Abstract
Regulators of G-protein signaling (RGS) constitute a family of proteins involved in the negative regulation of signaling through heterotrimeric G protein–coupled receptors (GPCRs). Several RGS proteins have been implicated in the down-regulation of chemokine signaling in hematopoietic cells. The chemokine stromal-cell–derived factor 1 (SDF-1) activates migration of hematopoietic progenitors cells but fails to activate mature megakaryocytes despite high levels of CXC chemokine receptor 4 (CXCR4) receptor expression in these cells. This prompted us to analyze RGS expression and function during megakaryocyte differentiation. We found that RGS16 and RGS18 mRNA expression was up-regulated during this process. Overexpressing RGS16 mRNA in the megakaryocytic MO7e cell line inhibited SDF-1–induced migration, mitogen-activated protein kinase (MAPK) and protein kinase B (AKT) activation, whereas RGS18 overexpression had no effect on CXCR4 signaling. Knocking down RGS16 mRNA via lentiviral-mediated RNA interference increased CXCR4 signaling in MO7e cells and in primary megakaryocytes. Thus, our data reveal that RGS16 is a negative regulator of CXCR4 signaling in megakaryocytes. We postulate that RGS16 regulation is a mechanism that controls megakaryocyte maturation by regulating signals from the microenvironment.
Introduction
Chemokines and their receptor(s) are broadly expressed in different tissues and regulate cell migration as well as several other important biologic processes.1 The chemokine stromal-cell–derived factor 1 (SDF-1) is a stromal-cell–derived factor that interacts with a specific receptor CXC chemokine receptor 4 (CXCR4) and plays a role in B lymphopoiesis and bone marrow myelopoiesis. Studies using mutant mice with targeted gene disruption have revealed that SDF-1 and CXCR4 are essential for B-cell differentiation, for colonization of bone marrow by hematopoietic stem cells (HSCs) and myeloid lineage during ontogeny as well as for blood-vessel formation in gastrointestinal tract, cardiac ventricular septum formation, and cerebellar differentiation.2,3 SDF-1CXCR4 signaling appears to be essential for the homing of hematopoietic stem/progenitor cells because treatment of immature human hematopoietic progenitor cells with anti-CXCR4 antibodies prevents their short-term engraftment into nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice.4-6 In addition, transplantation of CXCR4–/– fetal liver cells results in low numbers of B-lymphoid and myeloid lineage precursors in bone marrow but increased numbers in the peripheral blood compared with control animals.7 Proteolytic cleavage of the N-terminus of CXCR4 and of SDF-1 is one mechanism that has been identified for the regulation of CXCR4/SDF1 signaling in circulating and mobilized blood cells.8
The process of megakaryopoiesis occurs within a complex bone marrow microenvironment where chemokines, cytokines, and adhesive interactions play a major role. At the end of their maturation, polyploid megakaryocytes (MKs) migrate through bone marrow endothelial cells and release platelets directly into the marrow-intravascular sinusoidal space or the lung capillaries. The interactions of immature MKs with a permissive, endothelial-enriched microenvironment is promoted by chemokines, including SDF-1.9 Consistent with this process, CXCR4 is expressed all along the megakaryocyte differentiation from MK progenitors to platelets.10-14 However, during MK development, the outcome of CXCR4 signaling as measured by chemotaxis and extracellular signal regulated protein kinase (Erk) activation becomes markedly reduced, indicating the presence of possible negative regulators. Such mechanisms have remained elusive. Recent studies have shown that CXCR4 signaling is regulated by Regulators of G-protein signaling (RGS) proteins in mature B cells.15,16 Reminiscent of the MK differentiation, CXCR4 expression is maintained all along the B-cell maturation, but mature B cells fail to respond to SDF-1.17
RGS proteins have emerged as major modulators of signaling for heterotrimeric guanine nucleotide binding proteins (G proteins). Heterotrimeric G proteins are the link between many receptors, including chemokines, and downstream effectors. Heterotrimeric G proteins are composed of α, β, and γ subunits, each having multiple isoforms; that is, 4 isoforms for the α subunit: αi, αs, αq, and α12. Ligand-bound activated receptors catalyze the exchange of guanosine diphosphate (GDP) by guanosine triphosphate (GTP) on the α subunit, leading to dissociation of the α form from the βγ dimer, which both transduce the signal to effectors. RGS proteins function as guanosine triphosphatase-activating proteins (GAPs) for Gα subunits, accelerating the inactivation rate of Gα-GTP.18-20 So far, RGS proteins that have been identified interact with and activate members of the Gαi family, Gαq, and Gα12/13 but not Gαs.21 In addition to their GAP activity, RGS may also block signaling by acting as effector antagonists.22,23 Overexpression of RGS1, RGS3, and RGS4 in a pre–B-lymphoma cell line has been shown to reduce interleukin 8 (IL-8; CXCR1) and monocyte chemoattractant protein-1 (MCP-1; CCR2) induced activation.24 RGS16 inhibits IL-8 and RANTES (regulated on activation normal T cell expressed and secreted) (CCR5)–mediated signals in lymphocytes.25 Leukotriene B4, C5a, and fMLP (formyl-Met-Leu-Phe) responses are abolished by the introduction of RGS1 in the monocytic cell line THP1.26 RGS1 has been shown to be involved in the down-regulation of chemotaxis mediated by lymphoid chemokines, including SDF-1.27,28
Megakaryocytes express RGS16 and RGS18 proteins,29,30 but the pattern of expression during differentiation has not been examined in these studies. We herein describe an up-regulation of RGS16 and RGS18 expression during MK differentiation. Overexpression of RGS16 but not of RGS18 markedly diminished SDF-1–mediated migration, protein kinase B (AKT), and Erk activation of MO7e cells. Using RNA interference, we inhibited RGS16 in MO7e cells and in CD34+-derived MKs, leading to the increase of CXCR4 signaling. The RGS16 knocking down did not modify platelet formation and adhesion of MKs to either fibronectin or collagen I. Altogether, results suggest that RGS16 is a negative regulator of SDF-1/CXCR4 signaling in megakaryocytes.
Materials and methods
Cell culture
MO7e cells were cultured in α-minimal essential medium (αMEM) supplemented with 10% fetal calf serum, penicillin/streptomycin/glutamine (Invitrogen, Cergy-Pontoise, France) and with 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF; Novartis, Basel, Switzerland). 293 EBNA cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with fetal calf serum (FCS) and antibiotics.
Cord-blood samples from healthy full-term newborn infants were obtained from a cord-blood bank (Clinique Les Noriets, Vitry-sur-Seine, France) in agreement with national ethics guidelines. Low-density mononuclear cells (LDMCs) were prepared by centrifugation on Lymphoprep (Nyegaard, Oslo, Norway). CD34+ cells were separated using a magnetic cell-sorting system (miniMACS; Miltenyi Biotech GmbH, Bergisch Gladbach, Germany) according to the manufacturer's recommendations. The purity of the recovered CD34+ cells was greater than 90%. CD34+ cells were cultured in Iscoves modified Dulbecco medium (IMDM) with penicillin/streptomycin/glutamine and 11.5 mM α-thioglycerol (Sigma, Saint Quentin Fallavier, France) supplemented with 1.5% bovine serum albumin (BSA; Cohn fraction V; Sigma), sonicated lipids, and iron-saturated human transferrin (Invitrogen) and were stimulated with the combination of recombinant human thrombopoietin (TPO; 10 ng/mL) and stem-cell factor (SCF; 50 ng/mL) (generous gifts of Kerin Brewery, Tokyo, Japan and Amgen, Thousand Oaks, CA, respectively) (MK medium) as previously described.31 Cells were stained with anti-CD34–fluorescein isothiocyanate (FITC), anti-CD41–allophycocyanin (APC), and anti-CD42–phycoerythrin (PE) antibodies (BD Biosciences, le Pont de Claix, France). Different populations (CD34+CD41–CD42–, CD41+CD42low, and CD41+CD42+) were sorted on a fluorescence-activated cell sorting (FACS) DIVA cytometer (Becton Dickinson, Rungis, France).
Plasmid constructions
The complete coding region of RGS16 (1.3 kilobase [kb]) was amplified by polymerase chain reaction (PCR) from DAUDI cDNA using 5′ and 3′ primers each containing a KpnI site (underlined), as follows: sense, 5′-ACGGGTACCATGTGCCGCACCCTGGCCGC-3′, and antisense, 5′-TCCCCATGGTCAGGTGTGTGAGGGCTCGTC-3′. The amplified cDNA was subcloned into the PCR-TOPOII (topoisomerase II) expression vector (Invitrogen) and sequenced. RGS16 cDNA was then inserted at the C-terminal end of the enhanced green fluorescent protein (eGFP) into a retroviral vector pSF1N (kindly provided by Dr Ostertag, Germany). The RGS18 cDNA (Guthrie cDNA Resource Center, Sayre, PA) was inserted into the retroviral vector Migr-IRES-GFP.
To inhibit RGS16 expression, we designed small interfering RNA (siRNA) using Ambion software (Ambion, Huntingdon, United Kingdom). From these sequences, small hairpins were created as previously described.32,33 Oligonucleotides short hairpin (sh) RGS16 listed in Table 1 were synthesized (Invitrogen) and inserted into a pBlue Script containing the human H1 promoter. The H1-shRGS16 or H1-SCR (scramble sequence) unit was inserted into a HIV-derived lentiviral vector (pRRL sin PGK eGFP WPRE; Généthon, Evry, France).
Oligonucleotide short hairpin sequences (SEQs) used for RGS16 inhibition
. | shRGS16 . |
|---|---|
| SEQ2 | GACACGTCTGGGGATCTTTttcaagagaAAAGATCCCCAGACGTGTC |
| SEQ15 | GATCCGATCAGCTACCAAGttcaagagaCTTGGTAGCTGATCGGATC |
| SEQ24 | GAGAGGTTGAGTCACCCATttcaagagaATGGGTGACTCAACCTCTC |
| SEQ43 | GGGCTAATGATGAGGGTTGttcaagagaCAACCCTCATCATTAGCCC |
| SEQ68 | ATAAGTCTCTTGGCGGTCCttcaagagaGGACCGCCAAGAGACTTAT |
| SEQ73 | AGAGATGCCCCGAGATAGAttcaagagaTCTATCTCGGGGCATCTCT |
. | shRGS16 . |
|---|---|
| SEQ2 | GACACGTCTGGGGATCTTTttcaagagaAAAGATCCCCAGACGTGTC |
| SEQ15 | GATCCGATCAGCTACCAAGttcaagagaCTTGGTAGCTGATCGGATC |
| SEQ24 | GAGAGGTTGAGTCACCCATttcaagagaATGGGTGACTCAACCTCTC |
| SEQ43 | GGGCTAATGATGAGGGTTGttcaagagaCAACCCTCATCATTAGCCC |
| SEQ68 | ATAAGTCTCTTGGCGGTCCttcaagagaGGACCGCCAAGAGACTTAT |
| SEQ73 | AGAGATGCCCCGAGATAGAttcaagagaTCTATCTCGGGGCATCTCT |
Viral infectious particles production
The retrovirus-producing cell line 293 expressing Epstein-Barr virus nuclear antigen-1 (EBNA) was maintained in DMEM with 10% FCS and 250 μg/mL G418 (Invitrogen). Vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped retroviruses were produced by transient transfection of 293 EBNA with 3 different constructs. The expression plasmids, pCMV-G, which contained the VSV-G coding sequence, and pCMV-gag-pol, which contained the Gag-Pol coding sequence, were kindly supplied by Dr J. Morgenstern (Millenium, Boston, MA). The third vector was the retroviral vector pSF1N, containing individual GFP-RGS coding sequence or Migr for RGS18. Transfection was performed using the Exgen reagent (Euromedex, Mundolsheim, France) according to the manufacturer's protocol except that 500 ng of each plasmid and 7.5 μL Exgen were used for each 36-mm well. Supernatant containing infectious retroviral particles was recovered after 2 and 3 days of culture. Viral titers were determined by limiting dilution assay with NIH 3T3 cells, on the basis of GFP fluorescence.
Lentiviral stocks were prepared by transient cotransfection in human 293T cells of 3 plasmids (pRRL with SCR or shRGS16 sequence, the packaging plasmid pCMVΔR8,74, and the VSV-G protein envelope plasmid pMD.G). Lipofectamine 2000 (Invitrogen) was used for transfection according to the manufacturer's instructions. The concentration of viral particles was defined by limiting dilution assay with NIH 3T3 cells as for retroviruses.
Test of pBS-H1-shRGS16 on EBNA cells
EBNA cells were transfected using the Exgen technique as just described with 2 plasmids pBS-H1-shRGS16 and MIGR-IRES-eGFP as a marker of transfection efficiency. Cells containing plasmid with different sequences of shRGS16 were analyzed 48 hour after transfection and sorted on the eGFP level using a FACS DIVA flow cytometer (Becton Dickinson).
Cell infection
MO7e cells were infected with supernatant, using a multiplicity of infection (MOI) of 10 viruses per cell, in the presence of 4 μg/mL hexadimethrine bromide (Sigma). Cells were cultured for 24 hours and then reinfected in the same conditions. GFP-expressing cells were sorted using FACSVantage (Becton Dickinson) and maintained in culture in αMEM medium containing GM-CSF.
MKs were infected after 2 days of stimulation of CD34+ cells with TPO (10 ng/mL) and SCF (50 ng/mL) in MK medium. Cells were infected twice with a multiplicity of infection of 50, in the presence of 4 μg/mL hexadimethrine bromide (Sigma). Cells were stained with an anti-CD41–APC antibody and sorted to obtain CD41+GFP+ and CD41+GFP– populations for mRNA analysis and proplatelet formation. Cells were stained with an anti-CD34–PE antibody and sorted to obtain CD34+GFP+ populations for MK colony-forming unit (CFU-MK) assay.
mRNA analysis using real-time RT-PCR
Total RNA was isolated from sorted megakaryocytes, platelets, MO7e cells, and EBNA cells using the SV total RNA isolation kit (Promega, Charbonnières, France). cDNA was generated by reverse transcription using Superscript II RNase H-reverse transcriptase (Invitrogen). The expression levels of RGS and the internal reference β2-microglobulin (β2m) were measured by triplicate PCR which were performed using the ABI PRISM GeneAmp 5700 sequence Detection system and the TaqMan Universal PCR master Mix (PE Applied Biosystems, Courtaboeuf, France). The primers and probes were designed using Primer Express software and are listed in Table 2. A comparative threshold cycle (CT) method was used to determine gene expression. Indeed, the RGS CT value was normalized to the β2m CT value using the formula: 2–(CTRGS – CTβ2m). RNA samples were not used that exhibited fluorescein amidite (FAM)–labeled reverse transcription-quantitative polymerase chain reaction (RT-QPCR) products with CT values greater than 37. This is because inaccurate and unreliable relative expression values might be obtained. All samples demonstrated CT values less than 27 for FAM-labeled RT-QPCR products, indicating the samples contained undegraded cDNA that could be amplified.
Primers and probes designed with Primer Express
Human gene . | Sequence (5′→3′) . |
|---|---|
| RGS16 | |
| Forward | TCCAGGGCACACCAGATCTT |
| Reverse | TGGCAGTCTGCAGGTTCATC |
| FAM probe | CAGTGAGGCCCCTAAAGAGGTCAACATTG |
| RGS18 | |
| Forward | TGAATTCAGTGAGGAGAACATTGAAT |
| Reverse | TTTCTCATAGATTGCCTTTGCTTTTA |
| FAM probe | CTTCAAGAAATGCAAGGAACCTCAACAAATCAT |
| β2-Microglobulin | |
| Forward | GCGGCATCTTCAAACCTCC |
| Reverse | TGACTTTGTCACAGCCCAAGATA |
| FAM probe | TGATGCTGCTTACATGTCTCGATCCCACTT |
Human gene . | Sequence (5′→3′) . |
|---|---|
| RGS16 | |
| Forward | TCCAGGGCACACCAGATCTT |
| Reverse | TGGCAGTCTGCAGGTTCATC |
| FAM probe | CAGTGAGGCCCCTAAAGAGGTCAACATTG |
| RGS18 | |
| Forward | TGAATTCAGTGAGGAGAACATTGAAT |
| Reverse | TTTCTCATAGATTGCCTTTGCTTTTA |
| FAM probe | CTTCAAGAAATGCAAGGAACCTCAACAAATCAT |
| β2-Microglobulin | |
| Forward | GCGGCATCTTCAAACCTCC |
| Reverse | TGACTTTGTCACAGCCCAAGATA |
| FAM probe | TGATGCTGCTTACATGTCTCGATCCCACTT |
Chemotactic assay
Cell migration was quantified through 5-μm pore filters for the MO7e cell line and 8-μm pore filters for MKs (Transwell, 24-well cell clusters; VWR, Strasbourg, France). Serum-free medium (200 μL) containing 2 × 105 cells was placed in the upper well of the transwell. In the lower chamber, 600 μL medium containing 300 ng/mL SDF-1α (Abcys, Paris, France) was added. After 4 hours at 37°C in 5% CO2, the cells of the lower chamber and the starting population were recovered in equal volumes, and the percentage of migration was calculated according to the following formula: number of cells in the lower well divided by the total cells put in the transwell at the beginning. The number of cells and their GFP levels were analyzed on a FACSsort (Becton Dickinson). For megakaryocytes, cells in the lower chamber and the total cells were stained using anti-CD41–APC and anti-CD42–PE antibodies after migration and analyzed on a FACSsort.
Analysis of p42/44MAP kinase and AKT activation by Western blot
After 12 hours of serum and GM-CSF starvation, stably infected MO7e cells (RGS16, RGS18 or GFP alone) were stimulated or not with 300 ng/mL SDF-1 for 1 and 5 minutes. Equal amounts of proteins (30 μg) were transferred onto nitrocellulose. Endogenous mitogen-activated protein (MAP) kinase activity was detected with an antibody specific for the p42/44 phosphorylated forms of MAP kinase (Cell Signaling, Beverly MA). AKT activity was detected by an antibody against phospho-AKT (Cell Signaling). Goat anti–rabbit horseradish peroxidase (HRP)–conjugated antibodies (Santa Cruz Biotechnologies, Santa Cruz, CA) were used to detect bound primary antibodies by enhanced chemiluminescence. The membranes were reprobed with an antibody directed against total p42/44 MAP kinase or totalAKT (Cell Signaling) to check that protein amounts were present in each sample.
Cell adhesion
Fibronectin (50 μg/mL; Sigma) and collagen type I (100 μg/mL) were incubated in 48-well plates for 2 hours at room temperature. After washing, plates were blocked with BSA for 1 hour at room temperature. pRRL-SCR or pRRL-24 CD41+/GFP+ sorted cells were then plated (5000 cells/well in 200 μL MK medium) for 15 minutes at 37°C, and SDF-1 (300 ng/mL) was added or not for another 15 minutes at 37°C. Wells were emptied and washed.Adherent cells were then fixed in 2% paraformaldehyde and counted under an inverted microscope. The percentage of cell adhesion corresponds to the number of adherent cells versus the number of total cells plated (5000 cells/well).
CFU-MK assay
pRRL-SCR or pRRL-24 CD34+/GFP+ sorted cells were performed by the plasma clot technique, as previously described.34 Briefly, 2000 cells/dish were plated in MK medium supplemented with bovine plasma fibrinogen (1 mg/mL; Sigma), ϵ amino caproic acid (0.01 mol/L; Sigma), thrombin (6 mU/mL), TPO (10 ng/mL), and SCF (50 ng/mL). Petri dishes were cultured at 37°C for 12 days. Colonies were quantified by an indirect immunophosphatase alkaline labeling technique using an anti–glycoprotein III (GpIIIa) polyclonal antibody (Dako, Glostrup, Denmark). CFU-MK were counted under an inverted microscope.
Quantification of proplatelet formation
pRRL-SCR or pRRL-24 CD41+/GFP+ sorted cells were cultured in 96-well plates at the concentration of 200 cells/well in 100 μL MK medium plus TPO and SCF for 8 days at 37°C. Proplatelets were defined as cells exhibiting one or more cytoplasmic processes with constriction areas. MK and proplatelets were counted under an inverted microscope.
Statistical analysis
Results of experimental points obtained from 3 repeated experiments are reported as the mean plus or minus standard deviation (SD). Statistical significance was determined using Student t test.
Results
RGS16 and RGS18 mRNA expression is enhanced during megakaryocyte maturation
It has been previously reported that RGS16 and RGS18 mRNA are expressed by megakaryocytes.29,30,35 This prompted us to analyze whether these RGSs were regulated during the process of MK differentiation which is known to be associated with reduced SDF-1 response. MKs were derived from culture of cord-blood CD34+ cells, and cells at various stages of development were obtained from these cultures using flow cytometric cell sorting on the basis of the CD34, CD41, and CD42 markers (Figure 1A). CD34+ cells constitute progenitor cells, whereas CD41+CD42low cells are enriched in immature MKs and CD41+CD42+ cells in mature megakaryocytes. Total mRNA was isolated from each of these 3 populations, and the level of RGS16 and RGS18 was analyzed by quantitative RT-PCR. To control for differences in mRNA loading, the intensity of RGS16 and RGS18 signal was normalized to the housekeeping β2-microglobulin gene (Figure 1B). RGS18 transcripts were detected in all 3 populations. RGS18 expression increased 13-fold from immature progenitors (CD34+CD41–CD42–) to the more mature cells (CD41+CD42+). The amounts of RGS16 transcripts were markedly lower than RGS18 transcripts in all 3 populations. Similarly to RGS18, RGS16 mRNA expression increased 6-fold from immature progenitors (CD34+CD41–CD42–) to the more mature cells (CD41+CD42+). In contrast to RGS16, RGS18 mRNA expression remains high in platelets (Figure 1C). These results indicate that both RGS16 and RGS18 mRNA are up-regulated during MK differentiation.
Expression of RGS16 and RGS18 during MK differentiation. Cord-blood CD34+ cells were cultured in MK medium with TPO and SCF. (A) Flow cytometry analysis of MK culture. Different fractions were sorted using the CD34, CD41, and CD42 markers. Cells were acquired in a morphologic gate which excluded dying cells. CD34+CD41–CD42– subsets were sorted after staining cells with an anti-CD34–FITC antibody. Numbers in the quadrants represent the percentage of each population in the culture. (B) Level of mRNA expression in these fractions. □ indicates the immature CD34+-cell fraction (CD34+, CD41–, CD42–); ▦, the immature MKs (CD41+, CD42low); ▪, the mature fraction (CD41+, CD42+). *P < .01. Data are mean ± SD of 3 independent experiments performed in triplicate. (C) Level of RGS16 and RGS18 mRNA expression in platelets. mRNA was extracted from these cells, and quantitative RT-PCR with specific primers for RGS was performed as described in “Materials and methods.” β2-Microglobulin amplification was used to confirm that the samples contained similar amounts of cDNAs. Data represent the mean ± SD of 3 independent experiments.
Expression of RGS16 and RGS18 during MK differentiation. Cord-blood CD34+ cells were cultured in MK medium with TPO and SCF. (A) Flow cytometry analysis of MK culture. Different fractions were sorted using the CD34, CD41, and CD42 markers. Cells were acquired in a morphologic gate which excluded dying cells. CD34+CD41–CD42– subsets were sorted after staining cells with an anti-CD34–FITC antibody. Numbers in the quadrants represent the percentage of each population in the culture. (B) Level of mRNA expression in these fractions. □ indicates the immature CD34+-cell fraction (CD34+, CD41–, CD42–); ▦, the immature MKs (CD41+, CD42low); ▪, the mature fraction (CD41+, CD42+). *P < .01. Data are mean ± SD of 3 independent experiments performed in triplicate. (C) Level of RGS16 and RGS18 mRNA expression in platelets. mRNA was extracted from these cells, and quantitative RT-PCR with specific primers for RGS was performed as described in “Materials and methods.” β2-Microglobulin amplification was used to confirm that the samples contained similar amounts of cDNAs. Data represent the mean ± SD of 3 independent experiments.
RGS16 but not RGS18 inhibits SDF-1–induced chemotaxis, AKT and Erk activation in the MO7e cell line
RGS proteins are involved in the negative regulation of G protein–coupled receptors (GPCRs) and have been shown to modulate CXCR4 signaling.36-38 To probe the regulatory activity of RGS16 and RGS18 on CXCR4 signaling, we constructed a retroviral vector encoding an RGS16-GFP fusion protein according to previous studies which showed that RGS1, RGS3, and RGS4 fusion proteins have similar activities as the wild-type RGS proteins.24 This construct was inserted into the pSF1N retrovirus and the RGS18 cDNA into the MIGR retrovirus, which gave both efficient transduction and high transgene expression in hematopoietic cells. Because of both the MK phenotype and efficient migratory response to SDF-1, the MO7e cell line was chosen for further study. Infection rates were evaluated to be about 80% as assessed by GFP expression in populations of stably transduced cells. GFP+ polyclonal populations (MO7e GFP, MO7e RGS16, and MO7e RGS18) were isolated by flow cytometry, and levels of expression of the various RGS constructs were evaluated by RT-PCR. As shown in Figure 2A, RGS16 mRNA level in MO7e RGS16 cells was 200-fold higher than RGS16 mRNA level in MO7e GFP cells. Similarly, RGS18 mRNA was 150-fold higher than endogenous RGS18 mRNA level obtained in MO7e GFP cells (Figure 2B). In MO7e RGS16 cells, overexpression of RGS16 did not alter the expression of RGS18 and vice versa.
Then we studied the effects of RGS16 and RGS18 overexpressions on SDF-1–induced chemotaxis in MO7e cells. RGS18 overexpression did not affect SDF-1–induced chemotaxis (Figure 2C). In contrast, RGS16 overexpression in MO7e cells was very effective in inhibiting SDF-1–mediated migration (75% inhibition compared with control cells transduced with GFP only). We subsequently tested the effect of these RGS overexpressions on the SDF-1–induced MAPK and AKT activation by Western blot. As shown in Figure 2D, MO7e GFP cells exhibited a strong Erk and AKT phosphorylation in response to SDF-1. Activation of CXCR4 by SDF-1 also induced a significant Erk and AKT activation in MO7e RGS18 cells. In contrast, MO7e RGS16 exhibited a lower Erk and AKT phosphorylation in response to SDF-1 as compared with MO7e GFP and MO7e RGS18. Thus, RGS16 protein overexpression led to the inhibition of SDF-1–mediated migration and intracellular signaling.
Inhibition of RGS16 expression using lentiviral-delivered short hairpin RNA
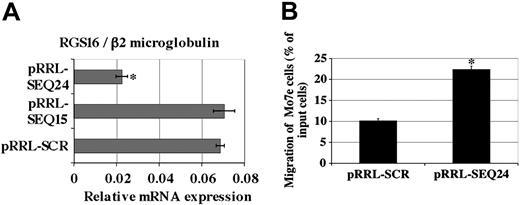
Results on the MO7e cell line prompted us to check whether RGS16 was implicated in SDF-1/CXCR4 signaling in primary megakaryocytes, using RNA interference. siRNA sequences specific for RGS16 mRNA were designed using the Ambion software available on the Internet and constructed as shRNA composed by forward and reverse sequences coding for siRNA linked by a short hairpin as described.33 The different cDNAshRNAs (shRGS16) were inserted into a pBlueScript vector containing the human H1 polymerase III promoter (Figure 3A). These different expression vectors were transfected into EBNA cells to assess endogenous RGS16 mRNA inhibition. EBNA cells were cotransfected with MIGR, a retrovirus vector containing the eGFP, which was used as a marker of transfection efficiency. GFP+ EBNA cells were sorted and analyzed by quantitative RT-PCR to assay the mRNA level of RGS16. Five of 6 shRGS16 sequences inhibited RGS16 mRNA levels by 2-fold or more (Figure 3 B). SEQ24 was the most effective (a 3-fold decrease) and was chosen, as well as the SEQ15 or a scramble control irrelevant siRNA, to construct lentiviral vectors to stably express anti-RGS16 siRNA into megakaryocytes.
Overexpression of RGS16 and RGS18 in MO7e cell line. MO7e cells were infected with retroviruses containing RGS16 or RGS18. These and the control cells, which contain only the GFP, were sorted and analyzed. (A) Level of overexpression of RGS16 in MO7e RGS16 cells. Quantitative RT-PCR was performed, and RGS mRNA level was compared with the β2m mRNA level. This histogram represents the mRNA quantity of exogenous RGS in MO7e RGS16 and in MO7e RGS18. (B) Level of overexpression of RGS18 in MO7e RGS18 cell line. Quantitative RT-PCR was performed, and RGS mRNA level was compared with the β2m mRNA level. This histogram represents the mRNA quantity of RGS in MO7e RGS16 and MO7e RGS18 cells. (C) SDF-1–induced migration of MO7e GFP, MO7e RGS16, and MO7e RGS18 cells. Data represent the percentage of migrated cells compared with total input cells. The graph shows the mean ± SD of 4 independent experiments performed in duplicate. *P < .01. (D) Effect of RGS protein overexpression on the Erk and AKT activation pathway. MO7e GFP, MO7e RGS16, and MO7e RGS18 cell lines were starved of serum and GM-CSF for 12 hours and were stimulated or not with 300 ng/mL SDF-1 for the times indicated. (D) Erk and AKT activation was analyzed by Western blot analysis using an anti–phospho-Erk (P-Erk) antibody and an anti–phospho-AKT (P-AKT) antibody. Protein loading was assayed by labeling with an anti-Erk antibody (Total Erk) and an anti-AKT antibody (Total AKT).
Overexpression of RGS16 and RGS18 in MO7e cell line. MO7e cells were infected with retroviruses containing RGS16 or RGS18. These and the control cells, which contain only the GFP, were sorted and analyzed. (A) Level of overexpression of RGS16 in MO7e RGS16 cells. Quantitative RT-PCR was performed, and RGS mRNA level was compared with the β2m mRNA level. This histogram represents the mRNA quantity of exogenous RGS in MO7e RGS16 and in MO7e RGS18. (B) Level of overexpression of RGS18 in MO7e RGS18 cell line. Quantitative RT-PCR was performed, and RGS mRNA level was compared with the β2m mRNA level. This histogram represents the mRNA quantity of RGS in MO7e RGS16 and MO7e RGS18 cells. (C) SDF-1–induced migration of MO7e GFP, MO7e RGS16, and MO7e RGS18 cells. Data represent the percentage of migrated cells compared with total input cells. The graph shows the mean ± SD of 4 independent experiments performed in duplicate. *P < .01. (D) Effect of RGS protein overexpression on the Erk and AKT activation pathway. MO7e GFP, MO7e RGS16, and MO7e RGS18 cell lines were starved of serum and GM-CSF for 12 hours and were stimulated or not with 300 ng/mL SDF-1 for the times indicated. (D) Erk and AKT activation was analyzed by Western blot analysis using an anti–phospho-Erk (P-Erk) antibody and an anti–phospho-AKT (P-AKT) antibody. Protein loading was assayed by labeling with an anti-Erk antibody (Total Erk) and an anti-AKT antibody (Total AKT).
Inhibition of RGS16 expression. (A) Construction of an expression plasmid bearing shRNA and a lentiviral expression vector coding for shRGS16. (B) Test of different sequences of shRGS16 in 293 EBNA cells. 293 EBNA cells were cotransfected with pBS-H1 bearing different sequences of shRGS16 and MIGR as a marker of transfection efficiency. EBNA cells were sorted as shown on the GFP level. GFP+ cells were analyzed for RGS16 expression by quantitative RT-PCR using β2m as reference. The graph shows the mean ± SD of 4 independent experiments.
Inhibition of RGS16 expression. (A) Construction of an expression plasmid bearing shRNA and a lentiviral expression vector coding for shRGS16. (B) Test of different sequences of shRGS16 in 293 EBNA cells. 293 EBNA cells were cotransfected with pBS-H1 bearing different sequences of shRGS16 and MIGR as a marker of transfection efficiency. EBNA cells were sorted as shown on the GFP level. GFP+ cells were analyzed for RGS16 expression by quantitative RT-PCR using β2m as reference. The graph shows the mean ± SD of 4 independent experiments.
Inhibition of RGS16 expression in megakaryocytes up-regulates SDF-1/CXCR4 signaling
MO7e cells were infected with different lentiviral vectors encoding shRNASEQ15, shRNASEQ24, or SCR control shRNA, together with a reporter GFP gene. Transduced GFP+ cells were purified and analyzed by quantitative RT-PCR. A 3-fold decrease in the expression of RGS16 mRNA was observed with the pRRL-SEQ24 in comparison to pRRL-SCR, whereas no inhibition was observed with pRRL-SEQ15 (Figure 4 A). The lack of efficacy of the SEQ15 constructed in the lentiviral vector was surprising compared with the inhibition obtained by transfection of SEQ15 plasmid in EBNA cells. This difference may be due to a lower number of SEQ15 siRNA obtained after transduction compared with transfection. Nevertheless, the lentiviral vector encoding SEQ24 was efficient and allowed the study of SDF-1–induced migration in MO7e cells (Figure 4B). Cell migration was increased by 68% in cells with pRRL-SEQ24, confirming that RGS16 modulates the SDF-1/CXCR4 signaling in MO7e cell line.
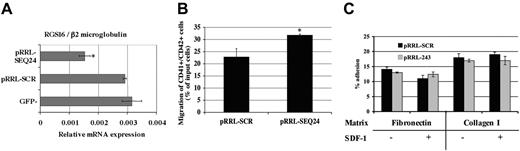
We have shown that RGS16 expression increased during MK differentiation. This correlates with the SDF-1 responsiveness of the mature MKs. Thus, the effect of an inhibition of RGS16 expression on SDF-1–induced migration of primary MKs was evaluated. Cord-blood CD34+ cells were stimulated 2 days with TPO and infected twice with the RGS16 siRNA. After 7 days of culture, pRRL-SCR GFP+/CD41+, pRRL-SEQ24 GFP+/CD41+, and GFP–/CD41+ were obtained by flow cytometry cell sorting. Analysis of RGS16 mRNA expression (Figure 5A) revealed a 2-fold decrease in pRRL-SEQ24 GFP+/CD41+ in comparison to the 2 control populations, which exhibited similar RGS16 mRNA levels. Moreover, the SDF-1–induced MK migration was investigated. Figure 5B illustrates the percentage of CD41+/CD42+ cells that migrated in response to SDF-1. SDF-1–induced migration of MK expressing RGS16 siRNA was increased by 57%, suggesting that RGS16 modulates negatively CXCR4 signaling in primary MKs.
Adhesion assays of pRRL-SCR GFP+/CD41+ and pRRL-SEQ24 GFP+/CD41+ cells on fibronectin and collagen I were performed as described in “Materials and methods.” As shown in Figure 5C, a significant fraction of megakaryocytes were shown to adhere to fibronectin and collagen I. However, we did not find any change in the adhesion to these extracellular matrix after RGS16 knocking down either in the presence or absence of SDF-1, suggesting that the level of RGS16 expression did not modify basal or SDF-1–induced adhesion to either fibronectin or collagen I.
Effects of RGS16 knocking down on megakaryopoiesis and platelet production
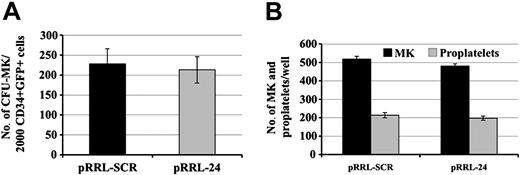
We studied whether inhibition of RGS16 expression affects CFU-MK formation by performing CFU-MK assays. pRRL-SCR GFP+/CD34+, pRRL-SEQ24 GFP+/CD34+ cells were obtained by flow cytometry cell sorting and grown in plasma clot in the presence of TPO and SCF to reveal their MK potential. Results of 3 experiments performed from cord-blood CD34+ cells are summarized in Figure 6A. Cloning efficiency of CFU-MK was quite similar in pRRL-SCR GFP+/CD41+ and pRRL-SEQ24 GFP+/CD41+.
We also analyzed the proplatelet formation from pRRL-SCR GFP+/CD41+ and pRRL-SEQ24 GFP+/CD41+ cells (Figure 6B). For this purpose, sorted cells were stimulated by SCF and TPO for 7 days, and proplatelet production was assessed. These analyses revealed comparable numbers of proplatelets between pRRL-SCR GFP+/CD41+, pRRL-SEQ24 GFP+/CD41+ cells, suggesting that RGS16 knocking down did not modify platelet formation in these culture conditions.
Discussion
SDF-1, also called pre–B-cell growth-stimulating factor-1 (PBSF), is a CXC chemokine constitutively produced by bone marrow stromal cells and is the major chemoattractant of HSCs and committed progenitors.39 This chemokine may be involved also in the retention of hematopoietic precursor cells in the bone marrow.5,6 The SDF-1 receptor CXCR4 is expressed all along the MK differentiation pathway from CFU-MK to platelets. However, the chemotaxis mediated by SDF-1 markedly decreases during MK differentiation.10-14 The negative regulation of CXCR4 signaling at the postreceptor level has also been described for mature B cells.15-17 In this cell lineage, it has been suggested that RGS proteins may be involved in the negative regulation of chemotaxis mediated by SDF-1 and other chemokines.
Inhibition of RGS16 expression in MO7e cells. MO7e cells were transduced with pRRL-SCR (control) or with pRRL-SEQ15 or with pRRL-SEQ24. (A) GFP+ cells were sorted and analyzed by quantitative RT-PCR for RGS16 expression. (B) SDF-1–induced migration of MO7e cell lines. MO7e pRRL-SCR (control) and MO7e pRRL-SEQ24 migrated in response to 300 ng/mL SDF-1. The percentage represents the proportion of migrated cells compared with input cells. The graph shows the mean ± SD of 4 independent experiments performed in duplicate. *P < .01
Inhibition of RGS16 expression in MO7e cells. MO7e cells were transduced with pRRL-SCR (control) or with pRRL-SEQ15 or with pRRL-SEQ24. (A) GFP+ cells were sorted and analyzed by quantitative RT-PCR for RGS16 expression. (B) SDF-1–induced migration of MO7e cell lines. MO7e pRRL-SCR (control) and MO7e pRRL-SEQ24 migrated in response to 300 ng/mL SDF-1. The percentage represents the proportion of migrated cells compared with input cells. The graph shows the mean ± SD of 4 independent experiments performed in duplicate. *P < .01
In this study we show that expression levels of RGS16 and RGS18 mRNA are up-regulated during MK differentiation. The observations reported here are consistent with the notion that dynamic control of specific RGS protein expression acts as a general mechanism for controlling the duration of G protein–coupled receptor signaling. Results of RGS16 and RGS18 protein overexpression in MO7e cells demonstrated that CXCR4 signaling was significantly inhibited by RGS16 but not by RGS18. This effect was further documented by reduced Erk and AKT signaling. Note that MAP kinase activation is constitutively (without agonist) higher in MO7e cells overexpressing RGS16 or RGS18 as compared with MO7e GFP cells, suggesting that RGS proteins may affect signaling events leading to activation of MAP kinase. The mechanisms leading to this high MAPK activation are presently unclear. In addition to this effect on basal MAPK signaling, MAPK activation by SDF-1 was also markedly inhibited by RGS16 overexpression but not by RGS18. MAP kinase activation is mediated by free Gβγ subunits.40 Thus, the decrease in CXCR4 signaling induced by RGS proteins is probably due to the GAP activity of RGS proteins themselves, increasing the rate of reassociation of G protein with βγ complexes and decreasing the amount of free βγ dimers. This is consistent with the most accepted model for RGS proteins, which bind directly to activated G proteins, stimulating GTP hydrolysis and promoting G-protein deactivation.18,41-46
Inhibition of RGS16 expression in primary MKs. Cord-blood CD34+ cells were cultured in MK medium with TPO and SCF. Cells were transduced with pRRL-SCR or pRRL-SEQ24 and sorted using the GFP and the CD41 marker. (A) Quantitative RT-PCR of CD41+/GFP+ and CD41+/GFP– sorted cells. RGS16 expression was normalized with the β2m one. (B) SDF-1–induced migration of MK pRRL-SCR and MK pRRL-SEQ24. Migrated cells were marked with anti-CD41 and anti-CD42 antibodies for flow analysis. The data represent the percentage of CD41+/CD42+ migrated cells compared with CD41+/CD42+ total input cells. The graph shows the mean ± SD of 4 independent experiments performed in duplicate. *P < .01 (C) Cell adhesion. pRRL-SCR and pRRL-SEQ24 CD41+/GFP+ sorted cells were plated in 48-well plates coated with collagen I or fibronectin in the presence or not of SDF-1 (300 ng/mL). Data represent the mean ± SD of 3 independent experiments.
Inhibition of RGS16 expression in primary MKs. Cord-blood CD34+ cells were cultured in MK medium with TPO and SCF. Cells were transduced with pRRL-SCR or pRRL-SEQ24 and sorted using the GFP and the CD41 marker. (A) Quantitative RT-PCR of CD41+/GFP+ and CD41+/GFP– sorted cells. RGS16 expression was normalized with the β2m one. (B) SDF-1–induced migration of MK pRRL-SCR and MK pRRL-SEQ24. Migrated cells were marked with anti-CD41 and anti-CD42 antibodies for flow analysis. The data represent the percentage of CD41+/CD42+ migrated cells compared with CD41+/CD42+ total input cells. The graph shows the mean ± SD of 4 independent experiments performed in duplicate. *P < .01 (C) Cell adhesion. pRRL-SCR and pRRL-SEQ24 CD41+/GFP+ sorted cells were plated in 48-well plates coated with collagen I or fibronectin in the presence or not of SDF-1 (300 ng/mL). Data represent the mean ± SD of 3 independent experiments.
Few studies have addressed the issue of RGS protein effects in CXCR4 signaling. RGS1 has been reported to be an effective inhibitor of chemotaxis toward SDF-1 in human or murine B lymphocytes.47,48 A short form of RGS3 was shown to impair CXCR4 signaling in a mature B-cell line.28 In another study, transiently transfected cells with constructs encoding RGS1, RGS3, and RGS4 but not RGS2 exhibited inhibition of chemoattractant-induced migration.24 Neill et al49 found that receptor-induced increases in inositol triphosphate levels were strongly reduced only in cells transfected with RGS3. In the present study, RGS16 and RGS18 proteins differentially affected CXCR4 signaling, suggesting that functional specificity of RGS proteins may depend on receptor and/or Gα-protein selectivity. In fact, we found that RGS18 is highly expressed in mature megakaryocytes and platelets. A report showed that RGS18 exhibits GAP activity toward Gαi.29,30,35 Binding assays of RGS18 with megakaryocytic-cell lysates demonstrated that RGS18 specifically binds Gαi and Gαq,35 suggesting that the dynamic control of this RGS acts as a mechanism for controlling the duration of one or more G-protein–coupled receptors expressed in megakaryocytes and platelets. Indeed, a recent work shows that RGS18 is phosphorylated in thrombin receptor-activating peptide (TRAP)–activated platelets,50 suggesting that RGS18 may be involved essentially in the protease-activated receptor 1 (PAR-1)/Gαi or Gαq signaling in megakaryocytes. RGS knock-out mice may help to precisely understand the role of this RGS on MK differentiation and platelet production.
Effect of RGS16 knocking down on CFU-MK and platelet production. Cord-blood CD34+ cells were cultured in MK medium with TPO and SCF. Cells were transduced with pRRL-SCR or pRRL-SEQ24 and sorted using GFP, CD34, CD41 markers. (A) CFU-MK assay. pRRL-SCR or pRRL-SEQ24 CD34+/GFP+ sorted cells were grown in plasma clot in the presence of TPO and SCF. The histograms show the number of CFU-MKs obtained after 12 days of culture for 2000 cells plated per dish. (B) Proplatelet production. pRRL-SCR or pRRL-SEQ24 CD41+/GFP+ sorted cells were plated in the presence of TPO and SCF. The histograms represent the number of MKs and proplatelets found per well. Data represent the mean ± SD of 3 independent experiments.
Effect of RGS16 knocking down on CFU-MK and platelet production. Cord-blood CD34+ cells were cultured in MK medium with TPO and SCF. Cells were transduced with pRRL-SCR or pRRL-SEQ24 and sorted using GFP, CD34, CD41 markers. (A) CFU-MK assay. pRRL-SCR or pRRL-SEQ24 CD34+/GFP+ sorted cells were grown in plasma clot in the presence of TPO and SCF. The histograms show the number of CFU-MKs obtained after 12 days of culture for 2000 cells plated per dish. (B) Proplatelet production. pRRL-SCR or pRRL-SEQ24 CD41+/GFP+ sorted cells were plated in the presence of TPO and SCF. The histograms represent the number of MKs and proplatelets found per well. Data represent the mean ± SD of 3 independent experiments.
To further demonstrate that RGS16 plays a key role in CXCR4 signaling, a lentiviral-delivered short hairpin RNA was used to inhibit RGS16 expression in MKs. A 3-fold decrease of RGS16 mRNA was achieved and led to an increase in CXCR4 signaling. Indeed, the SDF-1–induced migration of MKs was augmented in the presence of lentiviral vectors expressing anti-RGS16 siRNA. RGS16 has been already described to be important for lymphocyte activation and trafficking by attenuating the signaling of chemokine receptors such as CXCR4.36,51 Our experiment describes that RGS16 is also important in the MK lineage. Indeed, this may be part of the mechanism by which CXCR4 signaling is down-regulated during MK differentiation, leading to the release of mature MKs from the bone marrow microenvironment and to their interaction with the vascular endothelium for efficient platelet production. The up-regulation of RGS16 and RGS18 during MK differentiation suggests that RGS16 and RGS18 proteins are important targets for the biologic responses of MKs, and this is particularly true for RGS16 in CXCR4 signaling in these cells. Further experiments performed on RGS knock-out mice may help to precisely understand the role of RGS on MK differentiation and platelet production.
Prepublished online as Blood First Edition Paper, July 5, 2005; DOI 10.1182/blood-2005-02-0526.
Supported by grants from INSERM, the Gustave Roussy Institute (IGR) (grant CRI-SPS-2003-02) (F.L.) and the Association pour la Recherche contre le Cancer (grant 4309) (F.L.), by fellowships from La Ligue Nationale contre le Cancer (M.B. and C.R.) and Fondation pour la Recherche Médicale (D.C.), and by the Association pour la Recherche contre le Cancer (A.F. and Y.Z.).
M.B. designed and performed the experiments, analyzed the data, and wrote the paper; C.R. designed and performed the experiments and assisted in writing the manuscript; P.J., A.F., Y.Z., and D.C. designed and performed the experiments; A.G. designed the experiments and assisted in writing the manuscript; W.V. assisted in writing the manuscript; and F.L. designed the experiments, analyzed the data, and wrote the paper.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Frederic Larbret and Yann Lecluse (IFR54, Villejuif, France) for cell sorting experiments.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal