Abstract
The ataxia telangiectasia mutated (ATM) protein is the principal activator of the p53 protein in the response to DNA double-strand breaks. Mutations in the ATM gene have been previously found in B-cell chronic lymphocytic leukemias (B-CLLs) but their clinical significance is unknown. We analyzed 155 CLL tumors and found 12% with ATM mutations and 4% with TP53 mutations; 2 tumors contained mutations in both genes. Retrospective analysis on selected samples indicated that the ATM mutations were usually present at diagnosis. Compared with patients with wild-type ATM/TP53 genes, patients with ATM mutations had statistically significantly reduced overall and treatment-free survival. Although present in both IGVH mutation subgroups, ATM mutations were associated with unmutated IGVH genes and they provided independent prognostic information on multivariate analysis. Mutations in the ATM gene resulted in impaired in vitro DNA damage responses. Tumors with ATM mutations only partially correlated with tumors with loss of an ATM allele through an 11q deletion and, interestingly, those 11q-deleted tumors with a second wild-type ATM allele had a preserved DNA damage response. The majority of patients with ATM mutations were refractory to DNA damaging chemotherapeutic drugs and as such might benefit from therapies that bypass the ATM/p53 pathway.
Introduction
The clinical progression of B-cell chronic lymphocytic leukemia (B-CLL) is heterogeneous and prognostic markers are important both for patient management and understanding of disease biology. Established prognostic markers in CLL include the mutation status of the immunoglobulin (IG) VH genes and cytogenetic changes, but both have limitations.1-3 The IGVH mutation status stratifies risk for CLL populations but it is less predictive of outcome for the individual patient. In addition, although poor-risk cytogenetic abnormalities such as 17p or 11q deletions predict impaired patient survival, they only occur in a minority of patients with CLL.1-3 Furthermore, the mechanisms by which these prognostic markers impact on patient survival and/or the proliferation of CLL cells are not fully understood and their value in designing patient-tailored therapy is, therefore, limited.3,4
The response of a tumor to treatment is likely to influence the clinical outcome.5 Chemotherapeutic agents used in CLL mediate cell death through DNA damage and p53-dependent apoptosis.6,7 Consequently, loss of functional p53 is known to be associated with treatment resistance and shortened survival times in CLL.3,7 In contrast to many solid tumors, in CLL, mutations in the TP53 gene are uncommon and we have shown recently that mutations in ataxia telangiectasia mutated (ATM), the gene that acts upstream of p53, are more frequent.8 ATM is a protein kinase that coordinates an integrated cellular response to DNA damage in the form of double-stranded DNA (dsDNA) breaks.9,10 It is likely, therefore, that similar to TP53 genetic defects, ATM mutations could also identify patients with CLL who are refractory to chemotherapeutic drugs such as chlorambucil and fludarabine.
The ATM gene is located within the minimally deleted region on chromosome 11q. Deletions of chromosome 11q are associated with a severe clinical phenotype and reduced survival in CLL. The relationship between ATM mutations and 11q deletions in the pathogenesis of CLL has not been fully resolved.3 Furthermore, although ATM mutations have been reported in CLL, their prevalence in a large cohort and their impact on patient survival has not been assessed.8,11-13
We have, therefore, studied a cohort of 155 CLL tumors to establish the frequency and clinical consequences of ATM mutations and hence their potential value as a new prognostic marker in CLL. The study included an assessment of the timing and stability of ATM mutations and the pathogenic effects of combinations of ATM mutations and loss of an ATM allele. Finally, we performed a multivariate analysis that included age, stage, and IGVH mutation status in order to establish whether ATM mutations can provide independent prognostic information in CLL.
Patients, materials, and methods
Patients
Approval for the study was obtained from the local research ethics committee, South Birmingham, United Kingdom. We studied 155 patients with B-CLL diagnosed using standard immunophenotypic criteria. The study was performed according to local ethical guidelines and written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The patients were selected only by their attendance at the outpatient clinic between December 2001 and January 2003, and all patients who attended between these dates were included. The follow-up ranged from 25 to 217 months, with the median follow-up of 71 months (Table 1). Overall survival (OS) and treatment-free survival (TFS) were calculated as the date of diagnosis of CLL to the date of death, or analysis if alive, and the date of diagnosis to date of treatment or analysis if untreated, respectively. The data analysis was performed on December 1, 2004.
Characteristics of the 155 patients in the CLL cohort
. | Wild type, n = 133 . | ATM mutant, n = 16* . | TP53 mutant, n = 6† . |
|---|---|---|---|
| Male-female (M/F ratio) | 78:55 (1.4) | 8:8 (1.0) | 5:1 (5.0) |
| Median age at diagnosis, y | 66 | 68 | 81.5 |
| Stage at diagnosis | |||
| A | 88 | 11 | 5 |
| B/C | 26 | 4 | 1 |
| Stage at genetic analysis | |||
| A | 66 | 5 | 2 |
| B/C | 60 | 10 | 4 |
| VH gene | (n = 116) | (n = 16) | (n = 6) |
| Unmutated | 29 | 10 | 2 |
| Mutated | 87 | 6 | 4 |
| 11q deletion (n = 77) | (n = 60) | (n = 13) | (n = 4) |
| Present | 5 | 4 | 0 |
| Absent | 55 | 9 | 4 |
| Era of diagnosis | |||
| Before 1996 | 44 | 5 | 0 |
| 1997-2000 | 44 | 6 | 4 |
| 2001-2003 | 45 | 4 | 2 |
| Median follow-up, mo (range) | 68.5 (25-217) | 79 (26-156) | 60.5 (34-88) |
| Percentage treated | 43 | 73 | 66 |
| Median time to treatment, mo (n = 152) | 130 (n = 131) | 40 (n = 15) | 40 (n = 6) |
| Percentage died | 17 | 47 | 66 |
| Median survival, mo (n = 154) | > 217 (n = 133) | 85 (n = 15) | 45 (n = 6) |
. | Wild type, n = 133 . | ATM mutant, n = 16* . | TP53 mutant, n = 6† . |
|---|---|---|---|
| Male-female (M/F ratio) | 78:55 (1.4) | 8:8 (1.0) | 5:1 (5.0) |
| Median age at diagnosis, y | 66 | 68 | 81.5 |
| Stage at diagnosis | |||
| A | 88 | 11 | 5 |
| B/C | 26 | 4 | 1 |
| Stage at genetic analysis | |||
| A | 66 | 5 | 2 |
| B/C | 60 | 10 | 4 |
| VH gene | (n = 116) | (n = 16) | (n = 6) |
| Unmutated | 29 | 10 | 2 |
| Mutated | 87 | 6 | 4 |
| 11q deletion (n = 77) | (n = 60) | (n = 13) | (n = 4) |
| Present | 5 | 4 | 0 |
| Absent | 55 | 9 | 4 |
| Era of diagnosis | |||
| Before 1996 | 44 | 5 | 0 |
| 1997-2000 | 44 | 6 | 4 |
| 2001-2003 | 45 | 4 | 2 |
| Median follow-up, mo (range) | 68.5 (25-217) | 79 (26-156) | 60.5 (34-88) |
| Percentage treated | 43 | 73 | 66 |
| Median time to treatment, mo (n = 152) | 130 (n = 131) | 40 (n = 15) | 40 (n = 6) |
| Percentage died | 17 | 47 | 66 |
| Median survival, mo (n = 154) | > 217 (n = 133) | 85 (n = 15) | 45 (n = 6) |
ATM mutant tumors with wild-type TP53
TP53 tumors including 2 tumors with an additional mutation in ATM
Our method of recruitment led to a cohort that is biased toward a benign phenotype, particularly for those patients who were diagnosed in the earliest years. In an attempt to compensate for this, we adjusted for year of diagnosis in our multivariate survival analyses. Nevertheless, any detrimental effect on survival due to ATM mutations would be underestimated due to the selective survival bias, since patients who died prior to December 2001 would not have been included in the study.
Blood samples
Peripheral-blood samples were collected over the study period and mononuclear cells separated and samples stored in 90% fetal calf serum (FCS) and 10% dimethyl sulfoxide (DMSO) in liquid nitrogen.8 Samples were subsequently thawed and used for genetic and biochemical testing. Prior to biochemical testing, the cellular viability of samples was determined using trypan blue and shown to be more than 90% in all analyzed cases. In 5 patients with ATM mutations, samples collected in the years prior to the study period were available and these were used to determine if the mutations were stable over time and present early in the course of the disease. One additional tumor, previously identified with 2 ATM mutations, was also tested over serial samples spanning 14 years.
Mutational analysis of ATM and TP53 genes
Genomic DNA was obtained from 5 × 106 mononuclear cells using QIAamp DNA blood minikit (Qiagen, Crawley, West Sussex, United Kingdom). Polymerase chain reactions (PCRs) were performed for each of 62 ATM coding exons and flanking intronic sequences using 60 primer pairs modified from a previous protocol.14 TP53 exons 3 to 10 were amplified using 5 primer pairs.8 Reaction products were analyzed by denaturing high-performance liquid chromatography (DHPLC) using the WAVE DNA fragment analysis system (Transgenomic, Omaha, NE). Chromatograms were analyzed visually and the PCR product was sequenced if a variant pattern was detected. Analysis of sequences was performed using an ABI Prism sequencer and ABI software (Applied Biosystems, Foster City, CA). A sequence change was designated as a mutation if it was predicted to lead to a truncated protein, caused an amino acid substitution neither previously reported as polymorphism nor present in 200 normal alleles, or had been previously shown to produce abnormal exon splicing (T.S. and B.A., unpublished data, August 2004).15,16
IGVH mutation status
Amplification and sequencing of IGVH genes was performed using Framework1 and JPS primers, as previously described.8 Tumors were classified as IGVH unmutated if there was at least 98% concordance between the tumor DNA and the respective family sequence, and IGVH mutated if there was less than 98% concordance.1,2
Assessment of 11q status
Karyotyping was performed in a diagnostic cytogenetic laboratory. Interphase fluorescence in situ hybridization (FISH) for 11q status was performed on tumors found to have an ATM mutation (Qbiogene, Irvine, CA).
Assessment of ATM activity by Western blotting
Lymphocytes (2 × 106) were resuspended in RPMI with 10% FCS and irradiated with 5 Gy ionizing irradiation (IR). Cellular lysates were extracted at 0, 15, and 45 minutes after IR and proteins were separated on 6% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels as previously described.8 Antibodies used for Western blotting were a rabbit antibody to serine 15 phosphorylated p53, rabbit antibody to serine 1981 phosphorylated ATM, mouse monoclonal antibody to actin (all produced by Sigma, St Louis, MO), sheep antibody to p53 (donated by D. P. Lane, University of Dundee, Dundee, United Kingdom), rabbit antibody to serine 966 phosphorylated structural maintenance of chromosome 1 (SMC1) (Bethyl Laboratories, Montgomery), rabbit antibody to serine 343 phosphorylated Nbs1 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit antibody to Nbs1 (Novus Biologicals, Littleton, CO), and mouse monoclonal ATM antibody.17
The phosphorylation responses were classified as follows. An ATM mutant tumor was classified as functionally ATM defective if at 15 minutes following IR both the levels of p53 serine 15 phosphorylation and the ATM serine 1981 phosphorylation were less than 50% of the level in the representative wild-type tumor for the same exposure time, as assessed by optic density. An ATM mutant tumor was classified as having a partially deficient response if either the level of p53 serine 15 phosphorylation or ATM serine 1981 phosphorylation was less than 50% of the level in the wild-type tumor at 15 minutes after IR. Protein levels were corrected for actin.
Assessment of apoptosis induced by DNA damage
Cells were cultured and lysates extracted at 0, 8, and 24 hours following 5 Gy IR. Proteins were separated on 8% SDS-PAGE gels and blotted with mouse antibodies to poly adenosine diphosphate (ADP) ribose polymerase 1 (PARP1) (R&D Systems, Minneapolis, MN) and actin. The change in optic density of the uncleaved PARP1 protein (corrected for actin) was determined between 0 and 24 hours.
An annexin V apoptosis kit (BD Pharmingen, Oxford, United Kingdom) and Coulter Epics XL-MCL flow cytometer (Beckman Coulter, Fullerton, CA) were used to measure apoptosis in 15 representative ATM wild-type and 7 ATM mutant tumors.
Statistical methods
The Kaplan Meier method was carried out to compare OS and TFS between genetic subgroups, and the log rank test was used to assess the significance of differences in survival. Multivariate analysis (Cox proportional hazards regression) was undertaken to assess the interdependence of prognostic factors, including ATM status, IGVH status, age at diagnosis, stage at diagnosis, and year of diagnosis. Since advanced stage is an indication for treatment in CLL, patients with advanced stage disease were excluded from the multivariate analysis for TFS. The Fisher exact test was used to confirm any association between pairs of parameters and the Kruskall-Wallis test was used to compare median levels of PARP1 cleavage.
Results
Characteristics of the CLL cohort
The CLL cohort consisted of 91 men and 64 women. The age at diagnosis ranged between 28 and 92 years and the median age was 67 years. The cohort included 41 patients with unmutated and 97 patients with mutated IGVH genes. In 77 patients, 11q status was known and 9 patients had a deletion of 11q, comprising 4 patients with an ATM mutation and 5 patients with wild-type ATM on the remaining allele. Forty-nine patients were diagnosed prior to 1996, 54 between 1997 and 2000, and 51 between 2001 and 2003. At the time of data analysis, 43% and 17% of ATM/TP53 wild-type patients had been treated and had died, respectively, compared with 73% and 47% of patients with ATM mutations and 66% and 66% of patients with TP53 mutations. These characteristics according to genetic status are summarized in Table 1.
ATM mutations occur in 12% of B-CLL tumors and are associated with impaired overall and treatment-free survival in patients with B-CLL
We found 23 ATM mutations in 18 CLL tumors. This indicated a 12% prevalence of ATM mutations in our CLL cohort of 155 patients. In 7 tumors there was loss of both functional alleles, in 3 cases as a consequence of ATM mutations and in 4 cases as a consequence of an ATM mutation in combination with an 11q deletion. In contrast, 11 tumors had a single ATM mutation with no loss of the remaining allele (Table 2). We also detected heterozygous TP53 mutations in 6 tumors, indicating a prevalence of 4% (Table 3). In 16 tumors ATM mutations were associated with wild-type TP53, in 4 tumors a TP53 mutation was associated with wild-type ATM, but notably, 2 tumors (CLLs 84 and 124) had a mutation in both the ATM and TP53 genes.
ATM mutations in the 155 patients in the CLL cohort
Tumor ID . | Sequence change . | Protein change . | Present in diagnostic sample . | ATM function . | IGVH mutation status and family . | TFS, mo . | OS, mo . | Refractory to Flu/Chl . |
|---|---|---|---|---|---|---|---|---|
| Tumors with 2 mutant ATM alleles | ||||||||
| 77* | 1058DelGT | Truncated protein | Yes | Defective | 99%, VH1-2 | 0 | 83† | Yes |
| 77* | 5224G/C | A1742P | Yes | Defective | 99%, VH1-2 | 0 | 83† | Yes |
| 96* | 2929Ins9 | Inframe insertion | Yes | Defective | 96%, VH4-61 | 33 | 79 | Yes |
| 96* | 5041A/G | I1681V | Yes | Defective | 96%, VH4-61 | 33 | 79 | Yes |
| 96* | 5044G/T | D1682Y | Yes | Defective | 96%, VH4-61 | 33 | 79 | Yes |
| 11* | 5821G/C | V1941L | Not known | Defective | 98%, VH3-15 | 108 | 156† | Yes |
| 11* | 7313C/A | T2438K | Not known | Defective | 98%, VH3-15 | 108 | 156† | Yes |
| 11* | 8833Del34 | Truncated protein | Not known | Defective | 98%, VH3-15 | 108 | 156† | Yes |
| Tumors with 1 mutant ATM allele and loss of the second allele | ||||||||
| 57* | 2308G/T | Truncated protein | Not known | Partial defect | 97%, VH4-59 | 76 | 101† | No |
| 15* | 7047C/G | C2349W | Not known | Defective | 100%, VH1-69 | 40 | 75 | Yes |
| 07* | 8600G/A | G2867E | Yes | Defective | 99%, VH3-11 | 0 | 96 | Yes |
| 152* | 8839A/T | T2946S | At 1 year§ | Defective | 99%, VH1-6 | 35 | 79† | No |
| Tumors with 1 mutant ATM allele and 1 wild-type ATM allele | ||||||||
| 75 | 1009C/A | R337S | Yes | Defective | 98%, VH3-30 | 0 | 39 | Yes |
| 69 | 6815DelA | Truncated protein | Not known | Defective | 100%, VH3-15 | 39 | 72† | No |
| 119 | 9022C/T | R3008C | Not known | Defective | 97%, VH3-64 | 36 | 47† | No |
| 93 | 5290del C | Truncated protein | Yes | Defective | 100%, VH5-51 | 26† | 26† | NT |
| 92 | 5980A/G | K1994E | Yes | Partial defect | 99%, VH1-69 | 24† | 24 | NT |
| 124‡ | 5228C/T | T1743I | Not known | Partial defect | 85%, VH3-15 | 48† | 48† | NT |
| 84‡ | 5821G/C | V1941L | Yes | Partial defect | 93%, VH3-7 | 14 | 17 | Yes |
| 27 | 3383A/G | Q1127R | Not known | Normal | 99%, VH3-21 | 0 | 77 | Yes |
| 113 | 3964C/A | L1322F | Not known | Normal | 91%, VH3-7 | 24† | 24 | NT |
| 147 | IVS16-10T/G | Exon skip | Not known | Not done | 94%, VH5-51 | NA | NA | NA |
| 81 | 5882A/G | Y1961C | Not known | Not done | 93%, VH3-15 | 36† | 36† | NT |
Tumor ID . | Sequence change . | Protein change . | Present in diagnostic sample . | ATM function . | IGVH mutation status and family . | TFS, mo . | OS, mo . | Refractory to Flu/Chl . |
|---|---|---|---|---|---|---|---|---|
| Tumors with 2 mutant ATM alleles | ||||||||
| 77* | 1058DelGT | Truncated protein | Yes | Defective | 99%, VH1-2 | 0 | 83† | Yes |
| 77* | 5224G/C | A1742P | Yes | Defective | 99%, VH1-2 | 0 | 83† | Yes |
| 96* | 2929Ins9 | Inframe insertion | Yes | Defective | 96%, VH4-61 | 33 | 79 | Yes |
| 96* | 5041A/G | I1681V | Yes | Defective | 96%, VH4-61 | 33 | 79 | Yes |
| 96* | 5044G/T | D1682Y | Yes | Defective | 96%, VH4-61 | 33 | 79 | Yes |
| 11* | 5821G/C | V1941L | Not known | Defective | 98%, VH3-15 | 108 | 156† | Yes |
| 11* | 7313C/A | T2438K | Not known | Defective | 98%, VH3-15 | 108 | 156† | Yes |
| 11* | 8833Del34 | Truncated protein | Not known | Defective | 98%, VH3-15 | 108 | 156† | Yes |
| Tumors with 1 mutant ATM allele and loss of the second allele | ||||||||
| 57* | 2308G/T | Truncated protein | Not known | Partial defect | 97%, VH4-59 | 76 | 101† | No |
| 15* | 7047C/G | C2349W | Not known | Defective | 100%, VH1-69 | 40 | 75 | Yes |
| 07* | 8600G/A | G2867E | Yes | Defective | 99%, VH3-11 | 0 | 96 | Yes |
| 152* | 8839A/T | T2946S | At 1 year§ | Defective | 99%, VH1-6 | 35 | 79† | No |
| Tumors with 1 mutant ATM allele and 1 wild-type ATM allele | ||||||||
| 75 | 1009C/A | R337S | Yes | Defective | 98%, VH3-30 | 0 | 39 | Yes |
| 69 | 6815DelA | Truncated protein | Not known | Defective | 100%, VH3-15 | 39 | 72† | No |
| 119 | 9022C/T | R3008C | Not known | Defective | 97%, VH3-64 | 36 | 47† | No |
| 93 | 5290del C | Truncated protein | Yes | Defective | 100%, VH5-51 | 26† | 26† | NT |
| 92 | 5980A/G | K1994E | Yes | Partial defect | 99%, VH1-69 | 24† | 24 | NT |
| 124‡ | 5228C/T | T1743I | Not known | Partial defect | 85%, VH3-15 | 48† | 48† | NT |
| 84‡ | 5821G/C | V1941L | Yes | Partial defect | 93%, VH3-7 | 14 | 17 | Yes |
| 27 | 3383A/G | Q1127R | Not known | Normal | 99%, VH3-21 | 0 | 77 | Yes |
| 113 | 3964C/A | L1322F | Not known | Normal | 91%, VH3-7 | 24† | 24 | NT |
| 147 | IVS16-10T/G | Exon skip | Not known | Not done | 94%, VH5-51 | NA | NA | NA |
| 81 | 5882A/G | Y1961C | Not known | Not done | 93%, VH3-15 | 36† | 36† | NT |
Flu indicates fludarabine; Chl, chlorambucil; OS, overall survival; TFS, treatment-free survival; ND, not done; NT, not treated; NA, not available.
Patients with predicted loss of both functional ATM alleles
Treatment or death, respectively, has not occurred
Patients with ATM and TP53 mutations
See “ATM mutations are present early in disease ontogenesis and usually at diagnosis”
TP53 mutations in the 155 patients in the CLL cohort
Patient ID . | Sequence change . | Protein change . | IGVH mutation and IGVH family . | Treatment-free survival, mo . | Overall survival, mo . |
|---|---|---|---|---|---|
| 29 | 807ins12 | Inframe insertion | 99%, VH4-34 | 20 | 22 |
| 84* | 797G/A | G266Q | 93%, VH3-7 | 14 | 17 |
| 110 | 536A/T | H179L | 90%, VH3-7 | 88† | 88† |
| 124* | 403T/C | C135R | 85%, VH3-15 | 48† | 48† |
| 70 | 739A/T | N247Y | 86%, VH2-5 | 0 | 50 |
| 146 | 455C/T | P152L | 100% VH3-49 | 56 | 66 |
Patient ID . | Sequence change . | Protein change . | IGVH mutation and IGVH family . | Treatment-free survival, mo . | Overall survival, mo . |
|---|---|---|---|---|---|
| 29 | 807ins12 | Inframe insertion | 99%, VH4-34 | 20 | 22 |
| 84* | 797G/A | G266Q | 93%, VH3-7 | 14 | 17 |
| 110 | 536A/T | H179L | 90%, VH3-7 | 88† | 88† |
| 124* | 403T/C | C135R | 85%, VH3-15 | 48† | 48† |
| 70 | 739A/T | N247Y | 86%, VH2-5 | 0 | 50 |
| 146 | 455C/T | P152L | 100% VH3-49 | 56 | 66 |
Patients with ATM and TP53 mutations
Treatment or death, respectively, has not occurred
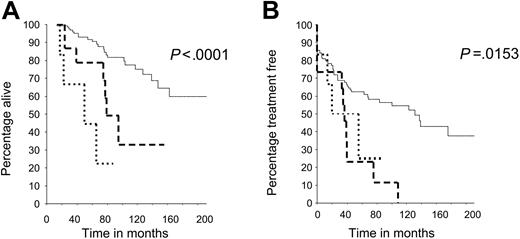
The overall survival (OS) and treatment-free survival (TFS) from diagnosis were determined in all patients and related to the mutational status of the ATM and TP53 genes (Table 1). Patients with an ATM mutation had a statistically significant reduction in both OS and TFS (log rank test, P < .001, P = .007, respectively) compared with patients whose tumors retained wild-type ATM and TP53 genes. Patients whose tumors carried TP53 mutations had the shortest OS. The survival differences among all 3 genetic subgroups were highly statistically significant (P < .001 for OS and TFS). The median OS and TFS were 85 months and 40 months, respectively, for patients with an ATM mutation and more than 217 months and 130 months for patients with ATM/TP53 wild-type tumors (Figure 1A-B). Furthermore, 8 of the 12 patients with an ATM mutation who had received chemotherapy were clinically refractory to treatment (Table 2).
ATM mutations reduce patient survival independently of age, stage, and IGVH status
The poorer OS in patients with ATM mutations could be due to the association of ATM mutations with other prognostic factors including age, stage, and IGVH mutation status. There was no difference in age and disease stage at diagnosis between patients with and without ATM mutations (Table 1). We found a significant association between ATM mutations and unmutated IGVH genes, 24% (10/41) of IGVH unmutated tumors showed ATM mutations, compared with 8% (8/97) of IGVH mutated tumors (P = .024, Fisher exact test).
As expected, regardless of the ATM mutation status, the TFS was significantly shorter in patients with IGVH unmutated genes than in patients with IGVH mutated genes (P < .001). The difference in OS between the IGVH unmutated and mutated subgroups did not reach significance but we observed a clear trend of poorer OS in patients with unmutated IGVH genes (P < .19). Interestingly, the 10-year OS for patients with mutated IGVH in our cohort was shorter (72% vs 85%) compared with a previously published report.1 This could be a reflection of the fact that mean and median ages in our cohort were slightly higher than previously reported1 and when we considered only younger patients (< 65 years), IGVH mutation status became significantly predictive of OS (P = .034).
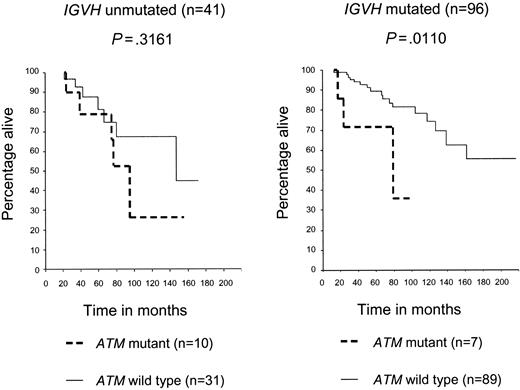
We next determined if the prognostic effect of an ATM mutation was independent of IGVH mutation status. The presence of an ATM mutation led to a significant reduction in OS in the IGVH mutated group (P = .011), but not in the IGVH unmutated CLL group (P = .31). The stratified log rank test for both graphs was significant (P < .001), therefore indicating that ATM mutations reduce survival after adjusting for IGVH status (Figure 2).
We then used multivariate Cox regression on 135 patients, for whom information on all parameters was available, to assess if ATM mutations were an independent predictor of OS. After adjustment for age, stage, and IGVH mutation status, ATM mutations were still significantly predictive of OS (P < .001). The adjusted relative hazard (risk of death) for patients with ATM mutations, as compared with those with wild-type mutations, was 3.0 (95% confidence interval [CI] 1.23-7.29). Furthermore, when the era of diagnosis was included in the model, this hazard ratio increased to 3.4, confirming that in this retrospective cohort, the effect of ATM mutations on OS is likely to be underestimated. We also applied multivariate analysis to assess if ATM mutations were an independent predictor of TFS. After adjustment for age and IGVH status, ATM mutations were also significantly predictive of reduced TFS (P = .035). The relative hazard (risk of commencing treatment) for patients with ATM mutations, compared with those with wild-type mutations, was 2.44 (95% CI 1.07-5.60). Thus, we confirmed that ATM mutations are an independent prognostic factor with respect to both OS and TFS in CLL.
ATM mutations are present early in disease ontogenesis and usually at diagnosis
In order to justify our retrospective approach, we performed horizontal mutation analysis 2 to 7 years back from the time of initial analysis in 5 tumors from our cohort of patients with ATM mutations. In 4 cases the analysis included diagnostic material. Here we were able to show that all 6 ATM mutations were present in the diagnostic sample of CLLs 77, 96, 07, and 75. In the fifth case (CLL 152) the earliest sample available was 1 year following diagnosis, but again the ATM mutation was present in this sample (Table 2). In addition, 3 further cases (CLLs 93, 92, and 84) had ATM mutations detected in their diagnostic sample as a part of the cohort analysis. Taken together, we could demonstrate that ATM mutations were present in 7 of 7 diagnostic samples analyzed.
Reduced survival in patients whose tumors have ATM and TP53 mutations, compared with patients with wild-type ATM and TP53 genes. (A) Overall survival (OS) in patients with CLL with ATM or TP53 mutations was reduced compared with patients with CLL with ATM/TP53 wild-type mutations, and these differences were significant (P < .001). (B) Treatment-free survival (TFS) in patients with ATM and TP53 mutations was reduced compared with patients with ATM/TP53 wild-type mutations, and these differences were also significant (P < .001).
Reduced survival in patients whose tumors have ATM and TP53 mutations, compared with patients with wild-type ATM and TP53 genes. (A) Overall survival (OS) in patients with CLL with ATM or TP53 mutations was reduced compared with patients with CLL with ATM/TP53 wild-type mutations, and these differences were significant (P < .001). (B) Treatment-free survival (TFS) in patients with ATM and TP53 mutations was reduced compared with patients with ATM/TP53 wild-type mutations, and these differences were also significant (P < .001).
ATM mutations reduce both OS and TFS, independently of IGVH status. Patients with CLL with ATM mutations had poorer OS than did patients with CLL with ATM wild-type mutations, and this was more marked in the IGVH mutated (P = .0110) than unmutated (P = .3161) subgroup. The stratified log rank test for both graphs was significant (P < .001), therefore, indicating that ATM mutations reduce survival after adjusting for IGVH status.
ATM mutations reduce both OS and TFS, independently of IGVH status. Patients with CLL with ATM mutations had poorer OS than did patients with CLL with ATM wild-type mutations, and this was more marked in the IGVH mutated (P = .0110) than unmutated (P = .3161) subgroup. The stratified log rank test for both graphs was significant (P < .001), therefore, indicating that ATM mutations reduce survival after adjusting for IGVH status.
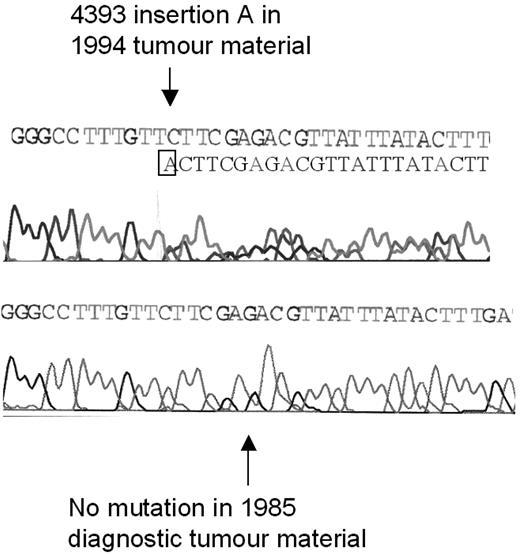
ATM mutation analysis was performed over multiple time points on an additional tumor that had previously been found to have 2 ATM mutations (2114insA, 4393insA).8 In this single CLL tumor, although one ATM mutation (2114insA) was present at diagnosis, interestingly, the second ATM mutation (4393insA) occurred during disease progression and following chlorambucil treatment (Figure 3).
Pathogenic effect of identified ATM mutations
ATM is a protein kinase that coordinates the response to DNA damage through phosphorylation of multiple target proteins. This results in arrest of the cell cycle and either repair of the DNA damage or activation of apoptosis.9,10,18,19 In this study, the pathogenic effect of ATM mutations was validated by 2 means. The functional integrity of the ATM kinase was determined by the assessment of the phosphorylation levels of ATM targets, principally ATM (via autophosphorylation) and p53, and by induction of apoptosis following IR.
Effect on ATM-dependent DNA damage response. Functional ATM-dependent DNA damage responses were studied in 16 of 18 tumors with ATM mutations where viable cells were available, including 7 tumors with loss of both functional alleles and 9 tumors with one mutant and one wild-type ATM allele, in 4 of 5 tumors with an 11q deletion but no ATM mutation, and in 14 ATM/TP53 wild-type tumors.
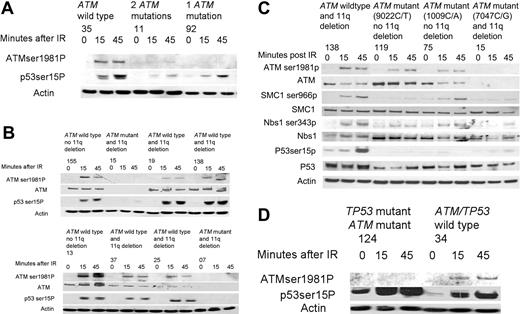
In the wild-type tumors, both ATM Ser1981 autophosphorylation and p53 Ser15 phosphorylation were induced by 15 minutes following IR (Figure 4A-D). As expected, 6 of 7 tumors with loss of both functional ATM alleles had deficient ATM kinase activity (CLLs 77, 96, 11, 15, 7, and 152). In the remaining tumor (CLL 57) there was a partially deficient response consistent with the possibility that loss of ATM occurred in a subclone of the total CLL tumor population (Figure 4A-B, Table 2).
In 4 of 4 tumors (CLLs 19, 138, 37, and 25) with loss of one ATM allele through an 11q deletion but with a second wild-type ATM allele, the induction of p53 phosphorylation was comparable to the wild-type tumors (Figure 4B). The percentage of cells in the CLL clone with an 11q deletion on FISH analysis for CLLs 19, 138, 37, and 25 were 95%, 80%, 75%, and 97%, respectively. Therefore, our results indicate that the ATM protein encoded from the single wild-type allele is sufficient for normal enzymatic activity.
In contrast, 7 of 9 analyzed tumors with one mutant and one wild-type ATM allele (CLLs 75, 69, 119, 93, 92, 124, and 84) exhibited reduced induction of phosphorylation of either one or both ATM targets compared with tumors with both wild-type ATM alleles (Table 2).
Two of these tumors (CLLs 84 and 124) had mutations in both the ATM and TP53 genes. Both tumors exhibited reduced levels of phosphorylated ATM following DNA damage, indicative of a reduction of ATM autophosphorylation. However, the preserved induction of phosphorylation of the abundant mutant p53 protein following DNA damage indicated retention of this aspect of ATM function (Figure 4D).8
In 3 tumors (CLLs 75, 119, and 92), we identified only a single missense ATM mutation (1009C/A, 9022C/T, and 5980A/G, respectively) in the presence of a wild-type allele, suggesting the potential for a dominant-negative effect. We, therefore, evaluated phosphorylation of further ATM targets in CLLs 75 and 119 and compared them with a tumor with 11q deletion and loss of ATM (CLL 138) as well as with a tumor with loss of both functional ATM alleles through ATM mutation and deletion of 11q (CLL 15). Consistent with a dominant-negative effect, phosphorylation of ATM, p53, SMC1, and Nbs1 were all reduced in CLLs 75 and 119 compared with CLL 138 with a single ATM allelelic deletion and were comparable to CLL 15 with loss of both functional ATM alleles (Figure 4C).
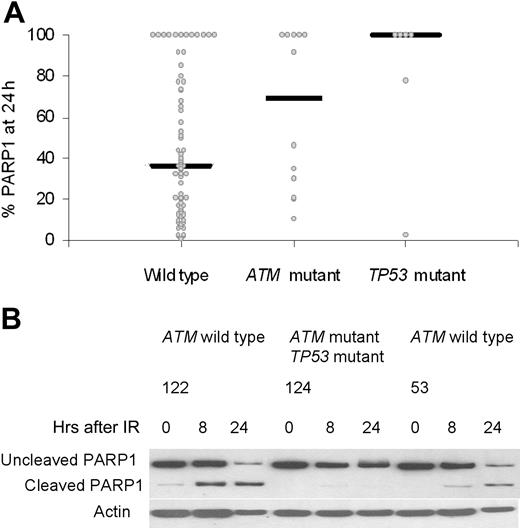
Effect on DNA damage apoptosis. Induction of apoptosis is one downstream effect of IR if the DNA damage response pathway is functional. We assessed apoptosis by caspase-dependent cleavage of PARP1 at 24 hours after DNA damage in 70 wild-type tumors, 12 ATM mutant tumors, and 6 TP53 mutant tumors (Figure 5A). The percentage of PARP1 that remained uncleaved after DNA damage was used as a measure of resistance to apoptosis. An unchanged level of uncleaved PARP1 (100%) at 24 hours indicated an absence of apoptosis in a tumor, and was observed in 67% (4/6) of tumors with TP53 mutations, 42% (5/12) of tumors with ATM mutations, and also 16% (11/70) of ATM/TP53 wild-type tumors. The median values for residual PARP1 levels at 24 hours after IR were 100% for TP53 mutant tumors, 69% for ATM mutant tumors, and 36% for ATM/TP53 wild-type tumors. These differences were statistically significant (Kruskal-Wallis test, P = .019; Figure 5B). Interestingly, 12 ATM/TP53 wild-type tumors showed resistance to PARP1 cleavage. These tumors had normal activity of the p53 pathway (data not shown), suggesting a defect in either a p53-dependent downstream target or an alternative apoptotic pathway.
Development of an ATM mutation during disease progression. The ATM mutation 4393 insertion A was not present at the time of diagnosis of CLL in 1985 (bottom sequence) but was present in tumor material from 1994, after treatment with chlorambucil (top sequence). After the insertion of an additional adenine (A) nucleotide in one allele, the reading frames of the 2 alleles become out of line, resulting in a double sequence; one corresponding to the mutated and one corresponding to the unmutated allele.
Development of an ATM mutation during disease progression. The ATM mutation 4393 insertion A was not present at the time of diagnosis of CLL in 1985 (bottom sequence) but was present in tumor material from 1994, after treatment with chlorambucil (top sequence). After the insertion of an additional adenine (A) nucleotide in one allele, the reading frames of the 2 alleles become out of line, resulting in a double sequence; one corresponding to the mutated and one corresponding to the unmutated allele.
The effects of ATM mutations on the phosphorylation of ATM targets after ionizing irradiation. (A) CLL 35, with no ATM mutations, exhibited induction of ATM and p53 phosphorylation at 15 and 45 minutes after irradiation (IR). CLL 11, with 2 ATM mutations, shows almost no induction of the phosphorylation of either ATM or p53 at both time points after IR. CLL 92, with one ATM mutation, showed no induction in the phosphorylation of ATM but does exhibit some induction in the phosphorylation of p53 after IR. (B) CLLs 155 and 13, with wild-type ATM and no 11q deletion, had a normal ATM-dependent response. CLLs 15 and 7, with an 11q deletion (detected in 95% and 92% of cells respectively) and with an ATM mutation, revealed no ATM protein and no induction of p53 phosphorylation following IR. CLLs 19, 138, 37, and 25, with an 11q deletion (detected in 95%, 80%, 75%, and 97% of cells respectively) and a wild-type ATM allele, all exhibited induction of p53 phosphorylation comparable to wild-type tumors. (C) The induction of phosphorylation of multiple ATM targets is compared between tumor subtypes. Baseline levels of SMC1, Nbs1, and p53 proteins are shown below each phosphorylated protein and are comparable in all tumors. CLL 138, with loss of one ATM allele through an 11q deletion but with a remaining wild-type ATM allele, demonstrated induction of phosphorylation of ATM, p53, SMC1, and Nbs1 at early time points after IR, consistent with normal ATM activity. In contrast, CLLs 199 and 75, with one ATM allele with a missense mutation ATM and one wild-type ATM allele, showed impaired phosphorylation of ATM targets (most marked for CLL 199) despite high ATM protein levels, consistent with a potential dominant-negative effect. CLL 15, with loss of one ATM allele and a mutation in the remaining ATM allele, failed to phosphorylate any of the target proteins consistent with absent ATM protein. (D) CLL 34 had a normal ATM-dependent response. CLL 124, with a mutation in both the ATM and TP53 genes, showed absence of phosphorylation of ATM in response to IR, but increased baseline phosphorylation of p53 that increased further during response to IR.
The effects of ATM mutations on the phosphorylation of ATM targets after ionizing irradiation. (A) CLL 35, with no ATM mutations, exhibited induction of ATM and p53 phosphorylation at 15 and 45 minutes after irradiation (IR). CLL 11, with 2 ATM mutations, shows almost no induction of the phosphorylation of either ATM or p53 at both time points after IR. CLL 92, with one ATM mutation, showed no induction in the phosphorylation of ATM but does exhibit some induction in the phosphorylation of p53 after IR. (B) CLLs 155 and 13, with wild-type ATM and no 11q deletion, had a normal ATM-dependent response. CLLs 15 and 7, with an 11q deletion (detected in 95% and 92% of cells respectively) and with an ATM mutation, revealed no ATM protein and no induction of p53 phosphorylation following IR. CLLs 19, 138, 37, and 25, with an 11q deletion (detected in 95%, 80%, 75%, and 97% of cells respectively) and a wild-type ATM allele, all exhibited induction of p53 phosphorylation comparable to wild-type tumors. (C) The induction of phosphorylation of multiple ATM targets is compared between tumor subtypes. Baseline levels of SMC1, Nbs1, and p53 proteins are shown below each phosphorylated protein and are comparable in all tumors. CLL 138, with loss of one ATM allele through an 11q deletion but with a remaining wild-type ATM allele, demonstrated induction of phosphorylation of ATM, p53, SMC1, and Nbs1 at early time points after IR, consistent with normal ATM activity. In contrast, CLLs 199 and 75, with one ATM allele with a missense mutation ATM and one wild-type ATM allele, showed impaired phosphorylation of ATM targets (most marked for CLL 199) despite high ATM protein levels, consistent with a potential dominant-negative effect. CLL 15, with loss of one ATM allele and a mutation in the remaining ATM allele, failed to phosphorylate any of the target proteins consistent with absent ATM protein. (D) CLL 34 had a normal ATM-dependent response. CLL 124, with a mutation in both the ATM and TP53 genes, showed absence of phosphorylation of ATM in response to IR, but increased baseline phosphorylation of p53 that increased further during response to IR.
The effects of ATM and TP53 mutations on damage-induced apoptosis measured by cleavage of PARP1. (A) The percentage of the initial PARP1 remaining at 24 hours after IR was plotted for each tumor in each genetic category. There was no activation of apoptosis, as indicated by the 100% remaining uncleaved PARP1. Median values for the percentage of PARP1 remaining at 24 hours are shown for all genetic subgroups and were statistically significantly different (P = .019). (B) In CLLs 122 and 53, with wild-type ATM and TP53 genes, there was cleavage of PARP1 at both 8 and 24 hours after irradiation, reaching a maximum at 24 hours. In CLL 124, with one ATM mutation and one TP53 mutation, PARP1 cleavage was absent even at 24 hours.
The effects of ATM and TP53 mutations on damage-induced apoptosis measured by cleavage of PARP1. (A) The percentage of the initial PARP1 remaining at 24 hours after IR was plotted for each tumor in each genetic category. There was no activation of apoptosis, as indicated by the 100% remaining uncleaved PARP1. Median values for the percentage of PARP1 remaining at 24 hours are shown for all genetic subgroups and were statistically significantly different (P = .019). (B) In CLLs 122 and 53, with wild-type ATM and TP53 genes, there was cleavage of PARP1 at both 8 and 24 hours after irradiation, reaching a maximum at 24 hours. In CLL 124, with one ATM mutation and one TP53 mutation, PARP1 cleavage was absent even at 24 hours.
The effect of ATM mutations on DNA damage–induced apoptosis was verified using annexin/propidium iodide staining and fluorescent activation cell sorting (FACS) analysis in 22 tumors from the cohort. We analyzed 15 ATM wild-type tumors and 7 ATM mutant tumors by this method. Consistent with reduced PARP1 cleavage, ATM mutant tumors showed less efficient apoptosis, and this effect was more prominent at the later time points. At 24, 48, and 72 hours after IR the mean levels of apoptosis among the ATM wild-type tumors were 36%, 53%, and 73% respectively, compared with 22%, 31%, and 39% in the ATM mutant tumors (P = .017, .015, and .04). Mean levels of apoptosis after 24 hours of culture without irradiation were similar in both genetic subgroups with 22% and 19% mean apoptosis among untreated ATM wild-type and mutant CLL tumors, respectively.
Discussion
In this study we showed a prevalence of 12% for ATM mutations in a large cohort of CLL tumors. This frequency is lower than estimated from previous smaller studies where higher proportions of patients had aggressive disease.8,12 Instead, our cohort included a high proportion of CLL tumors with favorable prognostic features and was biased toward a benign phenotype.1,2 We used a DHPLC approach with an estimated 90% sensitivity for mutation detection.14 It is possible that some mutations may not have been detected with our screening method as the large size of the ATM gene renders a comprehensive screen for mutations challenging. P53 acts downstream from ATM in the DNA damage response and here we demonstrate a prevalence of TP53 mutations in CLL of 4%.
Both low ATM protein expression and reduced p53 damage response have been previously shown to be associated with reduced survival in patients with CLL.8,20,21 However, the relationship among ATM mutations, ATM protein expression, and p53 DNA damage response is not straightforward. In some instances, ATM missense mutations may result in normal or negligibly reduced protein levels despite an impairment of ATM protein function.21 Unlike ATM mutations, a clear distinction between damage response–proficient and –deficient tumors is often lacking since damage-induced p53 activation encompasses a range of responses. Here, by analyzing a large cohort of CLL tumors we have been able to show, for the first time, that mutations in the ATM gene also impact on both a patient's overall and treatment-free survival. Furthermore, multivariate analysis confirmed that the effect of ATM mutations on OS was independent of age, stage, and IGVH mutation status and the effect on TFS was independent of age and IGVH status. After controlling for age, stage, and IGVH mutation status, patients with ATM mutations were 3 times more likely to die at any time than patients with wild-type ATM.
ATM mutations might occur prior to clonal development, at the time of clonal transformation, or during disease progression.22-24 The timing is crucial when considering the clinical impact of the mutations. ATM mutations have been previously found to be early events in the pathogenesis of CLL and in several cases had a germ line origin.8,11,12 In the present study, we confirm that the ATM mutations detected were present early in disease progression and usually at diagnosis. This suggests that detecting patients with ATM mutations would have prognostic value. However, from a single case, we also have evidence that an ATM mutation can occur after the presentation of CLL and during subsequent clonal progression. In this patient, although one ATM mutation was present at diagnosis, a second ATM mutation developed following treatment with chlorambucil. A reduction in ATM activity consequent of the first mutation may have contributed to the development of the CLL clone, and loss of the second functional ATM allele, leading to a further reduction in ATM activity, may have provided an additional cellular survival advantage associated with resistance to DNA-damaging drugs.6,7,25
ATM mutations were detected in 18 tumors, including the 2 tumors that also had a mutation in the TP53 gene, of which 11 carried heterozygous mutations and 7 had a loss of both functional ATM alleles. The pathogenicity of the observed mutations in this study was validated by assessment of ATM kinase activity and by DNA damage–induced apoptosis. ATM is likely to act as a classic tumor suppressor gene when inactivation in both alleles is present and, indeed, we detected a reduction in the phosphorylation of ATM targets in all tumors with loss of both functional ATM alleles. Interestingly, reduced ATM activity was seen in 7 of 9 cases analyzed with one mutant ATM allele and one wild-type allele. This suggested the possibility of a dominant-negative effect in some of the tumors with a single ATM mutation. The analysis of additional ATM phosphorylation targets confirmed deficient ATM activity in 2 tumors with missense ATM mutations compared with a case with loss of a single ATM allele via an 11q deletion. Such a dominant-negative effect has been previously reported with specific ATM missense mutations and is in keeping with the model of ATM dimerization in the resting cell.18,26 It is, however, also possible that a second ATM mutation was present but not detected by the DHPLC analysis. Taken together, the impairment of ATM activity in the majority of ATM mutant tumors was consistent with the poorer prognosis of patients with ATM mutations, which included patients with either one or both ATM alleles affected.
Two tumors had heterozygous mutations in both the ATM and TP53 genes. This combination has not been previously reported in CLL but has been reported in diffuse large B-cell lymphomas and in breast cancer.27,28 ATM acts upstream of p53 in the apoptotic pathway induced by DNA damage but also activates several p53-independent cellular pathways, and it is conceivable that a mutation in both genes could increase the survival advantage of a malignant clone.10 Indeed, murine studies demonstrate that ATM and p53 are not congruent but can cooperate in their tumor suppressive functions.29
In this study we found only a partial correlation between tumors carrying ATM mutations and those with 11q deletions. Less than one third of ATM mutant CLL tumors had a gross 11q deletion and, consistent with previous reports, we also identified CLL tumors with an 11q deletion but no ATM mutation.3 We were able to show that, in contrast to some tumors with a one mutant and one wild-type ATM allele, all the tumors tested with loss of one allele, through an 11q deletion, and a second wild-type ATM allele had normal phosphorylation of ATM-dependent targets. Therefore, it is unlikely that these tumors had a mutation in the remaining ATM allele that escaped our detection. Our data imply that in some tumors with an 11q deletion the poor outcome may be related to loss of ATM activity due to a mutation in the remaining ATM allele and hence an impaired response to DNA damage. In other 11q deleted cases, however, we show that ATM is functional, suggesting that loss of ATM is not contributing to their poor outcome and that other unknown factors may be at play.
Previous studies have indicated that tumors with impaired DNA damage responses including those with ATM mutations invariably have unmutated IGVH genes.8,30 Using the widely accepted 2% nucleotide variation from germ line sequence to classify IGVH genes as unmutated or mutated, in this study we found that ATM mutations can occur in both CLL subgroups but are statistically associated with tumors with IGVH unmutated genes.1,2 The differences between the current and the previous studies are likely to reflect the nature of the studied cohorts. One significant outcome of this study is that it further stratifies tumors with IGVH mutated genes. Compared with ATM mutant CLLs, tumors with TP53 mutations were more frequently associated with mutated IGVH genes. The differences between the cellular phenotypes of ATM and TP53 mutant tumors might be related to the timing of acquisitions of these mutations. For example, since ATM has a role in DNA repair and isotype class switch recombination,31 ATM mutations might contribute to CLL clonal transformation. A mutation in the TP53 gene, in contrast, might be a later event in CLL progression. However, in view of the low numbers, it is difficult to ascertain the true relationship between TP53 mutations and IGVH status.
In summary, we have been able to demonstrate that a single genetic event in the DNA damage response pathway, an ATM mutation, has a significant impact on the survival of patients with CLL. ATM mutations provided prognostic information that was independent of age, stage, and IGVH mutation status and identified a subset of tumors distinct from those with chromosomal losses. Furthermore, we validated the pathogenic effects of the ATM mutations, and showed that tumors with ATM mutations could be identified by their defective DNA damage–induced phosphorylation responses.
We believe that our findings have important clinical implications. ATM mutations may be important in predicting potential treatment failures, since chemotherapeutic drugs such as chlorambucil and fludarabine act via induction of DNA damage and apoptosis in target cells. Furthermore, drug treatment may potentially lead to the selection of tumor subclones that carry genetic mutations, rendering them resistant to apoptosis. Early treatment of CLL prior to disease progression has previously been shown to impair patient survival, although the mechanism underlying this has never been satisfactorily explained.32 Two thirds of the treated patients with ATM mutations in this study were clinically refractory to chemotherapy and in vitro resistance to fludarabine also has been shown previously in tumors with ATM mutations.30 Therefore, it is likely that patients with either mutations in ATM or TP53 will benefit from therapies that induce tumor killing by mechanisms independent of activation of the p53 pathway.33-37 Ideally, our findings should be consolidated in a prospective manner, preferably as part of a clinical trial, where treatment failures could be correlated with ATM mutation status.
Prepublished online as Blood First Edition Paper, July 12, 2005; DOI 10.1182/blood-2004-11-4516.
Supported by the Leukaemia Research Fund, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal