We report, for the first time, a replication-defective retroviral vector–associated neoplasia in a nonhuman primate. Five years after transplantation with CD34+ cells transduced with a retroviral vector expressing enhanced green fluorescent protein (eGFP) and a drug-resistant variant of the dihydrofolate reductase gene (L22Y), a rhesus macaque developed a fatal myeloid sarcoma, a type of acute myeloid leukemia. Tumor cells contained 2 clonal vector insertions. One insertion was found in BCL2-A1, an antiapoptotic gene. This event suggests that currently available retroviral vectors may have long-term side effects, particularly in hematopoietic stem and progenitor cells.
Introduction
Replication-defective retroviruses are attractive tools for hematopoietic stem cell (HSC) gene therapy because of their ability to integrate into genomic DNA. The risk for insertional mutagenesis by these vectors was thought to be low, assuming random integration and a single hit per cell. Reports of leukemia in mice1 and humans2 related to vector insertional proto-oncogene activation have raised serious safety concerns in the field of gene therapy. Here, we report for the first time a replication-defective vector-associated neoplasia carrying not a potentially transforming gene in a nonhuman primate.
Study design
Details about the materials and methods of this study have been included as supplemental material and are available on the Blood website; see the Supplemental Materials link at the top of the online article.
Results and discussion
The rhesus macaque 96E113 underwent a myeloablative transplantation with granulocyte–colony-stimulating factor (G-CSF)/stem-cell factor (SCF) mobilized autologous CD34+ cells transduced with a murine stem-cell virus (MSCV)–based RD114 pseudotyped retroviral vector. The vector expressed the marker gene enhanced green fluorescent protein (eGFP), and a drug-resistant variant of the dihydrofolate reductase gene (L22Y), under the control of the MSCV long terminal repeat (LTR).3 This animal was unusual in that it had myeloid marking of up to 80% during the first year after transplantation. Insertion site analysis at the time demonstrated that most marked cells derived from a single transduced clone containing 2 vector insertions, one in chromosome 15, the other in chromosome 9. Subsequently, the marking levels declined and stabilized at 1% to 4% eGFP+ cells in the peripheral blood for the next 4 years. The animal remained clinically well and had normal blood counts throughout.4 It received one cycle of chemotherapy with transient in vivo selection of drug resistant cells 2 years after transplantation.5
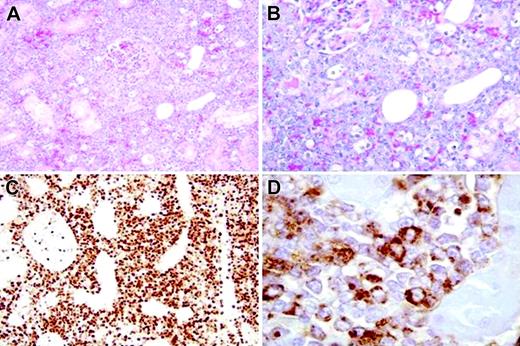
At 5 years after transplantation, 96E113 developed a left renal mass impinging on intestines and pancreas. At this time the peripheral blood showed 30% eGFP+ granulocytes, a left shift in circulating myeloid cells, but no circulating blasts. The kidney was removed; the animal died 5 days after the operation and was found to have immature highly proliferating (MIB-1++) myelomonocytic-cell infiltrates in kidneys, liver, pancreas, spleen, lymph nodes, and choroid plexus (Figure 1A-D). These immature cells were weakly positive for eGFP as demonstrated by immunohistochemistry (Figure S1) and confirmed by in situ hybridization. They expressed myelomonocytic but not lymphoid markers (CD45+/–, CD68+, CD3–, CD20–), classifying the tumor as a widespread myeloid sarcoma, a rare form of acute myeloid leukemia without circulating blasts or bone marrow infiltration.
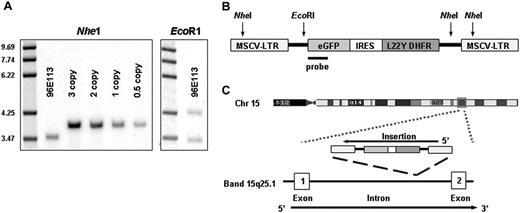
Universal vector Taqman quantitative polymerase chain reaction (PCR)6 demonstrated a vector copy number of at least one per cell in enriched blast cells (> 95%) from the kidney, confirmed by Southern blot on kidney DNA from an area infiltrated with tumor cells with bands after hybridization, using an eGFP probe indicating a copy number of 0.9 (Figure 2A-B). This copy number indicates that the tumor cells contained vector, since the sections used were composed of tumor infiltrates around normal renal tubules. Infection with replication-competent retroviruses was excluded on blast cells and mononuclear cells by PCR analysis using primers that amplify the gag region of wild-type virus (Figure S2), as well as via marker rescue analysis of the producer-cell line used to transduce the CD34+ cells.7
Tumor histology. Myelomonocytic tumor cells with large round nuclei and a variable amount of cytoplasm infiltrate extensively in the left kidney, intercalating between renal tubular cells, shown via hematoxylin and eosin staining, original magnification, × 20 (A) and × 40 (B). Immunohistochemical stains using EnVision Plus from Dako (Carpinteria, CA) as the detection system with chromogen diaminobenzidine (DAB), showed that these cells were uniformly strongly positive for the proliferation marker MIB-1 (original magnification, × 40) (C). The cells were positive for myeloperoxidase (MPO), supporting their myeloid origin (original magnification, × 100). Images were visualized under an Olympus BX41 microscope equipped with an Olympus UPlan Fl 20 ×/0.50 (∞/0.17) (A), 40 ×/0.75 (∞/0.17) (B,C), or 100 ×/1.30 (oil) (D) objective lens (Olympus, Melville, NY). A U-TV 0.5 × C connector was used. Images were captured with an Olympus DP12 digital camera, digitally acquired with an Olympus UBS SmartMedia Reader-Writer, and processed with Photoshop 7.0 software (Adobe Systems, San Jose, CA).
Tumor histology. Myelomonocytic tumor cells with large round nuclei and a variable amount of cytoplasm infiltrate extensively in the left kidney, intercalating between renal tubular cells, shown via hematoxylin and eosin staining, original magnification, × 20 (A) and × 40 (B). Immunohistochemical stains using EnVision Plus from Dako (Carpinteria, CA) as the detection system with chromogen diaminobenzidine (DAB), showed that these cells were uniformly strongly positive for the proliferation marker MIB-1 (original magnification, × 40) (C). The cells were positive for myeloperoxidase (MPO), supporting their myeloid origin (original magnification, × 100). Images were visualized under an Olympus BX41 microscope equipped with an Olympus UPlan Fl 20 ×/0.50 (∞/0.17) (A), 40 ×/0.75 (∞/0.17) (B,C), or 100 ×/1.30 (oil) (D) objective lens (Olympus, Melville, NY). A U-TV 0.5 × C connector was used. Images were captured with an Olympus DP12 digital camera, digitally acquired with an Olympus UBS SmartMedia Reader-Writer, and processed with Photoshop 7.0 software (Adobe Systems, San Jose, CA).
Mature blood cells and enriched blasts (> 95%) from kidney and liver infiltrates have been analyzed by linear amplification mediated (LAM)–PCR and inverse PCR, topoisomerase I–thymidine-adenine (TOPO TA) cloning and subsequent sequencing as described.4,8 The insertion sites were mapped against the current build of the human genome as well as the preliminary rhesus draft genome.9,10 In blood and tumor cells, the same vector insertions characterizing the dominant clone during the first year after transplantation were again identified. The genomic sequences around the first insertion retrieved at the 5′LTR junction via LAM-PCR and at the 3′ LTR junction via inverse PCR had 100% homology with Scaffold35893 from the current rhesus shotgun sequencing database, with a single best match (93%) in the human genome to the first intron of the BCL2-A1 gene (human chromosome 15, band 15q25.1; Figure 2C) and the second insertion mapped to sequences homologous to human chromosome 9, band 9q31.1, more than 20 kb from the closest gene, CDw92, encoding for a choline reporter-like protein found on leukocytes. The same 2 clonal bands previously identified as dominant during the first year after transplantation (Figure 2A-B), before becoming undetectable for the subsequent 4 years, were present in the tumor tissue. These 2 insertions were also identified by LAM-PCR analysis on DNA from individual blasts obtained via single-cell laser capture on kidney sections. PCR analysis using primers specific for the insertion into BCL2-A1 on chromosome 15q25.1 and the insertion into chromosome 9q31.1 revealed a similar pattern of high level contribution from the dominant clone in the blood during the first year after transplantation as previously reported,4 which then became undetectable, before dominating again in the blood at the time of myeloid sarcoma manifestation (Figure S3A-B). TaqMan reverse transcriptase (RT)–PCR was used to assess BCL2-A1 mRNA expression from bulk tumor RNA revealing levels comparable to granulocytes, where BCL2-A1 is highly expressed, and about 70 times higher than in the kidney tissue of an untreated control animal.
Molecular analysis of tumor and blood. (A) Southern blot analysis with an enhanced green fluorescent protein (eGFP) probe to determine copy number of the proviral genome in DNA extracted from the left kidney, which was heavily infiltrated with tumor cells. DNA (10 μg) from the tumor and from copy number control DNA obtained from Jurkat cells containing known copies per cell of the GFP gene were digested with NheI, which cuts once in each proviral LTR, and then hybridized with an eGFP probe. The copy number was calculated as 0.9 copies by phosphoimager scanning and comparison of band intensity to the Jurkat DNA copy number controls. The second panel shows 10μg tumor DNA digested with EcoR1, which cuts once within the proviral genome and therefore generates a unique band for each integration site based on the relative position of the first EcoR1 site in flanking genomic DNA. (B) Scheme of MgirL22Y retroviral vector (which contains eGFP), the internal ribosomal entry site (IRES), and the mutant dihydrofolate reductase gene (L22Y). Shown is the binding site of the probe as well as cutting sites of NheI and EcoRI. (C) BCL2-A1 is a small gene with one intron and 2 exons. The insertion occurred in the intron in the gene opposing DNA strand in chromosome band 15q25.1.
Molecular analysis of tumor and blood. (A) Southern blot analysis with an enhanced green fluorescent protein (eGFP) probe to determine copy number of the proviral genome in DNA extracted from the left kidney, which was heavily infiltrated with tumor cells. DNA (10 μg) from the tumor and from copy number control DNA obtained from Jurkat cells containing known copies per cell of the GFP gene were digested with NheI, which cuts once in each proviral LTR, and then hybridized with an eGFP probe. The copy number was calculated as 0.9 copies by phosphoimager scanning and comparison of band intensity to the Jurkat DNA copy number controls. The second panel shows 10μg tumor DNA digested with EcoR1, which cuts once within the proviral genome and therefore generates a unique band for each integration site based on the relative position of the first EcoR1 site in flanking genomic DNA. (B) Scheme of MgirL22Y retroviral vector (which contains eGFP), the internal ribosomal entry site (IRES), and the mutant dihydrofolate reductase gene (L22Y). Shown is the binding site of the probe as well as cutting sites of NheI and EcoRI. (C) BCL2-A1 is a small gene with one intron and 2 exons. The insertion occurred in the intron in the gene opposing DNA strand in chromosome band 15q25.1.
Our data indicate that a single stem-cell clone with 2 proviral inserts, one potentially activating BCL2-A1, a gene known to have antiapoptotic properties, dominated multilineage contribution to hematopoiesis after transplantation, became dormant for 4 years, and then re-emerged as the dominant clone contributing to myeloid hematopoiesis and a fatal myeloid sarcoma 5 years after transplantation. We hypothesize that the initial vector integrations in this clone produced a marked advantage during initial hematopoietic reconstitution, and that with time, possibly contributed to by cytotoxic chemotherapy administered in 2001, the clone evolved to a leukemia after acquiring additional mutations, although we have no direct cytogenetic or other evidence to date for specific additional mutational events. This hypothesis is supported by recent findings of enhanced cell survival and abnormal proliferative advantage due to insertions following retroviral gene marking in murine models.11-13 It is unknown whether our animal would also have developed leukemia without receiving one cycle of chemotherapy 2 years after transplantation. This animal was 1 of 7 animals that underwent transplantation using a similar vector and regimen, all followed for more than 5 years, and the other animals remain healthy and polyclonal in terms of proviral insertions to date. Overall, 82 large animals have been followed long-term and this is the only animal that developed a leukemia; however, nonrandom predominance of clones harboring vector insertions in genes possibly linked to hematopoietic immortalization has been documented.14,15
The BCL2-A1 protein blocks apoptosis and is associated with poor prognosis in human acute myeloid leukemia.16 The impact of a selective growth advantage on gene-marked cells needs to be addressed in long-term preclinical gene therapy studies to better assess the risk of leukemogenesis with standard retroviral as well as alternative integrating vectors. Our data might argue against the transfusion of a small number of stem cells prone to massive proliferation as this might favor an oligo- or monoclonal hematopoiesis.
Prepublished online as Blood First Edition Paper, January 26, 2006; DOI 10.1182/blood-2005-10-4108.
Supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, the National Institute of Mental Health, and the National Cancer Institute of the NIH/DHHS, Bethesda, MD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Uimook Chooi, Narda Theobald, John Tisdale, and Susan Wong for experimental suggestions and discussions. We thank the staff of 5 Research Court for animal care as well as the veterinary pathology staff, namely Michael Eckhaus and Lauren Brinster, for pathologic analysis. We thank L. Sorbara and Thomas Fountaine III from the Laboratory of Pathology in the National Cancer Institute for assistance with immunohistochemical analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal