hodgkin lymphomas, particularly DLBCLs and high-grade lymphomas, based on cytogenetic and loss-of-heterozygosity analysis.27-30 This observation raises the possibility that PRDM1 represents a tumor suppressor gene in this locus.
To investigate the role of PRDM1 as a tumor suppressor in DLBCLs, we began mutation and gene deletion analysis of PRDM1 in OCI-Ly3, a prototypic DLBCL line with an activated B-cell (ABC) phenotype,31 and subsequently extended our analysis to 35 de novo DLBCL cases. We identified PRDM1 alterations in OCI-LY3 and in 23% of the DLBCL cases, consistent with a tumor suppressor role in DLBCLs.
Materials and methods
Lymphoma cell lines and human tissues
OCI-Ly3 was grown in Iscove modified Dulbecco medium supplemented with 10% human serum (NABI Biopharmaceuticals, Boca Raton, FL), and U266 was cultured in RPMI medium 1640 with 10% heat-inactivated FCS (Invitrogen, Carlsbad, CA). Tissue samples of de novo DLBCLs were obtained according to the protocols approved by the Institutional Review Board of Weill Medical College of Cornell University. All samples were reviewed and classified according to World Health Organization classification.32 Nine cases have a germinal center B-cell (GCB) phenotype, and 23 cases have a nongerminal center B-cell (non-GCB) phenotype (3 cases are undetermined) as defined by immunohistochemical staining of paraffin-embedded tissue sections using antibodies against BCL-6, CD10, and IRF4/MUM1.33
Nucleic acid extraction
High-molecular-weight genomic DNA was extracted from tissue culture cells or frozen tissue sections using the salting-out method. Briefly, cultured cells or frozen tissue were lysed with lysis buffer (0.001 M Tris [tris(hydroxymethyl)aminomethane]/HCl, pH 8.2, 0.4 M NaCl, 0.002 M Na2 EDTA [ethylenediaminetetraacetic acid] and digested with proteinase K (Invitrogen) solution (0.5 × SSC, 0.5% sodium dodecyl sulfate [SDS], 5 mM Tris, 0.5 mM EDTA, 100 mg/mL proteinase K). High-molecular-weight genomic DNA was precipitated in the presence of 6 M sodium chloride.
Total RNA was isolated from cultured cells or cryostat tissue sections by TRIzol (Invitrogen) according to the manufacturer's protocol.
Primers
Oligonucleotide primers used in reverse-transcriptase polymerase chain reaction (RT-PCR), and genomic DNA PCR are summarized in Table 1. The positions for PRDM1 and c-myb primers are indicated with reference to ensemble transcript ID ENST00000071246/ENST00000344382 and ENST00000341545, respectively.
Summary of oligonucleotide primers
Primer name . | Position . | Sequence . | Experimental use . |
|---|---|---|---|
| PRDM1.E2.Probe.F | PRDM1 intron 1 nt 1490-1515 | AGCACTCTGCATCTGCTTTTAAAGTC | Probe generation for Southern blot (Figure 1B) |
| PRDM1.E2.Probe.R | PRDM1 intron 2 nt 74-54 | ACAAGGCCTCAAGGGCAAGTA | |
| c-myb.Int1.ProbeF | c-myb intron 1 nt 1144-64 | GAGGCAAAGCAAATCCATTCG | Probe generation for Southern blot (Figure 1B) |
| c-myb.Int1.ProbeR | c-myb intron 1 nt 1450-28 | CCTGTGAAGTCCTAAAGGCGAAA | |
| PRDM1.E2.75.F | PRDM1 exon 2 nt 75-95 | GGACATGGAGGATGCGGATAT | Analysis of PRDM1α expression (Figure 1C) and splicing aberration (Figure 3) |
| PRDM1.E4.60.R | PRDM1 exon 4 nt 60-36 | GTTGCTTTTCTCTTCATTAAAGCCG | |
| PRDM1.altE1.F | PRDM1 exon 1β 16-37 | TTTGCCATTCACTGCAGTAGCA | PRDM1β expression (Figure 1C) |
| PRDM1.E4.60.R | PRDM1 exon 4 nt 60-36 | GTTGCTTTTCTCTTCATTAAAGCCG | |
| c-myb.E1.195.F/ | c-myb exon 1 nt 195-212 | GCGCCATGGCCCGAAGAC | Analysis of c-myb expression from the regular promoter and alternative (beta) promoter (Figure 2A) |
| c-myb.altE1.98.F | c-myb exon 1β 7-27 | CAGGGGAAGAGAAACCCGACT | |
| c-myb.E2.98.R | c-myb exon 2 nt 98-78 | TTTTCCCCAAGTGACGCTTTC | |
| PRDM1.E2.75.F | PRDM1 exon 2 nt 75-95 | GGACATGGAGGATGCGGATAT | Forward primer for 3′ RACE (Figure 1C) |
| PRDM1.E2.126.F | PRDM1 exon 2 nt 126-95 | GAAGTGTACATACATTGTGAACGACCA | Forward nested primer for 3′ RACE (Figure 1C) |
| c-myb.E2.98.R | c-myb exon 2 nt 98-78 | TTTTCCCCAAGTGACGCTTTC | Reverse primer for 5′ RACE (Figure 2) |
| c-myb.E2.38.R | c-myb exon 2 nt 38-15 | TCTCAAAGTCCTCATCATCCTCGT | Reverse nested primer for 5′ RACE (Figure 2) |
| PRDM1.E2.1.F | PRDM1 exon 2 nt 1-23 | GCTGCCCCCAAGTGTAACTCCAG | cDNA generation for ORF sequencing |
| PRDM1.E4.60.R | PRDM1 exon 4 nt 60-36 | GTTGCTTTTCTCTTCATTAAAGCCG | |
| PRDM1.E4.52.F | PRDM1 exon 4 nt 52-73 | AAAAGCAACTGGATGCGCTATG | cDNA generation for ORF sequencing |
| PRDM1.E5.477.R | PRDM1 exon 5 nt 477-456 | AGGACGCGTTCAAGTAAGCGTA | |
| PRDM1.E5.454.F | PRDM1 exon 5 nt 454-475 | CCTACGCTTACTTGAACGCGTC | cDNA generation for ORF sequencing |
| PRDM1.E5.1077.R/ | PRDM1 exon 5 nt 1077-1057 | CGCAAACGTTGCATTCGTACT | |
| PRDM1.E7.67.R | PRDM1 exon 7 nt 67-47 | TCTCTCCAGAATGGAGTCGCA | |
| PRDM1.E5.1057.F | PRDM1 exon 5 nt 1057-1077 | AGTACGAATGCAACGTTTGCG | cDNA generation for ORF sequencing and analysis of splicing aberration (Figure 3A) |
| PRDM1.E7.290.R | PRDM1 exon 7 nt 290-268 | TCGATTTCTTCATTGATTCGGGT | |
| PRDM1.E7.32.F/ | PRDM1 exon 7 nt 32-52 | ATCTCAAGACCCACCTGCGAC | cDNA generation for ORF sequencing |
| PRDM1.E7.257.F | PRDM1 exon 7 nt 257-278 | TGGAAGATCTGACCCGAATCAA | |
| PRDM1.E7.585.R | PRDM1 exon 7 nt 585-559 | CTGAAAATCTTAAGGATCCATTGGTTC | |
| PRDM1.E3.79.F | PRDM1 exon 3 nt 79-102 | GACACAGTTCCTAAGAACGCCAAC | cDNA generation for ORF sequencing |
| PRDM1.E5.46.R | PRDM1 exon 5 nt 46-24 | TCATTTTTCTCAGTGCTCGGTTG | |
| PRDM1.E5.170.F | PRDM1 exon 5 nt 170-194 | GGACCTCGATGACTTTAGAAGACGT | cDNA generation for ORF sequencing |
| PRDM1.E5.575.R | PRDM1 exon 5 nt 575-552 | GAACTTGGGGTAGTGAGCGTTGTA | |
| PRDM1.E2.157.F | PRDM1 exon 2 nt 157-175 | TGGGATTCTGGTGCTGATG | cDNA generation for exon 2 frameshift (Figure 5) |
| PRDM1.E2.245.R | PRDM1 exon 2 nt 245-225 | TCACTGTTGGTGGCATACTTG | |
| PRDM1.5BR.F1 | PRDM1 exon 2 121-150 | GAAGAGAAGTGTACATACATTGTGAACGAC | Forward primers for cloning of 5′ breakpoint junction by LA-PCR (Figure 1B) |
| PRDM1.5BR.F2 | PRDM1 exon 2 nt 212-335 | GGAATCTGCTTTTCAAGTATGCCA | |
| PRDM1.3BR.F | PRDM1 intron 2 nt 666-641 | CCCCTTTGAAACAACAAAGAGTGTAC | Cloning of 3′ breakpoint junction (Figure 1B) |
| PRDM1.3BR.R | c-myb intron 1 nt 1181-1158 | AACAATCTAAGGAAAGGCGAATGG |
Primer name . | Position . | Sequence . | Experimental use . |
|---|---|---|---|
| PRDM1.E2.Probe.F | PRDM1 intron 1 nt 1490-1515 | AGCACTCTGCATCTGCTTTTAAAGTC | Probe generation for Southern blot (Figure 1B) |
| PRDM1.E2.Probe.R | PRDM1 intron 2 nt 74-54 | ACAAGGCCTCAAGGGCAAGTA | |
| c-myb.Int1.ProbeF | c-myb intron 1 nt 1144-64 | GAGGCAAAGCAAATCCATTCG | Probe generation for Southern blot (Figure 1B) |
| c-myb.Int1.ProbeR | c-myb intron 1 nt 1450-28 | CCTGTGAAGTCCTAAAGGCGAAA | |
| PRDM1.E2.75.F | PRDM1 exon 2 nt 75-95 | GGACATGGAGGATGCGGATAT | Analysis of PRDM1α expression (Figure 1C) and splicing aberration (Figure 3) |
| PRDM1.E4.60.R | PRDM1 exon 4 nt 60-36 | GTTGCTTTTCTCTTCATTAAAGCCG | |
| PRDM1.altE1.F | PRDM1 exon 1β 16-37 | TTTGCCATTCACTGCAGTAGCA | PRDM1β expression (Figure 1C) |
| PRDM1.E4.60.R | PRDM1 exon 4 nt 60-36 | GTTGCTTTTCTCTTCATTAAAGCCG | |
| c-myb.E1.195.F/ | c-myb exon 1 nt 195-212 | GCGCCATGGCCCGAAGAC | Analysis of c-myb expression from the regular promoter and alternative (beta) promoter (Figure 2A) |
| c-myb.altE1.98.F | c-myb exon 1β 7-27 | CAGGGGAAGAGAAACCCGACT | |
| c-myb.E2.98.R | c-myb exon 2 nt 98-78 | TTTTCCCCAAGTGACGCTTTC | |
| PRDM1.E2.75.F | PRDM1 exon 2 nt 75-95 | GGACATGGAGGATGCGGATAT | Forward primer for 3′ RACE (Figure 1C) |
| PRDM1.E2.126.F | PRDM1 exon 2 nt 126-95 | GAAGTGTACATACATTGTGAACGACCA | Forward nested primer for 3′ RACE (Figure 1C) |
| c-myb.E2.98.R | c-myb exon 2 nt 98-78 | TTTTCCCCAAGTGACGCTTTC | Reverse primer for 5′ RACE (Figure 2) |
| c-myb.E2.38.R | c-myb exon 2 nt 38-15 | TCTCAAAGTCCTCATCATCCTCGT | Reverse nested primer for 5′ RACE (Figure 2) |
| PRDM1.E2.1.F | PRDM1 exon 2 nt 1-23 | GCTGCCCCCAAGTGTAACTCCAG | cDNA generation for ORF sequencing |
| PRDM1.E4.60.R | PRDM1 exon 4 nt 60-36 | GTTGCTTTTCTCTTCATTAAAGCCG | |
| PRDM1.E4.52.F | PRDM1 exon 4 nt 52-73 | AAAAGCAACTGGATGCGCTATG | cDNA generation for ORF sequencing |
| PRDM1.E5.477.R | PRDM1 exon 5 nt 477-456 | AGGACGCGTTCAAGTAAGCGTA | |
| PRDM1.E5.454.F | PRDM1 exon 5 nt 454-475 | CCTACGCTTACTTGAACGCGTC | cDNA generation for ORF sequencing |
| PRDM1.E5.1077.R/ | PRDM1 exon 5 nt 1077-1057 | CGCAAACGTTGCATTCGTACT | |
| PRDM1.E7.67.R | PRDM1 exon 7 nt 67-47 | TCTCTCCAGAATGGAGTCGCA | |
| PRDM1.E5.1057.F | PRDM1 exon 5 nt 1057-1077 | AGTACGAATGCAACGTTTGCG | cDNA generation for ORF sequencing and analysis of splicing aberration (Figure 3A) |
| PRDM1.E7.290.R | PRDM1 exon 7 nt 290-268 | TCGATTTCTTCATTGATTCGGGT | |
| PRDM1.E7.32.F/ | PRDM1 exon 7 nt 32-52 | ATCTCAAGACCCACCTGCGAC | cDNA generation for ORF sequencing |
| PRDM1.E7.257.F | PRDM1 exon 7 nt 257-278 | TGGAAGATCTGACCCGAATCAA | |
| PRDM1.E7.585.R | PRDM1 exon 7 nt 585-559 | CTGAAAATCTTAAGGATCCATTGGTTC | |
| PRDM1.E3.79.F | PRDM1 exon 3 nt 79-102 | GACACAGTTCCTAAGAACGCCAAC | cDNA generation for ORF sequencing |
| PRDM1.E5.46.R | PRDM1 exon 5 nt 46-24 | TCATTTTTCTCAGTGCTCGGTTG | |
| PRDM1.E5.170.F | PRDM1 exon 5 nt 170-194 | GGACCTCGATGACTTTAGAAGACGT | cDNA generation for ORF sequencing |
| PRDM1.E5.575.R | PRDM1 exon 5 nt 575-552 | GAACTTGGGGTAGTGAGCGTTGTA | |
| PRDM1.E2.157.F | PRDM1 exon 2 nt 157-175 | TGGGATTCTGGTGCTGATG | cDNA generation for exon 2 frameshift (Figure 5) |
| PRDM1.E2.245.R | PRDM1 exon 2 nt 245-225 | TCACTGTTGGTGGCATACTTG | |
| PRDM1.5BR.F1 | PRDM1 exon 2 121-150 | GAAGAGAAGTGTACATACATTGTGAACGAC | Forward primers for cloning of 5′ breakpoint junction by LA-PCR (Figure 1B) |
| PRDM1.5BR.F2 | PRDM1 exon 2 nt 212-335 | GGAATCTGCTTTTCAAGTATGCCA | |
| PRDM1.3BR.F | PRDM1 intron 2 nt 666-641 | CCCCTTTGAAACAACAAAGAGTGTAC | Cloning of 3′ breakpoint junction (Figure 1B) |
| PRDM1.3BR.R | c-myb intron 1 nt 1181-1158 | AACAATCTAAGGAAAGGCGAATGG |
Southern blot analysis
Twenty micrograms of high-molecular-weight DNA isolated from cells was completely digested with EcoRI, and Southern blotting was performed. UV–cross-linked blots were prehybridized and hybridized with 2 to 4 × 106 cpm/mL of radiolabeled probe. High stringency washing was performed at 55°C for 30 minutes. A 439-bp PRDM1 fragment consisting of the entire exon 2 and flanking intronic sequence, and a 306-bp c-myb intron 1 fragment located distal to the 3′ breakpoint, were PCR amplified from genomic DNA, radiolabeled to high specific activity by random priming, and used as probes.
Reverse-transcriptase PCR
Two micrograms of DNaseI-treated total RNA was used for reverse transcription in a total volume of 40 μL with the Superscript First-Strand Synthesis system (Invitrogen). One twentieth of the RT reaction mix (= 100 ng) was then used for PCR amplification. The cycling conditions were as follows: 2 minutes at 94°C, 35 to 40 cycles at 94°C for 45 seconds, 58°C for 45 seconds, and 72°C for 1 minute, followed by a final extension step at 72°C for 5 minutes. Control PCR amplification was performed using β-actin primers. Rapid amplification of the 5′ and 3′ cDNA ends (5′ and 3′ RACE) were performed using the GeneRacer Kit (Invitrogen) according to the manufacturer's protocol. The primers used for RT-PCR and RACE reactions are listed in Table 1.
Sequence analysis of PRDM1 cDNA and genomic DNA
cDNA clones generated by RT-PCR (see Table 1 for primers) were purified by PCR product purification kit (Qiagen, Valencia, CA) and subjected to direct sequencing on both strands at the Cornell University Bioresource Center (Ithaca, NY). If necessary, the PCR products were cloned and sequenced. If cDNA sequencing revealed a nucleotide difference from the database, corresponding genomic DNA sequence was obtained by PCR amplification and subjected to sequencing. All RACE products were cloned and sequenced.
Cloning of 5′ breakpoint junction at PRDM1 locus by ligation-assisted PCR
Sequence spanning the PRDM1 5′ breakpoint junction in OCI-Ly3 was obtained from EcoRI-digested DNA using the ligation-assisted (LA) PCR in vitro cloning kit (TAKARA BIO, Otsu, Japan) according to the manufacturer's protocol. The 3′ breakpoint junction sequence was obtained by conventional PCR amplification. Primers used for these reactions are shown in Table 1.
Western blotting
Fifty micrograms of whole-cell extract from exponentially growing U266 and OCI-Ly3 cell lines was resolved by 6% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE) and transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, NH). The membrane was first blocked in 5% milk/Tris-buffered saline (TBS)-T at room temperature for 1 hour. It was then incubated with rabbit polyclonal antibodies against the N-terminal residues of PRDI-BF119 (1:1000) at 4°C overnight, followed by incubation with anti–rabbit IgG horseradish peroxidase antibody and visualization by enhanced chemiluminescence (ECL) (Amersham Pharmacia Biotech, Piscataway, NJ). The membrane was stripped and stained with amido black (Bio-Rad, Hercules, CA) as a protein loading control.
PRDM1 copy number detection
PRDM1 copy number was determined using the Taqman PCR reaction. Taqman probes and primers were designed using commercially available software. For normalization, we chose the SFRS gene on chromosome 6p, which allowed us to identify cases in which PRDM1 allelic loss is a result of specific gene or chromosomal deletion, for example, 6q deletion, and not a result of whole chromosome loss. PRDM1 and reference gene (SFRS) probes and primers are shown in Table 2.
Primers and probe used in Q-PCR for PRDM1 copy number determination
. | 5′ Primer . | 3′ Primer . | Probe . |
|---|---|---|---|
| PRDM1 Ex 3 | ATACCAAAGGGCACACGTT | CCAGCTCTTTTAGCTCCATTT | FAM-CACAGTTCCTAAGAACGCCAACAGGT-TAMRA |
| PRDM1 Ex 4 | ATGTGAATCCAGCACACTCTC | CCGACAATACCACACAAGAAG | FAM-CAAAACCTGGCTGCGTGTCAGA-TAMRA |
| PRDM1 Ex 5 | AGCACAAACACAGAGCAGTCTA | TTTAGGATTTCTTTCACGCTGT | FAM-TGCCCAAAGAATGTCCCAAAGAGA-TAMRA |
| PRDM1 Ex 7 | CCAGTGCCACAAGAACTACATC | CACTGATGTCAAACTTCTCGATT | FAM-CCCTTGGAAGATCTGACCCGAATC-TAMRA |
| SFRS | TTAGGATGAGCCTCTCCTAGACTT | TCCTACAGCCACTTCGAGAAC | FAM-CCCAAGGCCCACTTGGTCCAT-TAMRA |
. | 5′ Primer . | 3′ Primer . | Probe . |
|---|---|---|---|
| PRDM1 Ex 3 | ATACCAAAGGGCACACGTT | CCAGCTCTTTTAGCTCCATTT | FAM-CACAGTTCCTAAGAACGCCAACAGGT-TAMRA |
| PRDM1 Ex 4 | ATGTGAATCCAGCACACTCTC | CCGACAATACCACACAAGAAG | FAM-CAAAACCTGGCTGCGTGTCAGA-TAMRA |
| PRDM1 Ex 5 | AGCACAAACACAGAGCAGTCTA | TTTAGGATTTCTTTCACGCTGT | FAM-TGCCCAAAGAATGTCCCAAAGAGA-TAMRA |
| PRDM1 Ex 7 | CCAGTGCCACAAGAACTACATC | CACTGATGTCAAACTTCTCGATT | FAM-CCCTTGGAAGATCTGACCCGAATC-TAMRA |
| SFRS | TTAGGATGAGCCTCTCCTAGACTT | TCCTACAGCCACTTCGAGAAC | FAM-CCCAAGGCCCACTTGGTCCAT-TAMRA |
Our method of measuring copy number of PRDM1 is similar to the one described previously.34 Monoplex real-time quantitative PCR was carried out to determine the Ct value for 25 ng of template. To improve accuracy, a regression curve was first obtained for each sample by using serially diluted sample, and the Ct value was determined from each curve. Reactions were carried out in a total volume of 25 μL containing 1 × Taqman master mix (ABI, Foster City, CA), 20 ng template, and 0.2 μM of each primer and probe. The PCR conditions consist of 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 58°C for 30 seconds.
Standard curves were prepared for both the target (PRDM1) and endogenous reference (SFRS) using serially diluted DNA extracted from the cell line U266. They were formulated by plotting threshold cycle numbers (Ct) (y-axis) against template amount (x-axis). The standard curves for both PRDM1 and SFRS were included on each 96-well plate. In each run, all templates were assayed in duplicate, and each run was repeated at least twice for all the samples.
The amount of PRDM1 and SFRS present in each sample was determined from the appropriate standard curve based on threshold cycle numbers (Ct). Only standard curves with a correlation coefficient greater than 0.98 were used in the analysis. The PRDM1 amount was then normalized by dividing it by the SFRS amount. The haploid copy number was calculated as normalized PRDM1 in specimen divided by normalized PRDM1 in normal tonsil control, with the haploid copy number of the latter being 1. We used the mean value of the haploid copy number in OCI-Ly3 (repeated independently more than 3 times) plus 2 times the standard deviation plus 0.15 as the upper limit for scoring a deletion. This takes into account the presence of up to 30% contaminating cells in the sample as well as the measured variability of the assay. A value was considered a hemizygous deletion if the haploid copy number was lower than the cut-off value.
Results
Genetic alterations of PRDM1 in OCI-Ly3
We selected the OCI-Ly3 cell line for initial analysis of PRDM1 in DLBCL. OCI-Ly3 is a prototypic DLBCL cell line with an ABC phenotype,31 in which multiple cytogenetic abnormalities are detectable, including deletion of 6q15-27, identified by comparative genomic hybridization.35 We confirmed hemizygous deletion of PRDM1 by copy number determination using real-time quantitative PCR (Q-PCR) (see “PRDM1 mutations are associated with hemizygous deletions of PRDM1”).
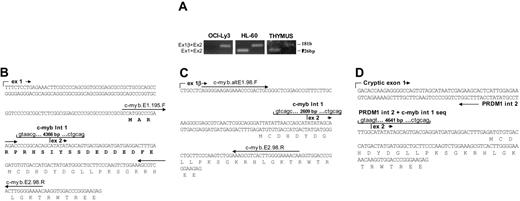
Genetic alterations of the PRDM1 allele in OCI-Ly3 are summarized in Figure 1A. Results of Southern analyses and ligation-mediated PCR revealed chromosomal inversion between PRDM1 and c-myb (Figure 2B). The 5′ breakpoint is located in intron 2 of PRDM1, 506 bp 3′ of exon 2; while the 3′ breakpoint is located in intron 1 of c-myb, 850 bp 3′ of exon 1. This chromosomal inversion dissociated exons 1 and 2 of PRDM1 from the other exons, leading to truncated PRDM1α transcripts generated by splicing of exon 2 to a novel 640-bp exon located approximately 20.5 kb 3′ of the 5′ breakpoint through a cryptic splice acceptor site (Figure 1A,C). However, they are predicted to encode a severely truncated PRDM1, which contains only the amino-terminal acidic domain and a portion of the PR domain (Figure 1A). Western blotting confirmed the absence of wild-type PRDM1α in OCI-Ly3 (Figure 1D).
Alterations of PRDM1 and c-myb in the DLBCL cell line OCI-LY3. (A) Diagrammatic representation of chromosomal inversion between the PRDM1 and c-myb loci and alterations in their transcript and protein products. Boxes indicate exons, with coding frame in black. The 5′ exons derived from the alternate promoters of PRDM1 and c-myb are denoted as 1β. The 5′ and 3′ breakpoints are shown by block arrows, and their positions relative to the 3′ ends of PRDM1 exon 2 and c-myb exon 1 are indicated. The expected sizes of the germ line and rearranged EcoRI fragments spanning the breakpoints and detected by Southern blotting (panel B) are marked. The cryptic exon located approximately 21 kb downstream of PRDM1 exon 2, as well as the c-myb cryptic exon 1, are indicated by asterisks. The 2-bp frameshift deletion (del) and sites of premature translation termination (UGA) are shown. Structures of the truncated PRDM1 and c-MYB are depicted with their relevant functional domains. PR indicates PR domain; ZnF, zinc fingers; ++, basic domain; —, acidic domain; S/T, serine/threonine-rich region. (B) Gene rearrangement and junction sequences at the proximal and distal breakpoints. Left: Southern blot analysis of EcoRI-digested genomic DNA detected gene rearrangement using the PRDM1 exon 2 probe (top) and the c-myb intron 1 probe (bottom). The germ line bands detected in U266 are absent in OCI-Ly3, consistent with deletion of the wild-type allele. Right: Junction sequences for the proximal breakpoint (top) and the distal breakpoint (bottom). Putative breakpoints are indicated by arrowheads. The germ line PRDM1 intron 2 and c-myb intron 1 sequences normally present adjacent to the breakpoints also are shown. The putative 5′ end of the PRDM1 intron 2 sequence translocated next to c-myb intron 1 and the putative 3′ end of the c-myb intron 1 sequence translocated next to PRDM1 intron 2 are marked by open and filled boxes, respectively. Nucleotides that appear to be duplicated next to the breakpoints are indicated in italics. The nucleotide at the proximal breakpoint junction that differs from the germ line sequence (g-to-c change) is underlined. (C) PRDM1 RNA expression analysis in OCI-Ly3. Top left: PRDM1α and PRDM1β transcripts were analyzed by RT-PCR using primers specific to exon 2 and exon 4, or exon 1β and exon 4, respectively. In OCI-LY3, no PCR products could be obtained using the exon 2/exon 4 primer pair as a result of chromosomal inversion (panel A), while normal splicing occurs between exon 1β and exon 4. Bottom left: A Northern blot was hybridized with a PRDM1 exon 4 to 7 cDNA probe, which detected both PRDM1α and PRDM1β transcripts in U266, but only PRDM1β mRNA in OCI-Ly3. The smaller (∼3.5 kb) transcripts present in OCI-Ly3 most likely are derived from alternative polyadenylation, as described previously.1 Right: Sequence of fusion transcripts consisting of PRDM1 exon 2 spliced to a cryptic exon approximately 21 kb downstream using consensus splice donor and acceptor sites (italics). These transcripts code for the first 61 amino acids of PRDM1α followed by a novel 36 amino-acid peptide sequence (italics). The consensus polyadenylation signal (AATAAA) is underlined. (D) Western blot analysis of PRDM1 in OCI-Ly3 and U266 using antibody to the N terminus of PRDM1. PRDM1α was detected in U266 but not in OCI-Ly3. Amido-black staining of the blot served as loading control. (E) Frameshift mutation of PRDM1. The sequence chromatogram of OCI-Ly3 genomic DNA shows a 2-bp deletion that leads to a frameshift and premature translation termination (bold). The numbering of the nucleotides corresponds to the nucleotide number starting from the 5′ end of exon 5.
Alterations of PRDM1 and c-myb in the DLBCL cell line OCI-LY3. (A) Diagrammatic representation of chromosomal inversion between the PRDM1 and c-myb loci and alterations in their transcript and protein products. Boxes indicate exons, with coding frame in black. The 5′ exons derived from the alternate promoters of PRDM1 and c-myb are denoted as 1β. The 5′ and 3′ breakpoints are shown by block arrows, and their positions relative to the 3′ ends of PRDM1 exon 2 and c-myb exon 1 are indicated. The expected sizes of the germ line and rearranged EcoRI fragments spanning the breakpoints and detected by Southern blotting (panel B) are marked. The cryptic exon located approximately 21 kb downstream of PRDM1 exon 2, as well as the c-myb cryptic exon 1, are indicated by asterisks. The 2-bp frameshift deletion (del) and sites of premature translation termination (UGA) are shown. Structures of the truncated PRDM1 and c-MYB are depicted with their relevant functional domains. PR indicates PR domain; ZnF, zinc fingers; ++, basic domain; —, acidic domain; S/T, serine/threonine-rich region. (B) Gene rearrangement and junction sequences at the proximal and distal breakpoints. Left: Southern blot analysis of EcoRI-digested genomic DNA detected gene rearrangement using the PRDM1 exon 2 probe (top) and the c-myb intron 1 probe (bottom). The germ line bands detected in U266 are absent in OCI-Ly3, consistent with deletion of the wild-type allele. Right: Junction sequences for the proximal breakpoint (top) and the distal breakpoint (bottom). Putative breakpoints are indicated by arrowheads. The germ line PRDM1 intron 2 and c-myb intron 1 sequences normally present adjacent to the breakpoints also are shown. The putative 5′ end of the PRDM1 intron 2 sequence translocated next to c-myb intron 1 and the putative 3′ end of the c-myb intron 1 sequence translocated next to PRDM1 intron 2 are marked by open and filled boxes, respectively. Nucleotides that appear to be duplicated next to the breakpoints are indicated in italics. The nucleotide at the proximal breakpoint junction that differs from the germ line sequence (g-to-c change) is underlined. (C) PRDM1 RNA expression analysis in OCI-Ly3. Top left: PRDM1α and PRDM1β transcripts were analyzed by RT-PCR using primers specific to exon 2 and exon 4, or exon 1β and exon 4, respectively. In OCI-LY3, no PCR products could be obtained using the exon 2/exon 4 primer pair as a result of chromosomal inversion (panel A), while normal splicing occurs between exon 1β and exon 4. Bottom left: A Northern blot was hybridized with a PRDM1 exon 4 to 7 cDNA probe, which detected both PRDM1α and PRDM1β transcripts in U266, but only PRDM1β mRNA in OCI-Ly3. The smaller (∼3.5 kb) transcripts present in OCI-Ly3 most likely are derived from alternative polyadenylation, as described previously.1 Right: Sequence of fusion transcripts consisting of PRDM1 exon 2 spliced to a cryptic exon approximately 21 kb downstream using consensus splice donor and acceptor sites (italics). These transcripts code for the first 61 amino acids of PRDM1α followed by a novel 36 amino-acid peptide sequence (italics). The consensus polyadenylation signal (AATAAA) is underlined. (D) Western blot analysis of PRDM1 in OCI-Ly3 and U266 using antibody to the N terminus of PRDM1. PRDM1α was detected in U266 but not in OCI-Ly3. Amido-black staining of the blot served as loading control. (E) Frameshift mutation of PRDM1. The sequence chromatogram of OCI-Ly3 genomic DNA shows a 2-bp deletion that leads to a frameshift and premature translation termination (bold). The numbering of the nucleotides corresponds to the nucleotide number starting from the 5′ end of exon 5.
The PRDM1β transcripts present in OCI-LY3 could be detected by Northern analysis and reverse-transcriptase PCR using specific primers (Figure 1C). While these transcripts are structurally normal, no wild-type PRDM1β is predicted to be generated from these transcripts because of a 2-bp frameshift deletion present in exon 5, which would result in premature translation termination and generation of a truncated PRDM1β (Figure 1A,E). Therefore, both wild-type PRDM1α and PRDM1β are absent in OCI-Ly3.
Alterations of c-myb transcripts in OCI-Ly3. (A) Absence of c-myb transcripts initiated from the conventional promoter. PCR amplification of cDNA prepared from OCI-Ly3, HL-60, and thymus total RNA was performed with specific primers (Table 1) to detect transcripts initiated from the conventional promoter (Ex 1 + Ex 2) or the alternate promoter (Ex 1β+ Ex 2), respectively. PCR products were analyzed by agarose gel electrophoresis. While HL-60 and normal thymus contain both types of c-myb transcripts, OCI-Ly3 lacks transcripts initiated from the conventional promoter (note absence of 126-bp band). (B-D) Partial sequence of c-myb transcripts with different exon 1 sequences (ex 1, panel B; ex 1β, panel C; cryptic exon 1, panel D). The exon 1 sequence depicted in panel B is not expressed in OCI-Ly3, but that shown in panel D is OCI-Ly3 specific. The size of the first intron in each case is indicated, and the consensus splice sites are shown. The positions of the primers used in panel A also are marked. Cryptic exon 1 (panel D) is derived from the nontranscribed strand of PRDM1 intron 2 (int 2). The “intron 1” sequence for this transcript is contributed by both PRDM1 intron 2 and c-myb intron 1 sequences (Figure 1A). The 20 amino acids truncated in c-MYB in panel C and panel D are marked in bold in panel B.
Alterations of c-myb transcripts in OCI-Ly3. (A) Absence of c-myb transcripts initiated from the conventional promoter. PCR amplification of cDNA prepared from OCI-Ly3, HL-60, and thymus total RNA was performed with specific primers (Table 1) to detect transcripts initiated from the conventional promoter (Ex 1 + Ex 2) or the alternate promoter (Ex 1β+ Ex 2), respectively. PCR products were analyzed by agarose gel electrophoresis. While HL-60 and normal thymus contain both types of c-myb transcripts, OCI-Ly3 lacks transcripts initiated from the conventional promoter (note absence of 126-bp band). (B-D) Partial sequence of c-myb transcripts with different exon 1 sequences (ex 1, panel B; ex 1β, panel C; cryptic exon 1, panel D). The exon 1 sequence depicted in panel B is not expressed in OCI-Ly3, but that shown in panel D is OCI-Ly3 specific. The size of the first intron in each case is indicated, and the consensus splice sites are shown. The positions of the primers used in panel A also are marked. Cryptic exon 1 (panel D) is derived from the nontranscribed strand of PRDM1 intron 2 (int 2). The “intron 1” sequence for this transcript is contributed by both PRDM1 intron 2 and c-myb intron 1 sequences (Figure 1A). The 20 amino acids truncated in c-MYB in panel C and panel D are marked in bold in panel B.
Exon 1 of c-MYB also is dissociated from its downstream exons by chromosomal inversion (Figure 1A). As predicted, 5′ RACE did not detect transcription initiation from the normal c-MYB promoter. Instead, we identified c-myb transcripts that are initiated from an alternative promoter not previously described (hereby referred to as β) or from a cryptic promoter in PRDM1 intron 2 (Figure 2). The exon 1β sequence of c-MYB has not been identified in the EST and Ensemble database. However, it could be detected by RT-PCR not only in cell lines (OCI-LY3 and HL-60) but also in normal thymus. Thus, these c-MYB transcripts are likely to represent endogenous c-MYB isoforms generated from alternative promoter usage. These transcripts, as well as those initiated from the cryptic promoter, encode c-MYB lacking the first 20 amino acids at the amino terminus (Figure 2). Interestingly, an N-terminal 20 aminoacid truncated form of c-MYB is oncogenic in chicken oncogenecity assays.36
Clonal mutations in clinical DLBCL cases
Thirty-five de novo DLBCL cases were first screened for PRDM1 mutations and splicing abnormalities by sequencing and agarose gel analysis of the cDNAs generated by RT-PCR using different primer pairs (Table 1). PRDM1 genomic sequence was obtained whenever applicable. The results of the mutation analysis are summarized in Table 3. Clonal mutations were detected in 8 of 35 cases. All 8 tumors have a non-GCB phenotype (7 cases BCL6+, CD10–, MUM1+; 1 case BCL6–, CD10–, MUM1+) as determined by immunohistochemical staining of paraffin-embedded tissue sections using antibodies against BCL-6, CD10, and IRF4/MUM1.33 The presence of mutations did not appear to correlate with histology, as both centroblastic and immunoblastic DLBCLs are represented approximately equally in the group (Table 3). These mutations were present in 5 tumors at first diagnosis and in 3 relapsed lymphomas.
Summary of PRDM1 alterations in DLBCL cases
PRDM1 gene mutations . | PRDM1 RNA aberrations . | . | PRDM1 protein alterations . | . | IHC . | . | . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | α . | β . | α . | β . | BCL6 . | CD10 . | IRF4 . | Non-GCB/GCB . | HS . | AD . | WT . | |||||
| 1 | NA | ‡525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | + | - | + | Non-GCB | IB | + | - | |||||
| 2 | Int 3 + 1G>T | Int 3 + 1G>T & §Ins Int 3 seq* | - | Del 102-789 | - | + | - | + | Non-GCB | IB | + | - | |||||
| §Δexon 3 | Del 62-101 (ΔPR) | - | |||||||||||||||
| 3 | Del 466-469 | Del 466-469 | - | Del 42-789 | - | + | - | + | Non-GCB | IB | + | - | |||||
| 4 | NA | ‡Int 6 +2U>A & | Same as α | Del 599-789 (ΔZF nos. 3-5) | Same as α | + | - | + | Non-GCB | IB | - | + | |||||
| §Ins 49 nt int 6 seq | |||||||||||||||||
| 5 | 525G>A | 525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | - | - | + | Non-GCB | CB | + | - | |||||
| 6 | Int 2 + 2U>G | Int 2 + 2U>G & | - | Del 62-789 | - | + | - | + | Non-GCB | CB | - | - | |||||
| §Ins 103 nt int 2 seq | |||||||||||||||||
| 7 | 525G>A | 525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | + | - | + | Non-GCB | CB | - | † | |||||
| 8 | 525G>A | 525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | + | - | + | Non-GCB | CB | + | - | |||||
PRDM1 gene mutations . | PRDM1 RNA aberrations . | . | PRDM1 protein alterations . | . | IHC . | . | . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | α . | β . | α . | β . | BCL6 . | CD10 . | IRF4 . | Non-GCB/GCB . | HS . | AD . | WT . | |||||
| 1 | NA | ‡525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | + | - | + | Non-GCB | IB | + | - | |||||
| 2 | Int 3 + 1G>T | Int 3 + 1G>T & §Ins Int 3 seq* | - | Del 102-789 | - | + | - | + | Non-GCB | IB | + | - | |||||
| §Δexon 3 | Del 62-101 (ΔPR) | - | |||||||||||||||
| 3 | Del 466-469 | Del 466-469 | - | Del 42-789 | - | + | - | + | Non-GCB | IB | + | - | |||||
| 4 | NA | ‡Int 6 +2U>A & | Same as α | Del 599-789 (ΔZF nos. 3-5) | Same as α | + | - | + | Non-GCB | IB | - | + | |||||
| §Ins 49 nt int 6 seq | |||||||||||||||||
| 5 | 525G>A | 525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | - | - | + | Non-GCB | CB | + | - | |||||
| 6 | Int 2 + 2U>G | Int 2 + 2U>G & | - | Del 62-789 | - | + | - | + | Non-GCB | CB | - | - | |||||
| §Ins 103 nt int 2 seq | |||||||||||||||||
| 7 | 525G>A | 525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | + | - | + | Non-GCB | CB | - | † | |||||
| 8 | 525G>A | 525G>A & §Ins 103 nt int 2 seq | - | Del 62-789 | - | + | - | + | Non-GCB | CB | + | - | |||||
Numbering of cDNA nucleotide and amino acid is based on sequence data NM_001198 and AAC33300.1, respectively.
Intronic sequence number refers to the nucleotide number from the 5′ end of the intron. For example, int 6 + 2 indicates the second nucleotide of intron 6.
Ins indicates insertion; Del, deletion; int, intron; IHC, immunohistochemistry; HS, histology; AD, allelic deletion; WT, wild-type allele expression; GCB, germinal center B-cell phenotype; IB, immunoblastic; and CB, centroblastic.
The exact length of intronic sequence included was not determined.
Very low levels detected, possibly from normal contaminating cells.
RNA editing.
Splicing aberrations.
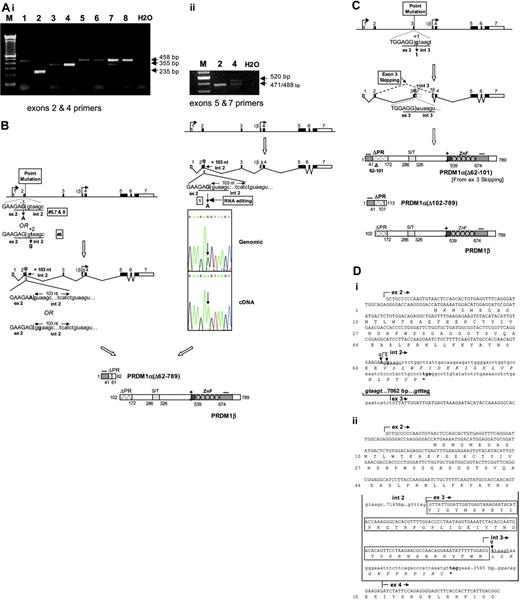
Of these 8 cases, 7 had point mutations in the consensus sequence of splice donor sites. Of these 7, 5 (nos. 1, 5, 6, 7, 8) mutations are located at the exon 2/intron 2 junction. Of these 5 mutations, 4 are recurrent, involving a G-to-A point mutation at the last nucleotide of exon 2 (Table 3). In one case (no. 2), a G-to-T mutation was identified in the first nucleotide of intron 3; and in another (no. 4), a T-to-G mutation was identified in the second nucleotide of intron 6. Interestingly, 2 of the 7 point mutations (tumor nos. 1 and 4) were detected in the cDNA but not in the genomic DNA (Figures 3A, 4), suggesting that these nucleotide alterations are due to RNA editing.
These point mutations inactivate the consensus splice donor sites, resulting in splicing aberrations detectable by RT-PCR spanning exons 2 and 4; or, for tumor no. 4, exons 5 and 7 (Figure 3A). In tumor nos. 1, 5, 6, 7, and 8, the RT-PCR products are larger than normal (355 bp) due to insertion of 103 nucleotides of intron 2 sequence (Figure 3B,Di). In tumor no. 2, the RT-PCR product is smaller than normal as a result of exon 3 skipping (Figure 3C). RT-PCR using exon 2 and intron 3 primers also demonstrated in this case transcripts retaining intron 3 sequence of undetermined length (data not shown; Figure 3Dii,iii). In all 6 cases with splicing abnormalities, the aberrantly spliced products are either exclusively present or predominate, and the normally spliced products are either absent or are detected at relatively low levels. The latter may be attributed to the presence of contaminating PRDM1-expressing normal cells in the tumor.
In tumor no. 4, which harbors a point mutation at the consensus splice donor site at the exon 6/intron 6 junction due to RNA editing (Figure 4), abnormal-sized as well as normal-sized PCR products were detected in similar amounts when primers from exon 5 and exon 7 were used (Figure 3A). Sequence analysis indicates, in addition to wild-type transcripts, the presence of abnormally spliced mRNAs that retain the first 49 nucleotides of intron 6 (Figure 4). The presence of both normal and mutant transcripts of approximately equal abundance implies that the extent of RNA editing in this tumor is approximately 50%, in contrast to tumor no. 1, where RNA editing approaches 100% efficiency.
In addition, a frameshift mutation associated with a 4-bp deletion in exon 2 was identified in one tumor (no. 3) (Figure 5). In this tumor, there is virtually no expression of the normal allele, based on agarose gel electrophoresis and sequence analysis of the PCR amplicons spanning the frameshift.
All mutations are predicted to lead to premature translation termination and synthesis of truncated PRDM1 (Figures 3, 4, 5; Table 3). Importantly, in all these tumors except no. 4, no significant amount of wild-type PRDM1α is expected to be produced because of the virtual absence of normal PRDM1α transcripts. In most cases, the truncated forms of PRDM1 consist of only the acidic N-terminal domain plus or minus part of the PR domain, and therefore are most likely nonfunctional. Although the Δ exon 3 PRDM1α transcripts present in tumor no. 2 are capable of generating a near full-length protein with internal deletion within the PR domain (del 62-101; Table 1), it is also likely to be functionally impaired.8 Both normal and truncated PRDM1 can be generated in tumor no. 4 (Figures 3E and 4). The latter retains 2 of the 5 zinc fingers, which have been shown to be sufficient to mediate DNA binding.37 However, its normal functions may be compromised because of the loss of the C-terminal portion of the protein.38 Thus, this less extensively truncated PRDM1 has the potential to interfere with normal PRDM1 function by a dominant-negative mechanism.
PRDM1 mutations are associated with hemizygous deletions of PRDM1
PRDM1 is located on chromosome 6q21, which is frequently deleted in human tumors.26 The absence of expression of wild-type transcripts in the tumors bearing PRDM1 mutations thus may be attributed to deleterious inactivation of the paired normal alleles. To test our hypothesis, we developed a real-time quantitative PCR (Q-PCR) assay to determine the PRDM1 copy number in tumors. This method has been used previously to accurately determine gene copy number and screen for gene deletion.34 Details of this Q-PCR approach are described in “Materials and methods.” We selected a suitable primer pair for exons 3, 4, 5, and 7 of PRDM1, which resulted in accurate copy number quantification as determined by analysis of deletion and normal control samples, which include OCI-LY3 and tonsil tissues pooled from several healthy individuals, respectively. For the deletion control OCI-Ly3, the haploid copy numbers (± SD) are 0.56 + 0.012 (exon 3), 0.56 + 0.027 (exon 4), 0.45 + 0.050 (exon 5), and 0.49 + 0.057 (exon 7) (expected value = 0.5, with value for normal control arbitrarily set as 1). For each exon measured, the upper limit for scoring a deletion was determined as the mean value of the haploid copy number in the deletion control OCI-Ly3 plus 2 times the standard deviation plus 0.15 (allowing up to 30% of contaminating benign cells in the tumor sample). A value was considered a hemizygous deletion if the haploid copy number was lower than the cut-off value. The results of the analysis are shown in Figure 6 with the cutoff limits. Based on these Q-PCR results, 4 cases (nos. 1, 2, 5, 8) clearly harbor hemizygous deletion of PRDM1. Tumor no. 3 has haploid copy numbers that are at or just below the cut-off levels and therefore also likely to have hemizygous PRDM1 deletion. On the other hand, tumors nos. 4, 6, and 7 have haploid copy numbers that are above the cut-off limit, which favor a normal copy number status. These Q-PCR results were corroborated by the relative representation of the wild-type and mutant alleles in sequence analysis, which showed that the mutant alleles were predominantly detected for tumor nos. 2, 5, and 8, while the wild-type and mutant nucleotides were present in approximately equal amounts for tumor nos. 6 and 7.
As previously mentioned, expression of normal PRDM1 transcripts was not detected in the DLBCL cases. The Q-PCR results for haploid copy number measurement imply that the paired allele is inactivated by allelic deletion in some cases with PRDM1 mutations. In other cases, the paired allele is retained and silenced by mechanism(s) other than genomic deletion, possibly by epigenetic modifications. Infrequently, the paired allele may still remain transcriptionally active (as in tumor no. 4), and the normal function of wild-type PRDM1 may be interfered with by the mutant through a dominant-negative mechanism.
Clonal mutations of PRDM1 in DLBCL clinical cases. (A) Expression analysis of PRDM1 transcripts in DLBCL by RT-PCR. (i) PCR amplification was performed on cDNAs using specific primers located in exons 2 and 4 (Table 1), which normally generate a 355-bp band upon agarose-gel electrophoresis. PCR amplicons of altered sizes were seen in most cases (1, 2, 5, 6, 7, 8), indicative of splicing aberrations. Note the absence of normal-sized bands in these lymphomas, which are consistent with the lack of expression of the normal PRDM1 transcripts. The relatively weak normal-sized bands seen in tumor no. 7 may be due to the presence of contaminating normal cells that express PRDM1. (ii) Using primers spanning exons 5 and 7, PCR amplification generated 2 major bands of approximately equal intensity in case no. 4, one normal-sized (471 bp), and the other larger than normal (520 bp). Upon sequencing, the normal-sized band is found also to contain a small amount of a slightly larger product (488 bp) in addition to the normal-sized product. The presence of abnormally sized products indicates splicing aberrations. (B) Splicing aberrations associated with point mutations at the exon 2/intron 2 junction. A schematic diagram illustrating splicing abnormalities in tumors 5, 6, 7, and 8. Point mutations at the consensus splice donor sequence (underlined) at the exon 2/intron 2 junction (G → A at position –1 in tumors 5, 7, and 8; t → g at position +2 in tumor no. 6) are indicated in bold. Right: Diagrammatic depiction of splicing abnormalities in tumor no. 1 as a result of RNA editing. Point mutation at the consensus splice donor sequence (underlined) at the exon 2/intron 2 junction (G → A at position –1) is indicated, along with the sequence chromatogram of cDNA showing the editing G-to-A conversion absent in genomic DNA. The cryptic splice donor site actually used in the tumors is in italics. The 103-bp intron 2 sequence inserted in the tumor transcripts and the resulting premature translation termination are marked. The predicted structures of PRDM1α and PRDM1β derived from the respective transcripts are shown, along with their functional domains. (C) A schematic diagram illustrating RNA splicing aberrations associated with point mutation at the exon 3/intron 3 junction, as seen in tumor no. 2. Point mutation (g → t) in position +1 of the consensus splice donor sequence (underlined) is shown in bold. Exon 3 skipping and retention of intron 3 sequence (of unknown length) are indicated. The former splicing abnormality causes an internal deletion in the PR domain, and the latter results in premature translation termination. The predicted structures of PRDM1α and PRDM1β are shown, along with their functional domains. (D) (i) cDNA nucleotide sequence of tumor transcripts retaining intron 2 sequence. The point mutations that inactivate the consensus splice donor site (underlined) are indicated in bold (G → A for tumors 1, 5, 7, and 8; t → g for tumor no. 6). The intron 2 cryptic splice donor site and the splice acceptor site at the intron 2/exon 3 junction used in the tumors are in italics. The retained 103-bp intron 2 sequence and the distance between its 3′ end and the 5′ end of exon 3 (7062 bp) is indicated. The premature translation termination codon UGA is in bold, and the novel peptide sequence predicted from translation of the intron 2 sequence is shown in italics. (ii) Partial cDNA nucleotide sequence showing exon 3 skipping or intron 3 sequence retention in transcripts from tumor no. 2. The point mutation (g → t) at the first nucleotide of intron 3 that inactivates the consensus splice donor site (underlined) is shown. The splice donor and acceptor sites used in splicing between exon 2 and exon 4 (or exon 3) are in italics, and the exon 3 nucleotide sequence and encoded amino acids that are internally deleted due to exon 3 skipping are boxed. Sizes of intron 2 and 3 are shown. The intron 3 sequence of indeterminate length retained in some of the tumor transcripts is partially shown, including the premature termination codon (in bold). The novel peptide sequence translated from the intron 3 sequence is shown in italics.
Clonal mutations of PRDM1 in DLBCL clinical cases. (A) Expression analysis of PRDM1 transcripts in DLBCL by RT-PCR. (i) PCR amplification was performed on cDNAs using specific primers located in exons 2 and 4 (Table 1), which normally generate a 355-bp band upon agarose-gel electrophoresis. PCR amplicons of altered sizes were seen in most cases (1, 2, 5, 6, 7, 8), indicative of splicing aberrations. Note the absence of normal-sized bands in these lymphomas, which are consistent with the lack of expression of the normal PRDM1 transcripts. The relatively weak normal-sized bands seen in tumor no. 7 may be due to the presence of contaminating normal cells that express PRDM1. (ii) Using primers spanning exons 5 and 7, PCR amplification generated 2 major bands of approximately equal intensity in case no. 4, one normal-sized (471 bp), and the other larger than normal (520 bp). Upon sequencing, the normal-sized band is found also to contain a small amount of a slightly larger product (488 bp) in addition to the normal-sized product. The presence of abnormally sized products indicates splicing aberrations. (B) Splicing aberrations associated with point mutations at the exon 2/intron 2 junction. A schematic diagram illustrating splicing abnormalities in tumors 5, 6, 7, and 8. Point mutations at the consensus splice donor sequence (underlined) at the exon 2/intron 2 junction (G → A at position –1 in tumors 5, 7, and 8; t → g at position +2 in tumor no. 6) are indicated in bold. Right: Diagrammatic depiction of splicing abnormalities in tumor no. 1 as a result of RNA editing. Point mutation at the consensus splice donor sequence (underlined) at the exon 2/intron 2 junction (G → A at position –1) is indicated, along with the sequence chromatogram of cDNA showing the editing G-to-A conversion absent in genomic DNA. The cryptic splice donor site actually used in the tumors is in italics. The 103-bp intron 2 sequence inserted in the tumor transcripts and the resulting premature translation termination are marked. The predicted structures of PRDM1α and PRDM1β derived from the respective transcripts are shown, along with their functional domains. (C) A schematic diagram illustrating RNA splicing aberrations associated with point mutation at the exon 3/intron 3 junction, as seen in tumor no. 2. Point mutation (g → t) in position +1 of the consensus splice donor sequence (underlined) is shown in bold. Exon 3 skipping and retention of intron 3 sequence (of unknown length) are indicated. The former splicing abnormality causes an internal deletion in the PR domain, and the latter results in premature translation termination. The predicted structures of PRDM1α and PRDM1β are shown, along with their functional domains. (D) (i) cDNA nucleotide sequence of tumor transcripts retaining intron 2 sequence. The point mutations that inactivate the consensus splice donor site (underlined) are indicated in bold (G → A for tumors 1, 5, 7, and 8; t → g for tumor no. 6). The intron 2 cryptic splice donor site and the splice acceptor site at the intron 2/exon 3 junction used in the tumors are in italics. The retained 103-bp intron 2 sequence and the distance between its 3′ end and the 5′ end of exon 3 (7062 bp) is indicated. The premature translation termination codon UGA is in bold, and the novel peptide sequence predicted from translation of the intron 2 sequence is shown in italics. (ii) Partial cDNA nucleotide sequence showing exon 3 skipping or intron 3 sequence retention in transcripts from tumor no. 2. The point mutation (g → t) at the first nucleotide of intron 3 that inactivates the consensus splice donor site (underlined) is shown. The splice donor and acceptor sites used in splicing between exon 2 and exon 4 (or exon 3) are in italics, and the exon 3 nucleotide sequence and encoded amino acids that are internally deleted due to exon 3 skipping are boxed. Sizes of intron 2 and 3 are shown. The intron 3 sequence of indeterminate length retained in some of the tumor transcripts is partially shown, including the premature termination codon (in bold). The novel peptide sequence translated from the intron 3 sequence is shown in italics.
RNA splicing aberrations associated with point mutation at the exon 6/intron 6 junction. Diagrammatic depiction of aberrant splicing in tumor no. 4 due to RNA editing. Both the wild-type (WT) and abnormally spliced transcripts (MUT) are shown (Figure 3Aii). The point mutation in the consensus splice donor sequence (underlined) at the exon 6/intron 6 junction (u → a at position +2) is indicated, along with the sequence chromatogram of cDNA showing the editing U(T)-to-A conversion absent in genomic DNA. The cryptic splice donor site in intron 6 used in the tumor is shown in italics. The 49-bp intron 6 sequence inserted in the tumor transcripts and the resulting premature translation termination are indicated. The predicted structures of PRDM1α and PRDM1β are shown along with their functional domains. Both wild-type and truncated PRDM1 proteins are expected to be produced. (B) Partial cDNA nucleotide sequence of transcripts showing retention of intron 6 sequence in tumor no. 4. The U(T) → A point mutation inactivating the normal consensus splice donor site (underlined) at the exon 6/intron 6 junction is indicated. The intron 6 cryptic splice donor site and the splice acceptor site at the intron 6/exon7 junction used in the tumors are in italics. The retained 49-nt intron 6 sequence is shown (solid arrow), and the distance between its 3′ end and the 5′ end of exon 7 is indicated (362 bp). The premature translation termination codon and the novel peptide sequence predicted from the intron 6 sequence are in bold and italics, respectively. The first 2 zinc fingers (ZF1 and ZF2) that remain in the truncated protein are boxed, with their boundaries marked by arrowheads. A minority of the transcripts used an upstream cryptic splice donor site (dotted underline) and retains only the first 18 nucleotides of intron 6 (see text). The encoded protein is not truncated and has an additional 6 amino acids (ERSIFW).
RNA splicing aberrations associated with point mutation at the exon 6/intron 6 junction. Diagrammatic depiction of aberrant splicing in tumor no. 4 due to RNA editing. Both the wild-type (WT) and abnormally spliced transcripts (MUT) are shown (Figure 3Aii). The point mutation in the consensus splice donor sequence (underlined) at the exon 6/intron 6 junction (u → a at position +2) is indicated, along with the sequence chromatogram of cDNA showing the editing U(T)-to-A conversion absent in genomic DNA. The cryptic splice donor site in intron 6 used in the tumor is shown in italics. The 49-bp intron 6 sequence inserted in the tumor transcripts and the resulting premature translation termination are indicated. The predicted structures of PRDM1α and PRDM1β are shown along with their functional domains. Both wild-type and truncated PRDM1 proteins are expected to be produced. (B) Partial cDNA nucleotide sequence of transcripts showing retention of intron 6 sequence in tumor no. 4. The U(T) → A point mutation inactivating the normal consensus splice donor site (underlined) at the exon 6/intron 6 junction is indicated. The intron 6 cryptic splice donor site and the splice acceptor site at the intron 6/exon7 junction used in the tumors are in italics. The retained 49-nt intron 6 sequence is shown (solid arrow), and the distance between its 3′ end and the 5′ end of exon 7 is indicated (362 bp). The premature translation termination codon and the novel peptide sequence predicted from the intron 6 sequence are in bold and italics, respectively. The first 2 zinc fingers (ZF1 and ZF2) that remain in the truncated protein are boxed, with their boundaries marked by arrowheads. A minority of the transcripts used an upstream cryptic splice donor site (dotted underline) and retains only the first 18 nucleotides of intron 6 (see text). The encoded protein is not truncated and has an additional 6 amino acids (ERSIFW).
Discussion
A tumor-suppressor role for PRDM1 in DLBCLs
This study provides evidence for recurrent inactivation of a PRDM transcription repressor gene family member, PRDM1, in DLBCL, the most common non-Hodgkin lymphoma in the Western world. It indicates a role for PRDM1 defects in the pathogenesis of DLBCLs and implies PRDM1 as a tumor suppressor gene in DLBCLs. Clonal deleterious mutations of PRDM1 were found in the prototypic ABC-like DLBCL cell line OCI-Ly3 and also in approximately 23% (8 of 35) of DLBCL cases, both at diagnosis and in relapse. The mutational spectrum is comprised predominantly of single-nucleotide mutations in the consensus splice donor site leading to splicing aberrations. However, frameshift deletions and chromosomal translocation occur as well. Interestingly, 5 of the 7 splice site mutations are located at the exon 2/intron 2 junction, suggesting the presence of a mutational hot spot. Except for tumor no. 4, all mutations we described are inactivating mutations predicted to generate a truncated protein lacking specific DNA-binding ability and with a disrupted PR domain. The paired alleles for the mutant PRDM1 alleles are either deleted or transcriptionally silenced by mechanism(s) yet to be determined. Thus, PRDM1 is inactivated primarily through the classic 2-hit model of a tumor suppressor gene.
In addition, based on our analysis of tumor no. 4, a dominant-negative mechanism of PRDM1 inactivation may be operating in a minority of DLBCLs. In this case, the PRDM1 mutant lacks the C-terminal acidic domain and zinc finger nos. 3 to 5. It retains the capacity to bind to DNA directly37 and presumably may interfere with wild-type PRDM1 function. However, it is unclear how the function of this PRDM1 mutant is affected. The zinc fingers of PRDM1 may have roles other than DNA binding, as suggested by the ability of the zinc fingers to interact with the histone methyltransferase G3a, although the latter function is apparently not affected in this PRDM1 mutant retaining the first 2 zinc fingers.5 In addition, studies of RIZ (PRDM2) suggest that the C-terminus may contain a region for intermolecular or intramolecular interaction with the PR domain.38 The deletion also removes part of a putative histone deacetylase (HDAC) association domain4 and may potentially affect the ability of PRDM1 to recruit HDACs.
Frameshift mutation in exon 2. (A) RT-PCR analysis of exon 2 sequence in tumor no. 3 (see Table 1 for primers). PCR products were resolved on a 10% polyacrylamide gel. The PCR product derived from the tumor is slightly smaller than that from U266. (B) Sequence chromatogram of cDNA derived from tumor no. 3 showed 4-bp deletion, which results in frameshift and premature translation termination. Note absence of expression of a normal PRDM1 allele in both (A) and (B).
Frameshift mutation in exon 2. (A) RT-PCR analysis of exon 2 sequence in tumor no. 3 (see Table 1 for primers). PCR products were resolved on a 10% polyacrylamide gel. The PCR product derived from the tumor is slightly smaller than that from U266. (B) Sequence chromatogram of cDNA derived from tumor no. 3 showed 4-bp deletion, which results in frameshift and premature translation termination. Note absence of expression of a normal PRDM1 allele in both (A) and (B).
Our findings provide additional evidence for PRDM family members as regulators of tumorigenesis. PRDM2/RIZ is a binding partner for the retinoblastoma tumor suppressor protein and is the frequent target for inactivation in a variety of human tumors, including breast, liver, and colon cancers.20 Its tumor suppressor function is directly confirmed by the tumorigenic phenotype of mice deficient for RIZ1, the PR-containing isoform of PRDM2. These mice demonstrated an increase in incidence of different tumor types, in particular DLBCLs, and accelerated tumor formation in a heterozygous p53 mutant background.21 PRDM3 (MDS1-EVI1) is a frequent target for chromosomal translocations in acute myeloid leukemia and myelodysplasia, which result in deregulation of its expression.39 PRDM4 and PRDM5 are mapped to putative tumor suppressor loci.40,41 Our findings further confirm the important role of the PRDM family in tumor development, especially as negative regulators of tumorigenesis.
Hemizygous deletion of PRDM1 in DLBCLs. Haploid copy number for PRDM1 exons 3, 4, 5, and 7 in tumor nos. 1 through 8 and OCI-Ly3 are shown. OCI-Ly3 was used as a control for hemizygous deletion of PRDM1. Calculations were performed as described in “Materials and methods.” The cut-off limit is indicated by a horizontal line. The value was considered a hemizygous deletion if the haploid copy number was lower than the cut-off. A value higher than the cut-off level supports a normal copy number status.
Hemizygous deletion of PRDM1 in DLBCLs. Haploid copy number for PRDM1 exons 3, 4, 5, and 7 in tumor nos. 1 through 8 and OCI-Ly3 are shown. OCI-Ly3 was used as a control for hemizygous deletion of PRDM1. Calculations were performed as described in “Materials and methods.” The cut-off limit is indicated by a horizontal line. The value was considered a hemizygous deletion if the haploid copy number was lower than the cut-off. A value higher than the cut-off level supports a normal copy number status.
Our analysis indicates that PRDM1α is consistently inactivated, while PRDM1β is truncated only in OCI-Ly3 and in tumor no. 4. These results suggest that PRDM1α is the protein isoform with a tumor suppressor role, the inactivation of which alone is necessary and sufficient to contribute to oncogenesis. Our findings for PRDM1 are rather similar to previous studies on PRDM2/RIZ, which implicated the larger, PR domain–containing isoform as the primary target for inactivation. For example, RIZ1 is selectively decreased or lost in breast, colon, and liver cancers.42-44 Moreover, inactivation of RIZ1 alone can promote tumorigenesis in mice.21 A possible alternative interpretation is that the tumor phenotype associated with PRDM1 and PRDM2 alterations is due to relative overexpression of the smaller protein isoform (PRDM1β or RIZ2). Indeed, overexpression of EVI, the PR-negative PRDM3 isoform, by chromosomal translocations appears to be linked to the pathogenesis of some acute myeloid leukemias.45 However, our findings in OCI-Ly3, which demonstrated deleterious mutations involving both PRDM1α and PRDM1β, support the notion that inactivation of PRDM1α, rather than an alteration in the relative expression of PRDM1α and PRDM1β, is primarily responsible for the oncogenecity associated with PRDM1 defects.
Apart from PRDM1, the chromosomal inversion present in OCI-Ly3 also affects the c-MYB locus by abolishing transcription from the normal promoter. c-MYB transcripts initiated from the alternative β promoter or the cryptic promoter (Figure 3) can only encode c-MYB with 20 amino-acid truncation at the amino-terminus. Interestingly, this truncated form of c-Myb has been shown to induce short-latency lymphomas in chickens.36 These results suggest that overexpression of the 20 amino-acid–truncated form of c-MYB relative to the full-length c-MYB may contribute to lymphomagenesis. The role of c-MYB alterations in human lymphoma development awaits further investigation.
Inhibition of terminal differentiation in DLBCL pathogenesis
Our studies give further support for a role of inhibition of terminal differentiation in the pathogenesis of DLBCLs. A deregulated BCL-6 may block plasmacytic differentiation,24,25 a mechanism that may be more applicable to DLBCLs with a GCB phenotype. Our findings propose PRDM1 mutation and deletion as an alternative mechanism of inhibition of terminal differentiation in DLB-CLs, which may be particularly relevant to ABC-like DLBCL pathogenesis. Consistent with this, PRDM1 defects were only identified in DLBCLs with an immunohistochemically defined non-GCB phenotype, which corresponds to the cDNA microarray-defined ABC phenotype,33 although we have not established a statistically significant difference in the incidence of PRDM1 defects between the GCB-like and non–GCB-like DLBCLs, owing to the small sample size. Interestingly, one of the characteristic genomic imbalances present in ABC-like DLBCLs is loss of 6q, where PRDM1 resides.46
Recent studies show that a disruption in Blimp1 causes a block early in the process of primordial germ-cell formation,47 indicating that the same molecule mediates germ-cell and plasma-cell specification and suggesting that the mechanisms of cell differentiation in germ line and somatic cells are well conserved.48 Analogous to Blimp1-deficient embryonic cells that fail to develop into primordial germ cells with their migration, proliferation, and gene expression characteristics, B cells without a functional PRDM1 fail to acquire a plasma-cell fate. Interestingly, all but one of the DLBCLs with PRDM1 alterations express BCL6. Continuous BCL6 expression may collaborate with PRDM1 inactivation to provide proliferative capacity to B cells blocked in the terminal differentiation pathway. Since BCL6 is normally down-regulated upon antigen activation49 and is not co-expressed with IRF4/MUM1 in germinal centers,50 BCL6 expression in these lymphomas is likely deregulated. However, we did not detect BCL6 rearrangements or somatic hypermutations at the negative autoregulatory BCL6 binding sites located at the first noncoding exon of the BCL6 gene51 (data not shown). The underlying mechanism of BCL6 expression in these lymphoma cells awaits further investigation. A hypothetical model illustrating the role of PRDM1 defects in DLBCL pathogenesis is shown in Figure 7.
Our data clearly indicate a role of PRDM1 inactivation in the pathogenesis of DLBCLs at first presentation. However, it is uncertain at this point whether PRDM1 inactivation is an initiation event, like BCL6 deregulation, in the development of DLBCLs or if it occurs at a later stage of tumor progression associated with clonal expansion subsequent to tumor initiation. We identified PRDM1 defects in relapsed samples, but the paired initial biopsies from these patients were not available for PRDM1 mutation analysis to determine the presence (or absence) of PRDM1 alterations at first presentation. For conclusive demonstration of the role of PRDM1 alterations in relapse of DLBCL, a cohort of paired samples consisting of first-diagnosis and relapse specimens need to be assembled and analyzed.
RNA editing and lymphomagenesis
The consensus splice site point mutations present in 2 DLBCL cases can be attributed to RNA editing. RNA editing is a rare form of post-transcriptional pre-mRNA processing whereby base-specific changes are enzymatically introduced at the RNA level. Only a few examples of RNA editing have been documented in mammalian cells, the most well-known of which are the A-to-I editing described in mRNA encoding subunits of glutamate-gated ion channels and the C-to-U editing found in human apoB lipoprotein mRNA, mediated respectively by adenosine deaminase 1 and 2 active on RNA (ADAR1 and ADAR2) and apoB editing catalytic subunit 1 (APOBEC-1).52,53 Tumor suppressor genes such as NF1 and WT1, as well as the hematopoietic-cell phosphatase PTPN6, also have been shown to be targets for RNA editing.53 In all 3 genes, RNA editing has been described in normal tissues, while hyperediting of NF1 and PTPN6 has been implicated in the pathogenesis of NF1 tumors54 and acute myeloid leukemia,55 respectively. In our studies, we documented 2 examples of PRDM1 inactivation that are likely caused by RNA editing, which raises the possibility of its involvement in lymphomagenesis. However, the true extent of contribution by RNA editing in lymphoma development remains to be determined by analysis of additional samples. Finally, the type of nucleotide conversions (G → A, U → A) encountered in our studies, though previously described in human cells,53,56 are uncommon and do not involve the deamination process involved in A → I and C → U conversions. The underlying mechanism for this unusual alteration awaits further investigation.
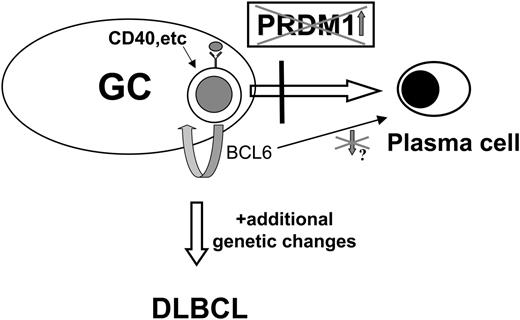
Hypothetical model of the role of PRDM1 in DLBCL pathogenesis. Germinal center (GC) B cells, upon activation through antigen binding to high-affinity receptor, undergo terminal differentiation to plasma cells as a result of PRDM1 induction. Genetic defects in PRDM1 prevent accumulation of functional PRDM1 and inhibit terminal differentiation. As a result, the GC B cells are forced to remain in an activated state and continue to proliferate, possibly driven by BCL6 deregulated expression (as described in “Discussion”). They may then acquire additional genetic alterations that lead to development of full-blown DLBCLs.
Hypothetical model of the role of PRDM1 in DLBCL pathogenesis. Germinal center (GC) B cells, upon activation through antigen binding to high-affinity receptor, undergo terminal differentiation to plasma cells as a result of PRDM1 induction. Genetic defects in PRDM1 prevent accumulation of functional PRDM1 and inhibit terminal differentiation. As a result, the GC B cells are forced to remain in an activated state and continue to proliferate, possibly driven by BCL6 deregulated expression (as described in “Discussion”). They may then acquire additional genetic alterations that lead to development of full-blown DLBCLs.
In summary, we provide in this study genetic evidence of PRDM1 as a tumor suppressor gene (TSG) in DLBCLs. The chromosomal location of PRDM1 suggests that it represents a TSG in the tumor suppressor locus previously identified at 6q21.27-30 Although we have demonstrated allelic deletion of PRDM1 by Q-PCR, we have not correlated these results with 6q21 deletion due to the unavailability of cytogenetic data, except in the OCI-Ly3 cell line. Thus, to confirm that PRDM1 indeed represents the tumor suppressor gene in the 6q21 tumor suppressor locus, a cohort of cases with 6q21 deletion needs to be assembled and analyzed for PRDM1 deletion and mutation. These studies are currently in progress. Preliminary results show the presence of PRDM1 mutations in DLBCL cases with chromosomal deletion encompassing 6q21 (W. T., unpublished data, December 2005). Our present data support the importance of defects in terminal differentiation pathway in the pathogenesis of DLBCLs and suggest that therapy with the potential to restore this pathway may be useful in the treatment of DLBCLs.
Prepublished online as Blood First Edition Paper, January 19, 2006; DOI 10.1182/blood-2005-09-3778.
W.T. designed and performed research, analyzed data, and wrote the paper; M.G. performed research and analyzed data; A.C. and J.W.L. performed research; W.C.C. contributed vital new reagents; and D.M.K. designed research.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal