Abstract
T lymphocytes and a subpopulation of B lymphocytes express the CD5 coreceptor. Its functional importance is evident from the multiple levels and developmental stages of the regulation of its expression. We here report the discovery of a novel regulatory exon upstream of the noncoding region of the CD5 gene in humans. This alternate exon 1 is designated E1B (with the conventional exon 1 renamed E1A) and was shown to regulate the expression of CD5. E1B-containing transcripts existed exclusively in B lymphocytes and encoded a protein that was truncated and retained intracellularly. As a consequence, the amount of E1A-containing transcripts was down-regulated and the membrane CD5 expression was diminished in the presence of E1B-containing transcripts. High levels of E1A transcripts were found in chronic lymphocytic leukemia, and there were no E1A transcripts in 697 pre-B cells, which have no membrane CD5. Introduction of E1B into Jurkat cells reduced their membrane expression of CD5, and sequence analysis revealed that the E1B motif is a defective human endogenous retrovirus. A balance between the 2 alternative exons 1 might be central to the regulation of membrane CD5 in human B cells, and, through CD5-associated SH2-containing phosphatase 1, to the modulation of B-cell antigen receptor-transduced signals.
Introduction
Originally thought to be exclusive to T cells, the CD5 marker distributes B lymphocytes into 2 compartments: (1) CD5– or B-2 cells and (2) CD5+ or B-1 cells.1 The B-1 cells are subdivided into B-1a that carry CD5, and B-1b that do not, but share other attributes of B-1a cells. B-1a lymphocytes lie at the crossroads between oncology and immunology because chronic lymphocytic leukemia (CLL) cells carry CD52 and leukemic patients display autoimmune phenomena.3 CD5 cross-linking influences antigen receptor-mediated signaling, and based on findings in CD5-deficient mice,4 its effect on B-cell receptor (BCR)–mediated signaling is negative5 through the association of CD56 with the SH2-containing phosphatase 1 (SHP-1).
The expression of CD5 is tightly controlled. Its membrane density is about 30-fold higher in T cells than in B cells.7 Given the higher membrane density of CD5 on mature T lymphocytes than on thymocytes,8 its expression is regarded as developmentally regulated. Furthermore, B-1a cells represent the majority of B-lineage cells during neonatal life but decline thereafter. In addition, when cultured in the presence of interleukin 4 (IL-4), B cells reduce their CD5 expression, whereas this is raised by phorbol myristic acetate (PMA) or cross-linking IgM in the presence of IL-6.9 A nuclear factor of activated T (NFAT) cell–dependent enhancer is necessary10 for the BCR-mediated induction of CD5 expression in B cells.
Also consistent with this regulation is the fact that B-1b cells retain CD5 mRNA, despite the absence of CD5 on their membranes.11 There is a feedback regulation of B-1a cells,12 and Epstein-Barr virus transformation inhibits CD5 expression.13 In the mouse, the H2 haplotype affects the distribution of B cells into B-1a, B-1b, and B-2 subsets,14 and IL-10 influences interrelationships between B-1a and B-1b cells within the B-1 population.15 Thus, regulatory mechanisms for CD5 expression exist in the mouse, but far less is known about such mechanisms in humans.
Understanding this modulation will help us to gain insights into the functional relevance of CD5. This glycoprotein is encoded by a single gene in T and B lymphocytes that maps to chromosome 11q12.2 in humans16 and consists of 11 exons. The level at which the CD5 gene is regulated remains elusive, although it is conserved in size and organization. Numerous transcription factors have thus far been characterized.
This investigation set out to examine mechanisms of CD5 expression in human lymphocytes. We report the discovery of an exon 1 that is exclusively transcribed in B lymphocytes and due to the integration of a human endogenous retrovirus (HERV) into the CD5 locus. Our data also provide evidence for a reciprocal expression of the alternative exon 1, designated E1B, with the conventional exon 1 renamed E1A. E1B transcripts are translated into a truncated isoform of the CD5 molecule devoid of the leader peptide. Consequently, products of E1B transcripts are not translocated to the membrane, but down-regulate membrane CD5 expression. This precludes the SHP-1–related signal inhibiting functions of CD5 and induces antibody production. A balance between the 2 alternative exons 1 might be central to the regulation of membrane CD5 in human B cells.
Materials and methods
Cellular protocols
B lymphocytes were isolated from tonsil single-cell suspensions, from cord blood (CB), and from healthy volunteer or CLL patient peripheral blood mononuclear cells (PBMCs) by centrifugation on Ficoll-Hypaque, followed by 2 rounds of rosetting with sheep erythrocytes and positive selection with anti-CD19–coated magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). T lymphocytes were isolated using anti-CD3–coated microbeads. Purity of B and T lymphocytes was verified by fluorescence-activated cell sorting (FACS) analysis using fluorescein isothiocyanate (FITC)–anti-CD19 or FITC–anti-CD3 monoclonal Abs (mAbs), with phycoerythrin (PE)–anti-CD5 (Beckman-Coulter, Villepinte, France). All B- and T-lymphocyte preparations were more than 99% pure CD19+ and CD3+ cells, respectively.
To sort B-1a (CD5+CD45RAinterm), B-1b (CD5–CD45RAdim), and B-2 (CD5–CD45RAbright) subpopulations, 108 tonsillar B lymphocytes were stained with PE–anti-CD5 (clone UCHT2) and FITC–anti-CD45RA (2H4) mAbs (Beckman-Coulter) at 4°C for 30 minutes, sorted by FACS, and reanalyzed to confirm purity. COS cells, Jurkat T cells (control CD5+ T), and Daudi B cells (control CD5– B) were purchased from the American Type Culture Collection (Manassas, VA). The human CD5– pre-B cell line 69717 was donated by Paul Guglielmi (INSERM, Montpellier, France).
Setting and ethical approval
The study protocol was approved by the Institutional Review Board of the University of Brest and informed consent of patients was obtained according to the Declaration of Helsinki.
FACS analysis
Cell preparations were adjusted to 5 × 105 cells/tube, fixed using 250 μL cytofix/cytoperm solution, washed, and incubated with control IgG2 or IgG2a, or either of 2 anti-CD5 mAbs, UCHT2 (gift from Peter Beverley, Compton, United Kingdom) or Leu1 (BD PharMingen, Grenoble, France), for 60 minutes at 4°C. FITC-goat anti–mouse IgG was added, and the cells were analyzed in an EPICS-XL FACS (Beckman-Coulter). In some experiments, they were permeabilized using 0.5% saponin, incubated with biotinylated (biot) goat anti–mouse IgG, and stained with horseradish peroxidase (HRP)–streptavidin (Vector Laboratories, Burlingame, CA). The mean fluorescence intensity for CD5 expression was recorded and the number of molecules per cell quantified by measuring the amount of antibody binding to the cells using the Quantum Simply Cellular kit (Flow Cytometry Standards, San Juan, Puerto Rico).
Cell culture
Cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and antibiotics. Dulbecco modified Eagle medium (DMEM) was substituted for RPMI 1640 medium in COS-1 cell cultures. In the induction experiments, lymphocytes were seeded at 107 cells/mL and cultured with 30 ng/mL PMA (Sigma, St Louis, MO), 10 U/mL IL-2, 10 μg/mL anti-IgM antibody coated onto Sepharose beads (Bio-Rad, Hercules, CA), or 0.001% Staphylococcus aureus Cowan (SAC) I.
Rapid amplification of cDNA
The 5′ transcript ends were amplified using 1 μg mRNA and switching mechanism at the 5′ end of the RNA transcript (SMART)–rapid amplification of cDNA ends (RACE) of cDNA performed using a kit (Clontech, Palo Alto, CA). The first strand of cDNA was synthesized using SMART IIA oligonucleotide and 5′RACE cDNA synthesis primer, and 5′RACE polymerase chain reaction (PCR) was performed using the sense UPM primer and the gene-specific antisense primer CD5 E5-6 with the Advantage 2 polymerase. The touchdown-PCR protocol included denaturation at 94°C for 5 minutes, starting 5 touchdown-PCR cycles of denaturation at 94°C for 30 seconds, and annealing at 72°C for 3 minutes. These were completed by another 5 cycles (denaturation at 94°C for 30 seconds, annealing at 70°C for 1 minute, and primer extension at 72°C for 3 minutes). Another 40 cycles consisted of denaturation at 94°C for 30 seconds, annealing at 68°C for 1 minute, and primer extension at 72°C for 3 minutes. The products were subcloned using the pTrueBlue ligation kit (Genomics One, Laval, QC, Canada), and sequenced using the BigDye Terminator Cycle Sequencing Kit and the ABI-310 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Reverse transcriptase–PCR for CD5 gene regulatory elements
One microgram mRNA was extracted by the RNAble method (Eurobio, Les Ulis, France), transcribed with reverse transcriptase (RT) and oligo-dT, and used for PCR amplification with either CD5-, CD3ζ-, CD79b-, or glyceraldehyde phosphate dehydrogenase (GAPDH)–specific primers (GenBank sequences: X04391, NM000 734, M80461, and BC014085, respectively). CD3ζ and CD79b amplifications established that B-cell preparations were devoid of T cells and T-cell preparations were devoid of B cells, respectively. PCR for CD5 mRNA comprised denaturation at 94°C for 5 minutes, starting with 5 touchdown cycles (94°C for 30 seconds, 65°C for 40 seconds, and 72°C for 1 minute), then with a decreasing temperature for 40 cycles (94°C for 30 seconds, 61°C for 40 seconds, and 72°C for 1 minute), followed by extension at 72°C for 10 minutes. For E1A and exon 2 of the CD5 mRNA in T lymphocytes, the protocol was the same, except that there were only 35 annealing cycles.
cDNA was separated on 2% agarose gel and visualized with 0.5 μg/mL ethidium bromide. The PCR products were checked by sequencing and digestions with BamHI (E1B), AccI (exon 3), or KpnI (exon 5). Band densities were evaluated using Molecular Analyst software (Bio-Rad), and the mean OD for each point was calculated.
Quantitative RT-PCR
Quantitative PCR was performed in 10-μL mixtures containing 50 ng template cDNA, 500 nM of each primer, and 1 × SYBR Green PCR Master mix (Applied Biosystems). Each assay included the reaction mixture with no template as a negative control and GAPDH mRNA as a positive control. Standard curves were constructed using the same PCR mixtures with 104 to 107 copies/mL mRNA from the CD5 gene insert in pTrueBlue. Numbers of CD5 transcript copies were inferred from the threshold cycle numbers and the standard curves normalized to GAPDH values. Results were exported as tab-delimited text files and imported into Microsoft Excel (Microsoft, Redmond, WA).
Expression of the CD5 protein product in rabbit reticulocytes
We used a TnT T7-Coupled Reticulocyte Lysate System (Promega, Charbonnières, France) for in vitro translation of CD5 transcripts. This kit supplied rabbit reticulocyte lysate, T7 RNA polymerase, and a mixture of amino acids. The 10-μL mixtures were made up of biot-lysine, 200 ng plasmid DNA, and pDNR, with the CD5 gene insert. A pDNR plasmid without an insert was the negative control and a plasmid containing the 61-kDa luciferase gene the positive control. The mixtures were incubated for 2 hours at 30°C and proteins separated by 5% to 20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride (PVDF) membranes that were stained with streptavidin-HRP, and revealed by enhanced chemoluminescence (ECL).
Transient transfection
COS-1 cells were plated out at 106 cell/well in DMEM and cultured until they reached 70% to 80% confluence. The medium was replaced with 500 μL optiMEM (Gibco, Paisley, United Kingdom) containing 6 μL Transfast (Promega) and 1 μg of the enhanced green fluorescent protein (EGFP) gene-containing plasmid (Clontech) or CD5-EGFP fusion protein gene-containing plasmid. After 3 hours of incubation, the medium was replaced with DMEM. Cells were incubated for 24 hours and harvested with 0.25% trypsin, and their transfection was verified by FACS. One cell aliquot was analyzed with a confocal microscope, and another adjusted to 107 cells/mL in lysis buffer (Tris [tris(hydroxymethyl) aminomethane]–HCl, pH 7.5, 1 mM EDTA [ethylenediaminetetraacetic acid], 140 mM NaCl, 1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 μM aprotinin, 3 μg/mL leupeptin, and 1μg/mL pepstatin). Its protein content was evaluated by Western blotting, using an HRP–anti-GFP antibody.
Establishment of cell lines expressing CD5-1B
Using the same method, Jurkat T cells were transfected with the plasmid LoxP (pLP) cytomegalovirus (CMV), with or without the full-length CD5-1B cDNA. Vector-containing lymphocytes were selected with G418 (Sigma). The CD5lo cells were sorted from the pLP CMV and the pLP–CD5-1B populations and placed in culture. After 7 days, they were analyzed by FACS with PE–anti-CD5 UCHT2. Cell extracts were also resolved by SDS-PAGE and probed with anti-CD5 Leu1. CD5-1A and CD5-1B mRNAs were quantified in pLP CMV and pLP–CD5-1B cells by quantitative PCR.
In situ hybridization
For the in situ hybridization, oligonucleotide primers were labeled with digoxigenin at the 3′ ends, using 50 U terminal transferase (Roche, Meylan, France). The cells were resuspended at 2 × 106/mL in Hanks balanced salt solution (HBSS), and 105 cells were attached to slides, dried at 25°C, and fixed with 4% paraformaldehyde.
Tonsil sections were deparaffinized and rehydrated. Cells and sections were permeabilized with 0.1% Triton X-100 and dehydrated. The slides were rehydrated, air-dried, treated with proteinase K incubated in the hybridization buffer (50% formamide, 2 × saline sodium citrate [SSC], 1 × Denhardt reagent, 10% dextran sulfate, 150 μg/mL tRNA, 100 μg/mL polyA, 100 μg/mL single-stranded DNA) for 1 hour at 37°C and incubated overnight at 37°C in the same buffer completed with 200 μL/mL of the digoxigenin primers. They were washed with a 2 × SSC, 0.1% SDS, followed by 1 × and 0.5 × SSC, and incubated in 1 × Roche Blocking Reagent for 30 minutes, and with alkaline phosphatase F(ab′)2 antidigoxigenin antibody (Roche) overnight at 25°C. Probes were localized by NBT/5-bromo-4-chloro-3-indolyl phosphate.
Sequence analyses
Production of recombinant CD5 vectors
The full-length CD5 cDNA clone extended from the known ATG1 initiation site at position –8129 to the stop codon at position +23550 in E11 of the genomic sequence of CD5 gene. It was generated by RT-PCR, using a sense primer for the –8129/–8105 region, and an antisense primer downstream of the stop codon (Table 1). Amplifications used a mixture of Pwo and Taq polymerases, and PCR products were subcloned into pTrueBlue vector (Genomics One). To generate CD5-1BpDNR dual, some of these were subcloned into pDNRdual, which has a T7 promoter for transcription in reticulocytes. The rest were subcloned into the pLP CMV vector using the Cre-recombinase system (Clontech), or served to generate fusion proteins with GFP. They were modified by PCR using a primer for the T7 promoter in the pTrueBlue, combined with an antisense primer. This anneals to the 3′ end of the original PCR product and substitutes an EcoRI restriction site for the stop codon at position +23002 of the gDNA. The fragments were cut with XhoI and EcoRI, purified, and subcloned into pEGFP-N3 cut with XhoI and EcoRI.
Synthetic oligonucleotides used in the experiments
Oligonucleotides . | Sequence . | Method . |
|---|---|---|
| CD5 1 sense (intron) | 5′-GTGAAGGGCTGCTTACTGCACACAGC-3′ | RT-PCR |
| CD5 2 sense (U3 HERV) | 5′-AGTGGGTCTAAGGGAGGAGACCAC-3′ | RT-PCR |
| CD5 3 sense E1B (leader HERV) | 5′-AGTCCGGACAACTGAAACTTGTTCGCT-3′ | RT-PCR and quantitative PCR |
| CD5 4 sense (gag HERV) | 5′-CGCTTGACAGGACTGGTCTTGGGA-3′ | RT-PCR |
| CD5 5 sense (promotor E1A) | 5′-GATGCATGGCCTTGTCCTGTGTG-3′ | RT-PCR |
| CD5 6 sense E1A | 5′-GAAGGCCAGAAACCATGCCCATGG-3′ | RT-PCR and quantitative PCR |
| CD5 7 sense (intron) | 5′-ATGTGTGTGAGTACTGTGTGTAGGGGA-3′ | RT-PCR |
| CD5 8 sense (intron) | 5′-ATGTGTGTGAGTACTGTGTGGGGGGA-3′ | RT-PCR |
| CD5 9 sense E2 | 5′-TCGGACGGCTCAGCTGGTATGAC-3′ | RT-PCR and quantitative PCR |
| CD5 E5-6 antisense | 5′-TGGAAACCTGAGCAAAGGAGGGCA-3′ | RT-PCR |
| CD5 E3 antisense | 5′-TGCCATCCGTCCTTGAGGTAGAC-3′ | Quantitative PCR |
| CD5 E1A antisense | 5′-CTGGCCGGGTATCTCGCTCTCAGAGAGGGA-3′ | In situ hybridization |
| CD5 E1A sense | 5′-TCCCTCTCTGAGAGCGAGATACCCGGCCAG-3′ | In situ hybridization |
| CD5 E1B antisense | 5′-ATCCTCCACAGGGGCCTGCCATGCACTGCT-3′ | In situ hybridization |
| CD5 E1B sense | 5′-AGCAGTGCATGGCAGGCCCCTGTGGAGGAT-3′ | In situ hybridization |
| CD5 E1B sense | 5′-GGTGTCTGAGGGGTTTTGTCTGTG-3′ | Cloning |
| CD5 E11 antisense | 5′-CGGGGTAGAAATAGGGACATGATTC-3′ | Cloning |
| CD5 E10 antisense | 5′-CATGGATCCCAGAATTCGCAGCCTCTG-3′ | Cloning |
| CD3ζ sense | 5′-TGCTGGATGGAATCCTCTT-3′ | RT-PCR |
| CD3ζ antisense | 5′-GTCTGGCCTTTGAGTGGTGA-3′ | RT-PCR |
| CD79b sense | 5′-AGAAGTGCAACAACACCTCGGAGGTCTACC-3′ | RT-PCR |
| CD79b antisense | 5′-CCATCCCATGTGTGGGGACGGATC-3′ | RT-PCR |
| GAPDH sense | 5′-GAAGGTGAAGGTCGGAGTC-3′ | RT-PCR and quantitative PCR |
| GAPDH antisense | 5′-GAAGATGGTGATGGGATTTC-3′ | RT-PCR and quantitative PCR |
Oligonucleotides . | Sequence . | Method . |
|---|---|---|
| CD5 1 sense (intron) | 5′-GTGAAGGGCTGCTTACTGCACACAGC-3′ | RT-PCR |
| CD5 2 sense (U3 HERV) | 5′-AGTGGGTCTAAGGGAGGAGACCAC-3′ | RT-PCR |
| CD5 3 sense E1B (leader HERV) | 5′-AGTCCGGACAACTGAAACTTGTTCGCT-3′ | RT-PCR and quantitative PCR |
| CD5 4 sense (gag HERV) | 5′-CGCTTGACAGGACTGGTCTTGGGA-3′ | RT-PCR |
| CD5 5 sense (promotor E1A) | 5′-GATGCATGGCCTTGTCCTGTGTG-3′ | RT-PCR |
| CD5 6 sense E1A | 5′-GAAGGCCAGAAACCATGCCCATGG-3′ | RT-PCR and quantitative PCR |
| CD5 7 sense (intron) | 5′-ATGTGTGTGAGTACTGTGTGTAGGGGA-3′ | RT-PCR |
| CD5 8 sense (intron) | 5′-ATGTGTGTGAGTACTGTGTGGGGGGA-3′ | RT-PCR |
| CD5 9 sense E2 | 5′-TCGGACGGCTCAGCTGGTATGAC-3′ | RT-PCR and quantitative PCR |
| CD5 E5-6 antisense | 5′-TGGAAACCTGAGCAAAGGAGGGCA-3′ | RT-PCR |
| CD5 E3 antisense | 5′-TGCCATCCGTCCTTGAGGTAGAC-3′ | Quantitative PCR |
| CD5 E1A antisense | 5′-CTGGCCGGGTATCTCGCTCTCAGAGAGGGA-3′ | In situ hybridization |
| CD5 E1A sense | 5′-TCCCTCTCTGAGAGCGAGATACCCGGCCAG-3′ | In situ hybridization |
| CD5 E1B antisense | 5′-ATCCTCCACAGGGGCCTGCCATGCACTGCT-3′ | In situ hybridization |
| CD5 E1B sense | 5′-AGCAGTGCATGGCAGGCCCCTGTGGAGGAT-3′ | In situ hybridization |
| CD5 E1B sense | 5′-GGTGTCTGAGGGGTTTTGTCTGTG-3′ | Cloning |
| CD5 E11 antisense | 5′-CGGGGTAGAAATAGGGACATGATTC-3′ | Cloning |
| CD5 E10 antisense | 5′-CATGGATCCCAGAATTCGCAGCCTCTG-3′ | Cloning |
| CD3ζ sense | 5′-TGCTGGATGGAATCCTCTT-3′ | RT-PCR |
| CD3ζ antisense | 5′-GTCTGGCCTTTGAGTGGTGA-3′ | RT-PCR |
| CD79b sense | 5′-AGAAGTGCAACAACACCTCGGAGGTCTACC-3′ | RT-PCR |
| CD79b antisense | 5′-CCATCCCATGTGTGGGGACGGATC-3′ | RT-PCR |
| GAPDH sense | 5′-GAAGGTGAAGGTCGGAGTC-3′ | RT-PCR and quantitative PCR |
| GAPDH antisense | 5′-GAAGATGGTGATGGGATTTC-3′ | RT-PCR and quantitative PCR |
Western blotting
EGFP expression in transfected COS-1 cells was evaluated by fluorescence microscopy and by Western blotting with anti-GFP (Clontech) and UCHT2 mAbs; 107 cells were washed in HBSS and lysed with 1% Triton X-100 in lysis buffer (20 mM Tris-HCl, 140 mM NaCl, 1 mM EDTA) with 1 mM PMSF, 10 μg/mL aprotinin, and 1 mM sodium orthovanadate. The lysates were centrifuged and the protein concentration determined in the supernatants. Protein (25 μg) was resolved on 12% SDS-PAGE, transferred to a PVDF membrane, incubated with antibody, and revealed with HRP-sheep anti–mouse IgG.
For immune precipitation, lysates from 107 cells were incubated with 50 μL protein G–coated magnetic beads for 30 minutes, and with 2 μg antiphosphotyrosine (PY) mAb for 30 minutes. Immune complexes were separated with 50 μL of the microbeads and washed 4 times with the lysis buffer and once with 20 mM Tris-HCl. Bound proteins were eluted by boiling the samples in SDS buffer containing 50 mM dithiothreitol (DTT), resolved on 5% to 20% SDS-PAGE gel, blotted onto PVDF membrane, and incubated with biot–anti-PY, biot-UCHT2, or biot-Leu1 mAbs, with HRP-streptavidin.
Results
Genetic and database analyses
To identify regulatory regions that modulate CD5 expression, gDNA and cDNA sequences of the CD5 gene were analyzed using the GENSCAN and the National Institutes of Health Web servers. Within the 5′ noncoding region, there was a defective HERV gene sequence, starting 8.2 kb upstream of the ATG1 initiation site in exon 1 (Figure 1A). This is identifiable based on the gag, pol, and env sequences, and long terminal repeats (LTRs). The HERV 5′ LTR encompasses the first 463 nucleotides, and a further 4319 nucleotides downstream is the 472-nucleotide 3′ LTR. The tRNA-glu primer-binding site signifies that this virus belongs to the HERV-E family.18 Its insertion generates a new promoter for exon 1, allows alternate splicing of the CD5 gene (Figure 1B), and lies 20.6 kb upstream of exon 2. It was designated E1B, and the known exon 1 was renamed E1A. E1B has the potential to splice to the initiation site on other exons of the CD5 gene. We were unable to identify any other sequences that could, potentially, be involved in the regulation of the gene.
To locate E1B, the cDNA of CD5 was mapped using 34 sense primers designed from gDNA and spanning regions in the 10 kb upstream of E1A and E1B (Figure 1A). E1A transcripts were found in both T and B cells, whereas E1B transcripts were specific for B cells. Restriction digestion patterns were identical to those predicted from the known cDNA sequence,19 and sequence analysis of E1B fully matched the 5′ sequence of the CD5 gene from the database. A model for independent splicing of E1B to downstream exon 2 is depicted in Figure 1B.
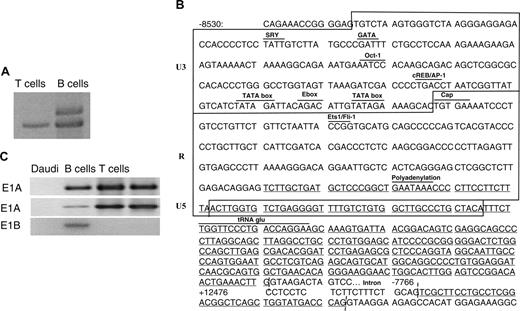
The 5′ end of the CD5 gene indicating location of the new exon 1. (A) The human CD5 locus depicting the 5′ end of the genomic map constructed from overlapping human CD5 genomic sequence and partial genomic sequences containing exons 1, 2, and 3. The 5.3-kb human endogenous retrovirus element lies upstream of the known exon 1 sequence (E1A). Distances between the exons are indicated from exon 1 ATG (+1). The bottom panel is a gel showing whether PCR products were obtained using the sense primers indicated above each gel lane (arrowheads, the numbers refer to the oligonucleotides listed in Table 1) and CD5 E5-6 as antisense primer. Sizes are 763 bp, 811 bp, and 730 bp, respectively. (B) Cartoon depicting the proposed splice sites in E1A and E1B-type transcripts and ATG starting coding region.
The 5′ end of the CD5 gene indicating location of the new exon 1. (A) The human CD5 locus depicting the 5′ end of the genomic map constructed from overlapping human CD5 genomic sequence and partial genomic sequences containing exons 1, 2, and 3. The 5.3-kb human endogenous retrovirus element lies upstream of the known exon 1 sequence (E1A). Distances between the exons are indicated from exon 1 ATG (+1). The bottom panel is a gel showing whether PCR products were obtained using the sense primers indicated above each gel lane (arrowheads, the numbers refer to the oligonucleotides listed in Table 1) and CD5 E5-6 as antisense primer. Sizes are 763 bp, 811 bp, and 730 bp, respectively. (B) Cartoon depicting the proposed splice sites in E1A and E1B-type transcripts and ATG starting coding region.
Transcription initiation sites of human CD5 gene in exon 1
To find transcriptional initiation sites of E1B, and demonstrate alternate use of E1A and E1B, 5′-RACE was performed on total mRNA from tonsillar B and T cells (Figure 2A). The products were subcloned in pTrueBlue plasmid and sequenced. Two types of plasmid products were generated: the known E1A linked with the full-length CD5 cDNA and the new E1B linked with the CD5 cDNA at an exon 2 site. In these plasmids, E1A was deleted and replaced with E1B (these sequence data are available from GenBank under accession nos. AY541703 and AY541704, respectively).
Figure 2B presents the position of the 5′ ends of sequenced products, in that E1B extends from –8133 through –7779. The RACE product across the E1B/exon 2 boundary is shown. It could be inferred, based on the U3-R-U5 organization of HERV-E, that the 213-nucleotide U3 promoter possesses 2 TATA boxes and several transcription factor–binding sites for E2A, Oct-1, and GATA. The 215-bp retroviral LTR starts with a TGTGAA cap site and ends with a polyA signal. There remains a 43-nucleotide U5-LTR sequence. There is also a tRNAglu primer-binding site. On the CD5-E1B cDNA, 2 initiation codons of the long open reading frame (ORF) were identified within exon 3, located 116 bp downstream from the E1B/exon 2 splice junction. This encodes a truncated form of CD5, whereas the 1299 nucleotide of exon 3 ORF encodes a 433-amino acid protein, with a molecular weight of about 52 kDa, and the 1116 nucleotides of exon 3 ORF a 372-amino acid protein, with a molecular weight of 44 kDa. The leader peptide and the N-terminal part of the most external domain are lacking in the novel CD5 molecule. These studies showed that E1B was expressed in transcripts from B lymphocytes, but not in those from T lymphocytes, and confirmed that the predicted alternative E1B splice site was on exon 2.
E1B is selectively transcribed in B lymphocytes
Primers annealed to E1A, E1B, or exon 2, together with an antisense primer specific for the junction between the rearranged exon 5 and exon 6. Whereas the PCR using the 5′ E1A primer yielded products in both B and T lymphocytes, the 5′ E1B primer was specific for B lymphocytes (Figure 2C). Quantitative PCR confirmed that T cells did not make E1B transcripts, whereas B cells did. Levels of PCR products were higher with the E1B primer than with the E1A in tonsillar B cells. Interestingly, E1B-containing transcripts were absent in other CD5-nonexpressing cells, such as monocytes, neutrophils, and endothelial cells.
To substantiate that transcription of E1B affects the density of surface CD5, B-1a, B-1b, and B-2 cells were sorted, and E1A and E1B transcripts enumerated (Figure 3A). The level of E1A was higher in B-1a than in B-1b and in B-1b than in B-2 cells. In situ hybridization with one pair of primers specific for E1A transcripts and another specific for E1B transcripts circumvented PCR artifacts. Cytospun cells (Figure 3B) showed that T cells had more E1A mRNA than B cells and that T cells did not contain E1B transcripts. In situ hybridization (Figure 3C) placed E1A transcript–containing cells in the T-lymphocyte zone and in the follicular mantle area of tonsil sections. In contrast, E1B transcript–containing cells were not seen in this T-lymphocyte zone but rather in the B-lymphocyte follicles and in the follicular mantle.
The 5′ RACE analysis of the CD5 gene initiation and expression in B and T cells. (A) CD5 5′ RACE analysis of exon 1 B (E1B). One band from T lymphocytes (899 bp) and 2 from tonsillar B cells (1096 bp, 899 bp) were separated, subcloned into pTrueBlue, and sequenced. The larger band amplified only from B-cell RNA contained E1B instead of E1A. (B) Regulatory motifs for transcription factor binding and TATA box location within the U3 region. Box delimits the U3/R/U5 parts. The 5′ RACE sequences are underlined. (C) RT-PCR analysis of cDNA from B or T cells using sense primers specific for CD5 exon 2, E1A, E1B, and antisense primers located at the exons 5-6 junction. Daudi B cells and Jurkat T cells are negative and positive controls.
The 5′ RACE analysis of the CD5 gene initiation and expression in B and T cells. (A) CD5 5′ RACE analysis of exon 1 B (E1B). One band from T lymphocytes (899 bp) and 2 from tonsillar B cells (1096 bp, 899 bp) were separated, subcloned into pTrueBlue, and sequenced. The larger band amplified only from B-cell RNA contained E1B instead of E1A. (B) Regulatory motifs for transcription factor binding and TATA box location within the U3 region. Box delimits the U3/R/U5 parts. The 5′ RACE sequences are underlined. (C) RT-PCR analysis of cDNA from B or T cells using sense primers specific for CD5 exon 2, E1A, E1B, and antisense primers located at the exons 5-6 junction. Daudi B cells and Jurkat T cells are negative and positive controls.
Effects of cell activation on the expression of E1A and E1B
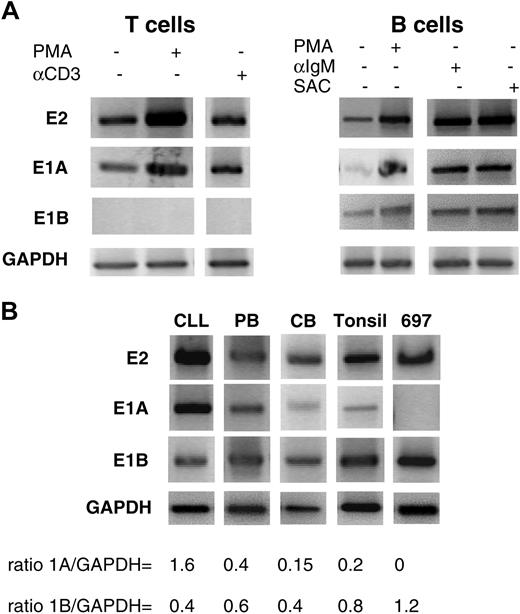
RT-PCRs of mRNA from T and B cells, before and after activation, explored the functional relevance of the alternative transcription of the 2 exons 1 (Figure 4A). RT-PCR of mRNA from T lymphocytes using the exon 2 and the E1A primers presented characteristic products. Using the E1B primer, no PCR products were obtained from activated T cells, whereas activation of B cells with PMA, anti-IgM, or SAC increased PCR products triggered by the E1A primer and the E1B primer.
E1A and E1B transcripts are differentially produced in B and T lymphocytes. (A) Tonsillar B-1a, B-1b, and B-2 cells were sorted based on their membrane expression of CD5 and CD45RA. The numbers of E1A- and E1B-containing transcripts were determined in these 3 subpopulations by quantitative PCR. (B) In situ hybridization analysis of cytospun tonsillar T and B cells (× 400) with sense and antisense probes for E1A (top) and E1B (bottom). The transcripts were revealed using digoxigenin-labeled probes. E1B transcript–containing cells are denoted by the arrows. (C) Localization of E1A and E1B transcript–containing cells (arrows) in tonsil sections (× 100). Germinal center (GC), follicular mantle (FM), and T-cell areas are indicated. Images were captured using a Zeiss Axioplan microscope (Zeiss, Oberkochen, Germany) using 40 ×/0.75 numeric aperture (NA) (B) and 10 ×/0.30 NA (C) objectives. Images were acquired with an Olympus Camedia C-4040 zoom digital camera and Camedia master 2.5 imaging solution, and were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
E1A and E1B transcripts are differentially produced in B and T lymphocytes. (A) Tonsillar B-1a, B-1b, and B-2 cells were sorted based on their membrane expression of CD5 and CD45RA. The numbers of E1A- and E1B-containing transcripts were determined in these 3 subpopulations by quantitative PCR. (B) In situ hybridization analysis of cytospun tonsillar T and B cells (× 400) with sense and antisense probes for E1A (top) and E1B (bottom). The transcripts were revealed using digoxigenin-labeled probes. E1B transcript–containing cells are denoted by the arrows. (C) Localization of E1A and E1B transcript–containing cells (arrows) in tonsil sections (× 100). Germinal center (GC), follicular mantle (FM), and T-cell areas are indicated. Images were captured using a Zeiss Axioplan microscope (Zeiss, Oberkochen, Germany) using 40 ×/0.75 numeric aperture (NA) (B) and 10 ×/0.30 NA (C) objectives. Images were acquired with an Olympus Camedia C-4040 zoom digital camera and Camedia master 2.5 imaging solution, and were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
E1A and E1B in resting and stimulated lymphocytes. (A) Stimulation of T and B cells. Sense primers specific for CD5 E1A, E1B, and exon 2, with antisense primers located at the exons 5-6 junction, were used in RT-PCR. (B) Expression of E1A, E1B, and exon 2 transcripts in B cells from different sources (CLL, chronic lymphocytic leukemia; PB, peripheral blood; tonsil, tonsillar cells; CB, cord blood).
E1A and E1B in resting and stimulated lymphocytes. (A) Stimulation of T and B cells. Sense primers specific for CD5 E1A, E1B, and exon 2, with antisense primers located at the exons 5-6 junction, were used in RT-PCR. (B) Expression of E1A, E1B, and exon 2 transcripts in B cells from different sources (CLL, chronic lymphocytic leukemia; PB, peripheral blood; tonsil, tonsillar cells; CB, cord blood).
E1B-containing transcripts in B-lymphocyte subpopulations
Were alternate transcripts differentially regulated in B-cell subpopulations? In contrast to tonsillar B lymphocytes, which mainly transcribed E1B (Figures 2C and 3B; Table 2), those from CLL patients expressed more E1A than E1B transcripts (Figure 4B; Table 2). We reasoned that such differences might relate to the membrane expression of the molecule, which is increased in CLL B cells.20 As we reported previously,21 malignant B lymphocytes from patients with CLL are heterogeneous in their membrane expression of CD5, and, therefore, heterogeneous in their transcription of CD5-E1A and CD5-E1B.
CD5 membrane expression relative to copy the total number of CD5 transcripts and to the ratio of E1A to E1B transcripts
Cells . | No. samples . | No. molecules, × 10-3 ABC units, mean ± SD* . | Total no. CD5 transcripts, range . | No. CD5-E1A transcripts, range . | No. CD5-E1B transcripts, range . |
|---|---|---|---|---|---|
| Blood T cells | 3 | 147.0 ± 7.9* | 450 000-610 000 | 450 000-610 000 | ND |
| CLL CD5+ B cells | 3 | 74.5 ± 5.9 | 150 000-200 000 | 133 845-188 950 | 5 080-42 120 |
| Blood B cells | 2 | 31.9 ± 4.1 | 20 000-30 000 | 12 840-20 300 | 4 400-11 340 |
| Cord blood B cells | 2 | 24.2 ± 2.1 | 10 000-25 000 | 10 350-19 625 | 3 950-5 425 |
| Tonsillar B cells | 3 | 14.7 ± 1.7 | 15 000-50 000 | 6 310-25 750 | 8 850-34 050 |
| 697 cell line | 4 | — | 12 000-16 000 | ND | 12 000-16 000 |
Cells . | No. samples . | No. molecules, × 10-3 ABC units, mean ± SD* . | Total no. CD5 transcripts, range . | No. CD5-E1A transcripts, range . | No. CD5-E1B transcripts, range . |
|---|---|---|---|---|---|
| Blood T cells | 3 | 147.0 ± 7.9* | 450 000-610 000 | 450 000-610 000 | ND |
| CLL CD5+ B cells | 3 | 74.5 ± 5.9 | 150 000-200 000 | 133 845-188 950 | 5 080-42 120 |
| Blood B cells | 2 | 31.9 ± 4.1 | 20 000-30 000 | 12 840-20 300 | 4 400-11 340 |
| Cord blood B cells | 2 | 24.2 ± 2.1 | 10 000-25 000 | 10 350-19 625 | 3 950-5 425 |
| Tonsillar B cells | 3 | 14.7 ± 1.7 | 15 000-50 000 | 6 310-25 750 | 8 850-34 050 |
| 697 cell line | 4 | — | 12 000-16 000 | ND | 12 000-16 000 |
ABC indicates antibody-binding capacity; —, not applicable; ND, not detected.
In this column, the number of molecules correlates directly to that of CD5-E1A transcripts (Spearman test, P < .01) and inversely to that of CD5-E1B transcripts (P < .01).
PCRs with B cells from the 697 pre-B cell line, PBMCs, and CBs revealed a spectrum of CD5 expression (Figure 4B), ranging from elevated levels in CLL B cells to no E1A transcripts in 697 pre-B cells. These PCRs were consistent with those obtained from tonsillar B cells, in that more products were obtained with the E1B than with the E1A primer. However, as predicted from the surface expression of CD5, there were more E1A than E1B transcripts in B lymphocytes from PBMCs than from CB samples.
Differences in the levels of the alternate transcript were documented by quantitative PCR; whereas transcripts with either of the exons 1 were seen in the CLL B cells at levels consistent with the PCR experiments, there were no E1A transcripts in the 697 pre-B cells. The numbers of total CD5, CD5-E1A, and CD5-E1B mRNA copies in the different B-cell populations are presented in Table 2. These data indicate that T lymphocytes have transcripts with E1A but not with E1B and that mature B lymphocytes express distinct transcripts with either of these exons 1. There was a correlation between the density of membrane CD5 and the amount of E1A transcripts: the higher the level of membrane CD5 expression, the more E1A transcripts (P < .01), and the lower the level of membrane CD5 expression, the more E1B transcripts (P < .01). Supporting this argument, B-1a cells, which express membrane CD5 (Figure 3A, left), had more E1A transcripts than B-1b and B-2 cells (Figure 3A, right). Figure 3A also confirms the finding by Kasaian et al11 that the total amount of transcripts for CD5 (ie, CD5-E1A plus CD5-E1B) was significantly higher in the B-1b than in the B-2 cells.
Nature of CD5 protein encoded by E1B transcripts
To demonstrate that E1B-type transcripts encode a functional protein, a gene construct containing cDNA from an E1B-type transcript was inserted into the pDNR plasmid. An insert-lacking pDNA plasmid and a plasmid containing the 61-kDa protein-encoding luciferase were the negative and positive controls, respectively. The reticulocyte system produced a 52-kDa protein in lysates from the plasmid containing the E1B-type transcript (Figure 5A). Based on the molecular weight of the protein product, one may assume that the initiation site of the protein is the distal 5′ ATG (Figure 1B).
With anti-PY antibody as a positive control (Figure 5B), immune precipitation analysis revealed that, whereas anti-CD5 mAb UCHT2 recognized the protein encoded by the E1B-type transcript in the 697 pre-B cells, this was not the case using anti-CD5 mAb Leu1, confirming that 2 distinct isoforms of the CD5 protein exist.
E1B-type transcripts translate into intracellularly retained protein
Our observation that UCHT2 recognizes both CD5 isoforms was exploited to localize these 2 proteins. Analysis of CD5 expression with UCHT2 showed that Jurkat T lymphocytes (which only have E1A transcripts) strongly expressed membrane CD5 (Figure 5C). In contrast, the 697 pre-B cells (which only have E1B transcripts) did not carry membrane CD5. However, when both cell lines were permeabilized, there was evidence for intracellular CD5 protein in the 697 pre-B cells, albeit at a low level, whereas Jurkat T-cells were similar to nonpermeabilized cells.
Alternative CD5 E1B motif encodes a truncated isoform of CD5 protein starting at exon 3 and retained intracellularly. (A) In vitro translation of CD5 using pLuc (C+), pDNRdual (C–), CD5-1B pDNRdual (CD5-1B) using the TNT T7-coupled reticulocyte system. (B) Precipitation of CD5 in T cells and 697 pre-B cells using antiphosphotyrosine (PY). MW indicates molecular weight; IP, immunoprecipitation. (C) FACS analysis of Jurkat T and 697 pre-B cells for CD5 protein expression in the absence (membrane CD5) or in the presence (intracellular CD5) of saponin. Filled histograms denote CD5 staining; open histograms, isotypic controls. (D) Immunohistochemical staining with UCHT2 of saponin-treated 697 pre-B cells. Images were acquired as in Figure 3B. (E) Western blot analysis of COS-1 cells transfected with EGFP-expressing vector, or fusion CD5 E1B transcript and EGFP. Top arrow indicates CD5-1B–EGFP; bottom arrow, EGFP. (F) Confocal microscopy analysis (× 1000) of the subcellular localization of fusion CD5 E1B transcript and EGFP in COS-1 cells. Confocal images were acquired using constant settings on a Leica TCS-NT fluorescence microscope with a 100 ×/1.40 NA oil objective using a FITC filter and Leica TCS-NT software version 1.6.587 (Leica Microsystems, Bensheim, Germany). Image sizes were adjusted using Adobe Photoshop.
Alternative CD5 E1B motif encodes a truncated isoform of CD5 protein starting at exon 3 and retained intracellularly. (A) In vitro translation of CD5 using pLuc (C+), pDNRdual (C–), CD5-1B pDNRdual (CD5-1B) using the TNT T7-coupled reticulocyte system. (B) Precipitation of CD5 in T cells and 697 pre-B cells using antiphosphotyrosine (PY). MW indicates molecular weight; IP, immunoprecipitation. (C) FACS analysis of Jurkat T and 697 pre-B cells for CD5 protein expression in the absence (membrane CD5) or in the presence (intracellular CD5) of saponin. Filled histograms denote CD5 staining; open histograms, isotypic controls. (D) Immunohistochemical staining with UCHT2 of saponin-treated 697 pre-B cells. Images were acquired as in Figure 3B. (E) Western blot analysis of COS-1 cells transfected with EGFP-expressing vector, or fusion CD5 E1B transcript and EGFP. Top arrow indicates CD5-1B–EGFP; bottom arrow, EGFP. (F) Confocal microscopy analysis (× 1000) of the subcellular localization of fusion CD5 E1B transcript and EGFP in COS-1 cells. Confocal images were acquired using constant settings on a Leica TCS-NT fluorescence microscope with a 100 ×/1.40 NA oil objective using a FITC filter and Leica TCS-NT software version 1.6.587 (Leica Microsystems, Bensheim, Germany). Image sizes were adjusted using Adobe Photoshop.
The results of the intracellular versus membrane expression of the presumed CD5 protein isoforms in the 2 cell lines was confirmed by immunocytochemistry. The CD5 molecules recognized by UCHT2 were expressed on the membrane of Jurkat T cells but confined to the cytoplasm of 697 pre-B cells (Figure 5D).
To confirm that E1B encodes an intracellular isoform of CD5, we fused the cDNA for E1B-type transcripts with the GFP gene and expressed the recombinant protein. SDS-PAGE analysis showed GFP expression (bottom arrow in Figure 5E) and the CD5-E1B-GFP recognized by anti-GFP mAb (top arrow in Figure 5E). In addition, COS-1 cells transfected with the construct containing the E1B transcript and the GFP gene showed that the E1B-containing construct directed intracellular protein synthesis.
Effect of E1B on the membrane expression of CD5
Introduction of CD5-1B cDNA into Jurkat cells reduced the membrane expression of the CD5 protein (Figure 6A). This could not be ascribed to a limited transcription of the CD5-1A–containing variant as established by quantitative PCR (Figure 6B). The anti-CD5 mAb Leu1 (Figure 6C) did not affect the amount of full-length CD5 protein. Our data suggest that truncated CD5 acts not to decrease the synthesis of the full-length variant of this protein but to prevent its translocation to the membrane.
Discussion
Expression of CD5 is regulated at multiple levels, among which is the protein level. We provide evidence for the latter mechanism in humans. E1B was discovered within the CD5 gene. This encodes a protein truncated and, therefore, retained in the cytoplasm, thus down-regulating the membrane expression of CD5. E1B lies 8.2 kb upstream of E1A. Because each of these 2 exons 1 possesses its own transcriptional promoter, they may be mutually exclusive. Sequence analyses indicate that a germline insertion of an HERV sequence underpins the new promoter within the CD5 gene, a setting similar to the expression of the growth factor pleiothrophin by trophoblasts.22 It is an important difference between the human and murine systems (Y.R. and P.Y., unpublished results, February 2005). Given the evidence for its integration within the CD5 locus after the divergence of New World from Old World monkey lineages, and prior to that of humans from Old World monkey lineages, it is not surprising that E1B was not found in the mouse B cells.
Transcription of E1A affects the translation initiation site of the gene and results in the expression of a full-length CD5 molecule translocated to the membrane. Because E1B transcripts splice out E1A, the initiation site shifts from ATG1 to ATG2 located 171 nucleotides downstream of ATG1 in exon 3. The new transcript is functional because the CD5 protein product is expressed in a plasmid construct with this exon. Its 52-kDa molecular weight matches the protein synthesized in rabbit reticulocytes. The most N-terminal domain of the membrane CD5 molecule exposes 2 epitopes, each recognized by a panel of CD5 mAbs classified according to their target epitope and exemplified by UCHT2 and Leu1.23 Unexpectedly, UCHT2 bound to the truncated variant of CD5, whereas Leu1 did not. This denotes that the epitope recognized by Leu1 is determined by the 11 N-terminal amino acids that are missing in the first domain of the E1B transcript–encoded protein. The inability to detect this incomplete isoform of CD5 might be explained by the use of the Leu1 CD5 mAb in similar studies.
Transfection of E1B-containing cDNA into Jurkat cells reduces their membrane expression of the CD5 protein. (A) The The pLP env vector, with or without E1B-containing cDNA, was introduced into Jurkat cells. The CD5lo cells were sorted and cultured and their membrane density of CD5 determined using PE-conjugated UCHT2 anti-CD5 antibody. (B) Quantitative RT-PCR demonstrates that the diminished expression of CD5 did not result from a reduction in the amount of E1A-containing transcripts. Data represents mean and SD of 3 experiments. (C) The E1A-related variant of CD5 was detected by blotting total cell extracts with the E1A-related variant-specific Leu-1 anti-CD5 antibody.
Transfection of E1B-containing cDNA into Jurkat cells reduces their membrane expression of the CD5 protein. (A) The The pLP env vector, with or without E1B-containing cDNA, was introduced into Jurkat cells. The CD5lo cells were sorted and cultured and their membrane density of CD5 determined using PE-conjugated UCHT2 anti-CD5 antibody. (B) Quantitative RT-PCR demonstrates that the diminished expression of CD5 did not result from a reduction in the amount of E1A-containing transcripts. Data represents mean and SD of 3 experiments. (C) The E1A-related variant of CD5 was detected by blotting total cell extracts with the E1A-related variant-specific Leu-1 anti-CD5 antibody.
Owing to the absence of a leader peptide, E1B-encoded CD5 molecules cannot reach the plasma membrane, as documented by immunohistochemical studies of the 697 pre-B cells that exclusively contain E1B-CD5 protein and analyses of cells transfected with a construct combining GFP and E1B-cDNA. Hence, differential usage of E1A and E1B regulates synthesis of full-length or truncated isoforms of CD5, and, thereby, its membrane expression level.
Whereas E1A was transcribed in both B and T cells, E1B transcripts were undetectable in T cells. This result suggests a relationship between the density of membrane CD5 and the amount of E1A transcripts in the B cells; high quantities of E1A transcripts were found in CLL cells, which express high levels of membrane CD5 compared with trace amounts in CB and adult B lymphocytes. Further, no E1A transcripts were found in 697 pre-B cells, which have no membrane CD5. In contrast to T lymphocytes, activation of B lymphocytes promoted the transcription of E1A transcripts and raised the membrane density of CD5. This observation is consistent with the expression of CD5 following BCR crosslinking, or engagement by T-independent type 2 antigens.24 Thus, up-regulation of CD5 may represent a physiologic response to attenuate signaling through the BCR-CD5 complex, which is crucial in preventing cells from responding to BCR engagement by self-antigen, or being killed by apoptosis.25
The CD5 expression regulation results from an interplay between several mechanisms. At the posttranslational level, shedding of membrane CD5 has been described.26 CD5 internalization, which is enhanced in T cells, but inhibited in B cells, is another mechanism of differential CD5 regulation.27 A signal transduction pathway promoting CD5 gene transcription has also been suggested to exist.28 Differences in CD5 gene expression in B-cell populations are ultimately determined by differences in activated transcription factors, and thus by the membrane receptors engaged. That such a regulation is unique to B cells implies that T cells do not express these receptors, do not activate the downstream transcription factors, and, therefore, do not transcribe CD5-E1B. Based on the B lymphocyte–restricted expression of E1B transcripts, one expects that factors regulating such transcripts are selectively activated in these cells, implying specificity of transcription factors for B cells. This selection results in the usage of one of the 2 exons 1. Comparison of the promoter sequences for the CD5 E1A in humans and mice has revealed a conserved binding site for the Ets-1 transcription factor.29 Other Ets-related transcription factors, such as PU.1, could be involved in the tuning of CD5 expression in B lymphocytes,30 whereas KE2-related transcription factors, such as E47, reduce CD5 expression in thymocytes.31 Other potential binding sites, including SRY, Oct-1, E2A, and AP-1, exist in the promoter region of E1B. Such mechanisms are involved in the expression of protein 4.1R, heme biosynthetic enzymes and its transcription factor.32-34 The NFAT-dependent enhancer, which is necessary for BCR-mediated induction of CD5 expression in murine B cells, might operate equally on human E1A and E1B.10 A critical role for SHP-1 in the regulation of BCR signal transduction has been inferred from hyperresponsiveness of B cells of moth-eaten mice that have a mutation in SHP-1.35
When E1A is selected, the full-length CD5 protein carries SHP-1 to the membrane, increases the concentration of phosphatase in the vicinity of the BCR, and encourages tolerance in autoreactive B lymphocytes. Conversely, when E1B is selected, a truncated CD5 protein is synthesized and SHP-1 is blocked within the endoplasmic reticulum. In that case, the strength of the BCR-mediated signaling increases, enabling the expansion of B lymphocytes. Not only does truncated CD5 protein not bind SHP-1, but it also reduces the expression of the full-length molecule in Jurkat T lymphocytes transfected with E1B-cDNA. As a consequence, following their release and internalization, the membrane CD5 molecules would not be replaced. Current studies are focused on analyzing more accurately how this mechanism regulates the level of membrane CD5 expression.
Prepublished online as Blood First Edition Paper, July 5, 2005; DOI 10.1182/blood-2005-02-0597.
Supported by the Communauté Urbaine de Brest, the Conseil Régional de Bretagne; the Académie Nationale de Médecine; and Catherine Mazzaco, director of Abbott France, Paris, France. R.A.M. is supported by the Arthritis Research Campaign of the United Kingdom. P.Y. and R.A.M. contributed equally to the preparation of this manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Paul Guglielmi and Peter Beverley for the gift of reagents, Catherine Mazzacco (director of Abbott France, Paris, France) for financial support, and Cindy Séné and Simone Forest for secretarial assistance.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal