Abstract
Acute lymphoblastic leukemia (ALL) in infants is characterized by rearrangements of the mixed lineage leukemia (MLL) gene, drug resistance, and a poor treatment outcome. Therefore, novel therapeutic strategies are needed to improve prognosis. Recently, we showed that FLT3 is highly expressed in MLL rearranged ALL (MLL). Here we demonstrate FLT3 expression in infants with MLL (n = 41) to be significantly higher compared to both infant (n = 8; P < .001) and noninfant patients with ALL (n = 23; P = .001) carrying germline MLL genes. Furthermore, leukemic cells from infants with MLL were significantly more sensitive to the Fms-like tyrosine kinase 3 (FLT3) inhibitor PKC412 (N-benzoyl staurosporine) than noninfant ALL cells, and at least as sensitive as internal tandem duplication-positive (ITD+) AML cells. Surprisingly, activation loop mutations only occurred in about 3% (1 of 36) of the cases and no FLT3/ITDs were observed. However, measuring FLT3 phosphorylation in infants with MLL expressing varying levels of wild-type FLT3 revealed that high-level FLT3 expression is associated with ligand-independent FLT3 activation. This suggests that infant MLL cells displaying activated FLT3 as a result of overexpression can be targeted by FLT3 inhibitors such as PKC412. However, at concentrations of PKC412 minimally required to fully inhibit FLT3 phosphorylation, the cytotoxic effects were only fractional. Thus, PKC412-induced apoptosis in infant MLL cells is unlikely to be a consequence of FLT3 inhibition alone but may involve inhibition of multiple other kinases by this drug. (Blood. 2005;106: 2484-2490)
Introduction
Translocations involving the mixed lineage leukemia (MLL, HRX, or ALL1) gene on chromosome band 11q23 are most frequently found in acute lymphoblastic leukemia (ALL) in infants (< 1 year of age), with an incidence as high as about 80%.1,2 The presence of MLL gene rearrangements is an independent prognostic factor for an adverse outcome.2-5 Hence, the prognosis for infant ALL is exceedingly poor with an event-free survival (EFS) of approximately 35%.6 This poor treatment outcome is largely due to cellular drug resistance. Leukemic cells from infants with ALL are significantly more resistant to most chemotherapeutic drugs both in vitro and in vivo, as compared to cells from older children with ALL.7-9 This is especially true for prednisolone and l-asparaginase. Because the clinical outcome for infants with ALL bearing germline MLL genes appears to be much more favorable,9-11 novel therapeutic targets specific for MLL-gene-rearranged infant ALL (infant MLL) are urgently needed.
In search for suitable targets, we recently compared gene expression profiles from MLL-gene-rearranged ALL patients with profiles from both ALL and acute myeloid leukemia (AML) patients carrying germline MLL genes. This study demonstrated that MLL-gene-rearranged ALL is characterized by high-level expression of the gene encoding Fms-like tyrosine kinase 3 (FLT3, STK-1, FLK-2, or CD135).12 FLT3 is a membrane-bound receptor for the hematopoietic growth factor FLT3 ligand (FLT3L or FL) and is important in early hematopoietic development.13 On binding of FLT3L, wild-type FLT3 receptors dimerize and become activated by phosphorylation, positively affecting several signal transduction pathways all of which favor cell survival and proliferation.14 In the absence of ligand binding, FLT3 only has minimal kinase activity as a consequence of auto-inhibition by the juxtamembrane (JM) domain of the receptor.15
With an incidence of approximately 30%, FLT3 is the most frequently mutated gene in AML.16 Mutations in FLT3 appear to activate the receptor in a ligand-independent manner, constitutively promoting proliferation and survival, thus providing the leukemic cell with a growth advantage and transforming capacity. Distinct types of such activating mutations within 2 separate regions of the FLT3 gene have been described. The first are in-frame internal tandem duplications (ITDs) within the JM domain-coding sequence of FLT3.17 These FLT3/ITDs disrupt the auto-inhibitory activity of the JM domain, leading to receptor dimerization and subsequent auto-phosphorylation in the absence of FLT3L.18 The second type of activating mutations affect either Asp835 or Ile836 within the second tyrosine kinase domain of the FLT3 receptor. At first, point mutations were described that resulted in alternative amino acids at Asp835.19 Recently, insertions after Asp835 as well as deletions of the adjacent codon Ile836 (Δ836) have been reported.20,21 Analogous to point mutations at Asp816 within a corresponding domain of the receptor tyrosine kinase c-KIT,22 these mutations alter the conformation of the activation loop from an inactive to an active state, allowing auto-phosphorylation and thus activation of FLT3 again in the absence of its ligand.14
The identification of activating mutations and their high incidence in patients with AML have led to the development of several small-molecule inhibitors to selectively target the constitutive FLT3 signal, inducing leukemic cells to undergo programmed cell death (apoptosis). The potential of several FLT3 inhibitors as therapeutic drugs has been or is currently being tested in phase 1/2 clinical trials in adults with relapsed or refractory AML or myelodysplastic syndromes (MDS).23-27 Recently we found the staurosporine derivative PKC412 (N-benzoyl staurosporine), a known inhibitor of FLT3,28 to be cytotoxic to acute lymphoblastic leukemia cell lines carrying translocations involving MLL and activated FLT3 receptors as a consequence of either mutation or overexpression of wild-type FLT3.21 Moreover, PKC412 also appeared to be active in vivo, efficiently targeting human MLL rearranged ALL cells overexpressing wild-type FLT3, in mice.21 The present study was designed to explore FLT3 as a therapeutic target in primary patient samples from infants with MLL. Therefore, we measured FLT3 as well as FLT3L mRNA expression levels in a large cohort of infants with MLL and compared these to the expression levels of these genes in both infants and older children (noninfants) with ALL carrying germline MLL genes. Additionally, the cytotoxic effects of PKC412 were determined in primary infant MLL and noninfant ALL samples. Finally, we assessed the infant MLL samples for the presence of activating mutations in FLT3 and analyzed the level of FLT3 receptor phosphorylation in patients carrying either mutated or wild-type FLT3.
Patients, materials, and methods
Patient samples
Primary bone marrow or peripheral-blood samples from untreated infants (< 1 year of age) initially diagnosed with ALL were collected at the Erasmus Medical Center (MC)/Sophia Children's Hospital and other hospitals participating in the INTERFANT-99 treatment study. Samples from pediatric ALL patients older than 1 year of age were obtained either from the German Cooperative ALL (COALL) study group or the Erasmus MC/Sophia Children's Hospital. Approval was obtained from the Erasmus MC Institutional Review Board for these studies. Informed consent was provided according to the Declaration of Helsinki. Within 24 hours after sampling, mononuclear cells were isolated by density gradient centrifugation using Lymphoprep (density 1.077 g/mL; Nycomed Pharma, Oslo, Norway), centrifuged at 480g for 15 minutes at room temperature. Isolated mononuclear cells were washed twice in phosphate-buffered saline (PBS) and resuspended in RPMI 1640 medium (Dutch modification without l-glutamine; Invitrogen Life Technologies, Breda, The Netherlands) supplemented with 20% fetal calf serum (FCS; Integro, Zaandam, The Netherlands), 2 mM l-glutamine (Invitrogen Life Technologies), 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL sodium selenite (ITS media supplement; Sigma, St Louis, MO), 100 IU/mL penicillin, 100 μg/mL streptomycin, 0.125 μg/mL amphotericin B, and 0.2 mg/mL gentamicin (Invitrogen Life Technologies). Contaminating nonleukemic cells were removed using immunomagnetic beads as described by Kaspers et al.29 All samples used contained more than 90% leukemic cells, as determined morphologically on cytospins stained with May-Grünwald-Giemsa stain (Merck, Darmstadt, Germany).
Patient characteristics were collected in reference laboratories of the INTERFANT-99 and the COALL study groups. As part of the INTERFANT-99 study, the infant ALL samples were screened for the presence of MLL rearrangements by fluorescent in situ hybridization (FISH) analysis, and the type of translocation determined using polymerase chain reaction (PCR). In the present study 35% of the infant ALL samples carried a t(4;11), 28.3% a t(11;19), 10% a t(9;11), and 13.3% had germline MLL genes. The remaining 13.3% of the samples either carried other less frequently found translocations involving the MLL gene, or PCR analysis could not be performed due to lack of material.
RNA and DNA extraction
Total RNA and gDNA were extracted from a minimum of 5 × 106 leukemic cells using TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer's instructions with minor modifications. Quantification of both RNA and DNA was performed using a spectrophotometer and the integrity of the extracted RNA was assessed on 1% agarose gels.
Quantitative real-time PCR
Extracted RNA was reverse transcribed and the obtained cDNA was used to quantify FLT3 and FLT3L mRNA expression relative to the endogenous housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), using quantitative real-time PCR (TaqMan) as described previously.30 Primer and probe combinations used to amplify and detect FLT3, FLT3L and GAPDH expression are listed in Table 1.
Primer and probe combinations used for quantitative real-time PCR
Target gene . | Oligonucleotide sequence . |
|---|---|
| FLT3 | |
| Forward | 5′-AGC ATC CCA GTC AAT CAG-3′ |
| Reverse | 5′-CTG GCT GGT GCT TAT GA-3′ |
| Probe | 5′- (FAM) -TTA AAG CCT ACC CAC AAA TCA GAT GT- (TAMRA) -3′ |
| FLT3L | |
| Forward | 5′-GAG CCC AAC AAC CTA TCT C-3′ |
| Reverse | 5′-GGA CGA AGC GAA GAC A-3′ |
| Probe | 5′- (FAM) -ATG GAG CGG CTC AAG ACT GT- (TAMRA) -3′ |
| GAPDH | |
| Forward | 5′-GTC GGA GTC AAC GGA TT-3′ |
| Reverse | 5′-AAG CTT CCC GTT CTC AG-3′ |
| Probe | 5′- (FAM) -TCA ACT ACA TGG TTT ACA TGT TCC AA- (TAMRA) -3′ |
Target gene . | Oligonucleotide sequence . |
|---|---|
| FLT3 | |
| Forward | 5′-AGC ATC CCA GTC AAT CAG-3′ |
| Reverse | 5′-CTG GCT GGT GCT TAT GA-3′ |
| Probe | 5′- (FAM) -TTA AAG CCT ACC CAC AAA TCA GAT GT- (TAMRA) -3′ |
| FLT3L | |
| Forward | 5′-GAG CCC AAC AAC CTA TCT C-3′ |
| Reverse | 5′-GGA CGA AGC GAA GAC A-3′ |
| Probe | 5′- (FAM) -ATG GAG CGG CTC AAG ACT GT- (TAMRA) -3′ |
| GAPDH | |
| Forward | 5′-GTC GGA GTC AAC GGA TT-3′ |
| Reverse | 5′-AAG CTT CCC GTT CTC AG-3′ |
| Probe | 5′- (FAM) -TCA ACT ACA TGG TTT ACA TGT TCC AA- (TAMRA) -3′ |
FAM indicates 6-carboxyfluorescein; TAMRA, 6-carboxytetramethyl-rhodamine.
Detection of FLT3/ITDs
Detection of ITDs of the JM domain of FLT3 was performed as described by Kiyoi et al.31 PCRs were carried out in a total reaction volume of 50 μL containing TaqMan buffer II (Applied Biosystems, Foster City, CA), 2 mM MgCl2, 200 μM of each dNTP (Amersham Pharmacia Biotech, Uppsala, Sweden), 300 nM forward and reverse primer,31 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems), and 500 ng gDNA as a template. Samples were heated for 10 minutes at 95°C to activate the AmpliTaq Gold polymerase and amplified during 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C. gDNA extracted from the MV4-11 cell line, which has been shown to posses a FLT3/ITD,32 was used a as positive control.
Detection of FLT3 activation-loop mutations
Detection of the activating mutations affecting either Asp835 or Ile836 within the activation loop of the FLT3 gene was performed essentially as described by Yamamoto et al.19 However, to fit our standard PCR procedure, a different set of primers (FLT3 forward: 5′-TCA CCG GTA CCT CCT ACT G-3′; reverse: ACT G-3′, 5′-AAA TGC ACC ACA GTG AGT G-3′) was designed to amplify the region of interest. To detect mutations, PCR products amplified as described (see “Detection of FLT3/ITDs”), were digested overnight at 37°C using the restriction enzyme EcoRV. Incomplete digested PCR products, visualized on 2% agarose gels, were extracted from the gel using the Wizard SV gel and Clean-up system (Promega, Leiden, The Netherlands). Clean undigested PCR fragments were cloned into pCR2.1 plasmids using a TA cloning kit (Invitrogen Life Technologies) and transformed into competent Escherichia coli (DH5α) cells by heat shock. Individual clones were recovered from overnight cultures using the Wizard Plus SV Minipreps DNA purification system (Promega) and sequenced on a 310 Genetic Analyzer (Applied Biosystems) using the BigDye Terminator v1.1 cycle sequencing protocol (Applied Biosystems) to confirm the presence of a mutation.
Detection of FLT3 gene amplification using FISH analysis
The presence of FLT3 amplification was determined with dual-color FISH analysis on cytospin preparations. Two BAC clones, 153M24 and 179F17, isolated from the human Bacteria Artificial Chromosome (BAC) library RPCI-11 (Children's Hospital Oakland Research Institute, BACPAC Resources, Oakland, CA), containing FLT3 sequences were used as probes. Probes were labeled by nick translation using digoxigenin-11-deoxyuridene triphosphate (dUTP) for 179F17 and biotin-16-dUTP for 153M24 and were hybridized and detected as previously described.33 The hybridization mixture contained 50 ng of each labeled probe and 5 μg human Cot-1 DNA. In all cases 2 independent observers examined 100 to 200 nuclei.
In vitro PKC412 cytotoxicity using MTT assay
In vitro cytotoxicity to PKC412 was determined using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazoliumbromide (MTT) assay as described previously.34 Briefly, leukemic cells were cultured in round-bottomed 96-well microtiter plates in the presence of 6 concentrations of PKC412 (N-benzoyl staurosporine; kindly provided by Thomas Meyer, Novartis Pharma, Basel, Switzerland), with the highest concentration of 10 μM and a 3-fold dilution factor. Control cells were cultured in 8 wells in the absence of PKC412. Four wells containing 100 μL culture medium were used as blanks. After incubating the plates for 4 days at 37°C in humidified air containing 5% CO2, 10 μL MTT, (5 mg/mL; Sigma) was added and the plates were incubated for an additional 6 hours under the same conditions. During this final 6-hour incubation, the yellow MTT tetrazolium salt is reduced to purple-blue formazan crystals by viable cells only. Formazan crystals were dissolved by adding 100 μL acidified isopropanol (0.04 N HCl-isopropyl alcohol) and the optical density, which is linearly related to the number of viable cells,35 was measured at 562 nm on a spectrophotometer. Assay results were deemed successful when a minimum of 70% leukemic cells was present in the control wells after 4 days of incubation and when the control optical density exceeds 0.050.34
Immunoprecipitation and Western blot analysis of FLT3 phosphorylation
Leukemic cells were cultured both in the absence and presence of 500 nM PKC412. After 4 hours of exposure to PKC412, cells were washed twice in ice-cold PBS and resuspended in 100 μL lysis buffer composed of 25 mM Tris (tris(hydroxymethyl)aminomethane) buffer, 150 mM NaCl, 5 mM EDTA (ethylenediaminetetraacetic acid), 10% glycerol, 1% Triton X-100, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 10 mM glycerolphosphate, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% aprotinin (Sigma), 10 mM sodium fluoride, and 20 μL freshly prepared sodium pervanadate. Accordingly, cell lysis was allowed for 30 minutes on ice. Cell lysates were cleared by centrifugation for 15 minutes at 10 000 g (13 000 rpm) and 4°C. Protein concentration was determined using the bicinchoninic acid (BCA) protein assay (Pierce Biotechnology, Rockford, IL) with different concentrations of bovine serum albumin (BSA) as standards.
For immunoprecipitation (IP), aliquots of whole cellular lysates containing 500 μg protein were precleared with 10 μL G-plus Agarose (Santa Cruz Biotechnology, Santa Cruz, CA) and accordingly IP was performed with 10 μL G-plus Agarose supplemented with 3 μg rabbit polyclonal antibodies specific for the human FLT3 (Santa Cruz Biotechnology). Following denaturation in Laemmli buffer, precipitates were separated on 4% to 12% polyacrylamide gradient gels and separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes on which tyrosine phosphorylation was detected using 4G10 mouse antibodies (Upstate Biotechnology, Lake Placid, NY) and visualized using peroxidase-labeled secondary rabbit anti-mouse antibodies and chemiluminescence (ECL kit; Amersham, Little Chalfont, United Kingdom). To confirm FLT3 receptor expression, blots were stripped and reprobed with anti-rabbit polyclonal antibodies against FLT3 (Santa Cruz Biotechnology).
Statistical analysis
Differences in FLT3 and FLT3L expression between patient groups were statistically evaluated using the Mann-Whitney U test. Differences in mean cytotoxicity responses between patient groups were statistically analyzed using the Student t test. All analyses were 2-tailed, and differences were considered statistically significant at P values of less than .05.
Results
Using quantitative real-time PCR we confirmed our earlier gene expression profiling data12 ; elevated FLT3 mRNA expression levels were detected in MLL-gene-rearranged infant ALL (infant MLL) as compared to both infants and older children (noninfants) carrying germline MLL genes (Figure 1A). Infant MLL patients (n = 41) expressed significantly (P < .001) about 16-fold higher levels of FLT3 mRNA as compared to infant ALL patients (n = 8) and about 2-fold higher than noninfant ALL patients (n = 22; P = .001). No differences in FLT3L expression were observed between infant MLL (n = 33), infant ALL (n = 4), and noninfant ALL (n = 10) patients (Figure 1B).
Relative FLT3 and FLT3L expression in childhood ALL with and without MLL gene rearrangements. (A) Relative mRNA expression of the Fms-like tyrosine kinase 3 (FLT3) as determined by quantitative real-time PCR (TaqMan) in infants with MLL (n = 41) and both infants (n = 8) and noninfants (n = 22) with ALL harboring germline MLL genes. (B) Relative FLT3 ligand (FLT3L) expression in primary infant MLL (n = 33), infant ALL (n = 4), and noninfant ALL (n = 10) samples. ○ indicates individual patients; and —, median expression values. Differences in FLT3 and FLT3L expression between 2 patient groups were statistically analyzed using the Mann-Whitney U test. NS indicates that P was higher than .05 in all comparisons.
Relative FLT3 and FLT3L expression in childhood ALL with and without MLL gene rearrangements. (A) Relative mRNA expression of the Fms-like tyrosine kinase 3 (FLT3) as determined by quantitative real-time PCR (TaqMan) in infants with MLL (n = 41) and both infants (n = 8) and noninfants (n = 22) with ALL harboring germline MLL genes. (B) Relative FLT3 ligand (FLT3L) expression in primary infant MLL (n = 33), infant ALL (n = 4), and noninfant ALL (n = 10) samples. ○ indicates individual patients; and —, median expression values. Differences in FLT3 and FLT3L expression between 2 patient groups were statistically analyzed using the Mann-Whitney U test. NS indicates that P was higher than .05 in all comparisons.
PKC412 has been shown to be cytotoxic to lymphoblastic leukemia cell lines carrying MLL gene rearrangements.20 In the present study we investigated the effects of PKC412 on primary infant MLL cells. In vitro PKC412 cytotoxicity was determined in 29 infant MLL and 19 noninfant ALL patients, as well as in 5 AML patients carrying FLT3/ITDs. Figure 2A shows the mean cytotoxic response at 4 increasing concentrations of PKC412. At PKC412 concentrations ranging from 40 to 375 nM, no response is observed in the noninfant ALL samples, whereas these concentrations of PKC412 increasingly induced leukemic-cell death in the infant MLL samples (Figure 2A). The differences in cytotoxic response to PKC412 between infant MLL and noninfant ALL samples were statistically significant at PKC412 concentrations of 125 nM and higher (P ≤ .01). Moreover, the cytotoxic response to PKC412 in infant MLL was comparable to the response observed in FLT3/ITD+ AML samples (Figure 2A). Using the median FLT3 expression from Figure 1 as the cut-off value, the infant MLL samples for which both FLT3 expression and PKC412 cytotoxicity data were obtained were divided into 2 groups expressing either low or high levels of FLT3 transcripts. As shown in Figure 2B, the cytotoxic response to PKC412 is more pronounced in infant MLL samples expressing high FLT3 mRNA levels as compared to infant MLL samples expressing lower levels of FLT3, although these differences did not reach statistical significance.
In vitro PKC412 cytotoxicity in childhood ALL with and without MLL gene rearrangements. (A) MTT dose-response curves showing the mean cytotoxic response to PKC412 in primary leukemic cells from infants with MLL (n = 29), noninfants with ALL (n = 19), and FLT3/ITD+ AML patients (n = 5). (B) MTT dose-response curves showing the mean cytotoxic response to PKC412 in infant MLL samples expressing high levels of FLT3 (n = 9) compared to infant MLL samples expressing lower FLT3 levels (n = 8). Error bars represent SEM.
In vitro PKC412 cytotoxicity in childhood ALL with and without MLL gene rearrangements. (A) MTT dose-response curves showing the mean cytotoxic response to PKC412 in primary leukemic cells from infants with MLL (n = 29), noninfants with ALL (n = 19), and FLT3/ITD+ AML patients (n = 5). (B) MTT dose-response curves showing the mean cytotoxic response to PKC412 in infant MLL samples expressing high levels of FLT3 (n = 9) compared to infant MLL samples expressing lower FLT3 levels (n = 8). Error bars represent SEM.
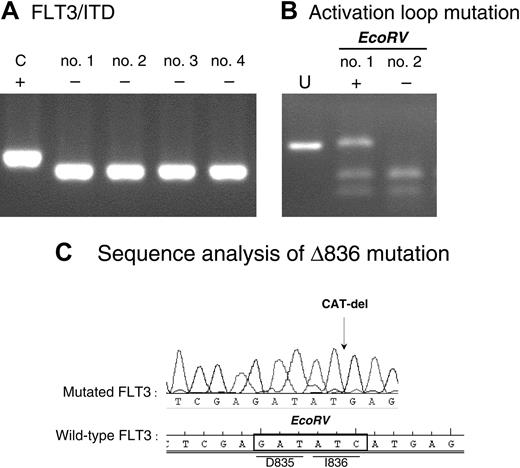
To investigate whether activating mutations in FLT3 could explain the observed sensitivity to PKC412, we next assessed the infant MLL samples for the presence of both ITDs within the JM domain and activation loop mutations affecting either Asp835 or Ile836. None of the 39 infant MLL samples tested appeared to exhibit a FLT3/ITD. Figure 3A shows the amplified region surrounding the FLT3/ITD in MV4;11 cells as a positive control and 4 examples of FLT3/ITD- infant MLL patients. Of 36 infant MLL samples tested only one patient (∼3%) harbored a mutation within the activation loop FLT3. Figure 3B shows incomplete EcoRV digestion of the amplified region harboring codons Asp835 and Ile836 (encompassing the EcoRV recognition sequence) within the FLT3 activation loop from this patient. Complete digestion of this PCR product from a patient sample negative for this type of mutation is also shown. Sequence analysis of the mutation revealed that this patient carried a 3-nucleotide (CAT) deletion that results in a deletion of isoleucine 836 (Δ836), as shown in Figure 3C.
FLT3-activating mutations in infant MLL. (A) ITD of the JM domain of FLT3 in the MV4-11 cell line (C indicates positive control) and 4 examples of infant MLL patients negative for FLT3/ITDs as determined by PCR. (B) Amplified PCR product covering the EcoRV site within the activation loop of FLT3, undigested (U) and digested in both a patient positive (+) and negative (-) for this type of mutation. (C) Sequence analysis of the activation loop mutation found in 1 of 36 patients tested.
FLT3-activating mutations in infant MLL. (A) ITD of the JM domain of FLT3 in the MV4-11 cell line (C indicates positive control) and 4 examples of infant MLL patients negative for FLT3/ITDs as determined by PCR. (B) Amplified PCR product covering the EcoRV site within the activation loop of FLT3, undigested (U) and digested in both a patient positive (+) and negative (-) for this type of mutation. (C) Sequence analysis of the activation loop mutation found in 1 of 36 patients tested.
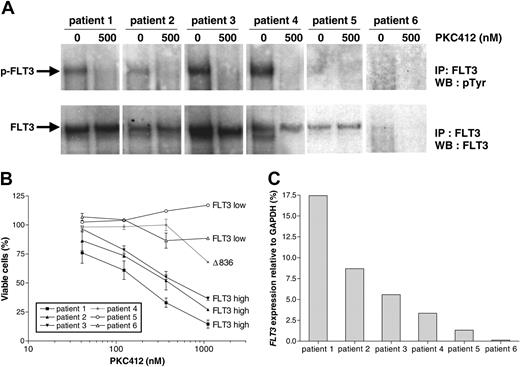
Because the frequency of known activating FLT3 mutations in our cohort of infant MLL patients appeared to be very low, whereas the infant MLL cells are more sensitive to the FLT3 inhibitor PKC412 as compared to leukemic cells from noninfant ALL patients (Figure 2), we asked whether high-level expression of FLT3 is sufficient to auto-phosphorylate and thus activate FLT3 in the absence of activating mutations. To study this, FLT3 phosphorylation was assessed in several infant MLL patients displaying varying levels of FLT3 expression as well as in a noninfant ALL patient (Figure 4). The first 3 patients express high levels of wild-type FLT3 (as determined by sequence analysis of the entire FLT3 gene) and exhibit significant levels of FLT3 phosphorylation and thus activation, which was completely reversed on exposure of the leukemic cells to 500 nM PKC412 for 4 hours (Figure 4A). Patient no. 4 represents the one infant MLL patient in whom the Δ836 activation loop mutation was identified. As expected, reversible FLT3 phosphorylation was also detected in this sample (Figure 4A). Patients no. 5 and no. 6 are samples from an infant MLL and a noninfant ALL patient, respectively, expressing low levels of FLT3 (Figure 4C). No FLT3 phosphorylation could be observed in these samples (Figure 4A). Figure 4B clearly demonstrates that the patients expressing high levels of wild-type FLT3 are sensitive to PKC412, whereas the samples from patients expressing low FLT3 levels that lack FLT3 phosphorylation do not respond to this drug. Patient no. 4, carrying the Δ836 activation loop mutation, seems to respond only at concentrations of PKC412 above 300 nM (Figure 4B).
Relation between high-level FLT3 expression, FLT3 activation, and in vitro sensitivity to PKC412. (A) Immunoprecipitation (IP) analysis of FLT3 in primary infant MLL cells carrying wild-type (patient nos. 1-3 and 5) or mutated (patient no. 4) FLT3, cultured for 4 hours in the absence and presence of 500 nM PCK412. Patient no. 6 represents a noninfant ALL patient. To determine the phosphotyrosine (pTyr) content of FLT3, immunoblots were probed with antiphosphotyrosine (4G10) and with anti-FLT3 to assess FLT3 loading. (B) MTT dose-response curves show the mean cytotoxic response to PKC412 for the individual patients. Error bars represent SEM of duplicate wells. (C) Representation of the FLT3 expression levels for the individual patients. WB indicates Western blot.
Relation between high-level FLT3 expression, FLT3 activation, and in vitro sensitivity to PKC412. (A) Immunoprecipitation (IP) analysis of FLT3 in primary infant MLL cells carrying wild-type (patient nos. 1-3 and 5) or mutated (patient no. 4) FLT3, cultured for 4 hours in the absence and presence of 500 nM PCK412. Patient no. 6 represents a noninfant ALL patient. To determine the phosphotyrosine (pTyr) content of FLT3, immunoblots were probed with antiphosphotyrosine (4G10) and with anti-FLT3 to assess FLT3 loading. (B) MTT dose-response curves show the mean cytotoxic response to PKC412 for the individual patients. Error bars represent SEM of duplicate wells. (C) Representation of the FLT3 expression levels for the individual patients. WB indicates Western blot.
In the MLL rearranged ALL cell line SEMK2-M1, overexpression of FLT3 has been shown to be due to amplification of the FLT3 gene on chromosome 13q12.21 To investigate whether gene amplifications of FLT3 also explained high-level FLT3 expression as observed in primary MLL rearranged ALL cells, we screened infant MLL (n = 39) samples for FLT3 amplifications using FISH analysis. However, none of the 39 samples tested showed amplified FLT3.
Discussion
The class 3 receptor tyrosine kinase Fms-like tyrosine kinase 3 (FLT3) is one of the most frequently mutated genes in hematologic malignancies, including both AML and ALL.14 ITDs of the JM domain and mutations within the activation loop of the receptor appeared to constitutively activate the receptor in a ligand-independent manner, consequently promoting proliferation and survival.19,31 It was recently shown by microarray analysis that FLT3 is overexpressed in patients with MLL-gene-rearranged ALL (designated MLL), when compared to patients with both conventional ALL and AML lacking chromosomal abnormalities involving the MLL gene.12 Furthermore, human leukemia cell lines carrying chromosomal translocations involving MLL and activated FLT3 are sensitive to FLT3 inhibition in vitro and in a mouse model of MLL.21 In the present study, we show that overexpression of wild-type FLT3 is associated with sensitivity to PKC412 in primary infant MLL cells.
Using quantitative real-time PCR analysis we confirmed high-level FLT3 expression in leukemic cells from patients carrying MLL gene rearrangements as previously shown by gene expression profiling.12 Infant MLL patients significantly expressed higher levels of FLT3 mRNA as compared to both infant ALL and noninfant ALL samples harboring germline MLL genes. Screening the cohort of infant MLL patients for the presence of described activating mutations in FLT3, however, showed these mutations to be extremely rare in this group of patients. Nevertheless, comparing the mean cytotoxic response of the FLT3 inhibitor PKC412 in leukemic cells from infant MLL and noninfant ALL patients, we observed that infant MLL patients are markedly more sensitive to this drug. To evaluate the relative sensitivity of infant MLL samples to PKC412 with respect to patient samples harboring activating mutations, we also assessed the cytotoxic response of PKC412 to several FLT3/ITD+ AML samples. Infant MLL patients appeared at least equally as sensitive to PKC412 as AML patients carrying FLT3/ITDs. Interestingly, infant MLL patients displaying high-level FLT3 expression tended to be more sensitive to PKC412 as compared to infant MLL patients expressing lower levels of FLT3, suggesting a relation between the level of FLT3 expression and sensitivity to PKC412. In addition, we observed that high-level FLT3 expression is associated with FLT3 phosphorylation, which is in concordance with previous data.36 This suggests that high-level expression of wild-type FLT3 may indeed be sufficient to activate the receptor in the absence of ligand binding, thereby sensitizing these leukemic cells to FLT3 inhibition.
The most frequent type of activating mutations found in AML are FLT3/ITDs, which occur in approximately 24% of the adult cases and in about 10% to 15% of the childhood cases.14 In adult ALL, tandem duplications of the JM domain are rarely observed.14 Recently Armstrong et al described novel deletions within a 7-amino acid region of the JM domain of FLT3 in 3 of 25 (12%) children with hyperdiploid ALL.37 In the present study, no FLT3/ITDs were observed as primary in infant MLL samples, which is in concordance with data reported by Xu et al.38 In contrast to MLL rearranged ALL, FLT3/ITD is rather frequently observed in AML patients carrying intragenic abnormalities within the MLL gene such as partial tandem duplications.39,40
In comparison to previously reported frequencies, the incidence of activation loop mutations in this cohort of infant MLL samples (∼3%) seems rather low. We have previously reported the incidence of activation loop mutations in another group of MLL samples to be approximately 17% (5 of 30).21 Taketani et al reported 8 of 44 (∼18%) infant MLL cases (< 1 year of age) to harbor mutations within the activation loop.41 Despite this discrepancy it can be concluded that the incidence of activating mutations in infant MLL is less than 20% (3%-18%); thus, activating mutations do not fully explain the relative sensitivity of primary leukemic cells from infant MLL patients to the FLT3 inhibitor PKC412. Recently Zheng et al reported evidence that in primary AML samples expressing both wild-type FLT3 and FLT3L, constitutively activated FLT3 can be detected as a consequence of autocrine signaling.42 Theoretically, this could sensitize these cells to FLT3 inhibition, creating the possibility that the sensitivity to PKC412 as observed in primary infant MLL samples might be explained by elevated FLT3L expression resulting in increased ligand-dependent receptor activation. However, we show that FLT3L expression does not statistically differ between primary infant MLL, infant ALL, and noninfant ALL samples, suggesting that it is unlikely that infant MLL cells are more sensitive to PKC412 than noninfant ALL cells due to elevated autocrine receptor activation. Therefore, the fact that infant MLL patients expressing high amounts of wild-type FLT3 mRNA exhibit pronounced levels of phosphorylated FLT3 that was completely inhibited by 500 nM PKC412 within a 4-hour exposure period, suggests that indeed overexpression of wild-type FLT3 is sufficient to activate FLT3 in a ligand-independent manner.
Thus, overexpression of FLT3 identifies groups of patients sensitive to FLT3 inhibition as is shown in the present study in infant MLL, as well as previously reported in AML patients.43 The reason for FLT3 overexpression in MLL patients, however, remains unclear. One explanation might be that the immunophenotype of infant ALL cells usually is that of a very immature early B-lineage progenitor in which FLT3 expression has been shown to be the highest.15,45 Leukemic cells from infant ALL patients carrying germline MLL genes most often show common or pre-B phenotypes, expressing lower levels of FLT3. The consistently high expression of FLT3 specifically in MLL may also suggest that MLL translocations influence the expression of this gene. In search for a mechanism by which FLT3 expression is elevated in infant MLL, we screened 39 infant MLL samples for the presence of FLT3 gene amplification as found in the MLL rearranged ALL cell line SEMK2, which expresses exceedingly high levels of wild-type FLT3.21 However, none of the patient samples appeared to harbor amplified FLT3, excluding gene amplification as the etiology for FLT3 overexpression.
After the discovery of the high incidence of activating FLT3 mutations in AML, several small-molecule tyrosine kinase inhibitors were developed to target the constitutive FLT3 signal as a potentially novel therapeutic drug. The efficacy of several of the most promising FLT3 inhibitors including PKC412,28 CEP-701,32 and SU541644 is currently being tested in phase 2 clinical trials in adult AML, and preliminary results are encouraging.23,24,26,27 Interestingly, sensitivity toward these inhibitors seems to vary between the different types of activating mutations.45 PKC412, a staurosporine derivative originally identified as a inhibitor of protein kinase C (PKC),46 has been shown to be effective against activated FLT3 resulting from both ITD and mutations in the activation loop of the receptor.45 Moreover, PKC412 has also been shown to display inhibiting activity against several other class 3 receptor tyrosine kinases such as KDR, c-KIT, and platelet-derived growth factor receptor (PDGFR).46 In the present study, we demonstrate that PKC412 exhibits antileukemic activity against primary infant MLL cells with activated FLT3 receptors, whereas these effects were not observed in infant MLL cells lacking activated FLT3. These observations imply that leukemic-cell death induced on exposure to PKC412 in these cells specifically seems due to inhibition of FLT3. However, PKC412 has been shown almost completely inhibit FLT3 phosphorylation at a concentration of 100 nM.21 Although the present study does show a statistically significant difference in mean PKC412 cytotoxicity between infant MLL (∼10% leukemic-cell death) and noninfant ALL (no response) patients at this dose level, the absolute difference is rather small. At higher dosages of PKC412 the differences are more pronounced, and substantial percentages of leukemic-cell death are observed within the infant MLL patient group. Given that FLT3 activation should fully be inhibited at PKC412 concentrations of approximately 100 nM, the increasing amounts of cell death as observed at higher PKC412 concentrations may not specifically be a consequence of FLT3 inhibition, but may rather be due to nonspecific inhibition of any of the multiple other kinases inhibited by PKC412. Therefore, these data suggest that infant MLL samples are significantly more sensitive to multitarget kinase inhibition that includes the inhibition of FLT3. Moreover, our data indicate that FLT3 inhibition alone is not sufficient to induce substantial degrees of leukemic-cell death in primary leukemia samples, including ITD+ AML samples. This latter may suggest that the greater degree of cytotoxicity that has been reported in similar primary MLL samples using the FLT3 inhibitor CEP-70136 may be due to inhibition of additional targets by this agent at concentrations similar to those required to inhibit FLT3. Nevertheless, initial results from phase 1/2 clinical trials using PKC412 or CEP-701 as single agents in refractory adult AML (and MDS) patients show that both inhibitors exhibit potential clinical activity. PKC412, administered orally at a dose of 75 mg 3 times daily, decreased peripheral-blood counts by 50% in 14 of 20 patients (70%).23 CEP-701, given orally at a dose of 60 mg twice daily, significantly reduced bone marrow and peripheral-blood blasts in 5 of 14 patients (36%).24 Thus, receptor tyrosine kinase inhibition using small-molecule FLT3 inhibitors may be a potential therapeutic approach for innovative treatments for MLL-gene-rearranged infant ALL. Based on these findings, a phase 1/2 study using FLT3 inhibitors in childhood leukemia is currently in preparation (R.P.). Possible strategies are to use FLT3 inhibitors in relapsed MLL or selected MLL rearranged cases at very high risk of relapse, either as single agents or in combination with conventional chemotherapeutic drugs, because preclinical studies demonstrating synergistic effects of FLT3 inhibitors with cytarabine (Ara-C) and daunorubicin have been reported.47
In conclusion, the present study shows that FLT3 expression is high in leukemic cells from infants with MLL rearranged ALL and that high FLT3 expression likely is associated with auto-phosphorylation (activation) of FLT3. Infant MLL samples were more sensitive to PKC412 than noninfant ALL samples. However, the concentration of PKC412 required to induce substantial levels of leukemic-cell death in these samples is higher than the concentration needed to completely inhibit FLT3 phosphorylation. Thus, PKC412-induced cell death in infant MLL samples likely is a consequence of multitarget kinase inhibition, including inhibition of FLT3. Therefore, inclusion of multitarget tyrosine kinase inhibitors in current treatment regimens may be a potential novel therapeutic strategy to improve outcome for infant MLL patients.
Prepublished online as Blood First Edition Paper, June 14, 2005; DOI 10.1182/blood-2004-09-3667.
Supported by a grant from the Sophia Foundation for Medical Research (SSWO grant 296). P.N. was supported by Roggenbuck-Stiftung Hamburg. M.H. was supported by Kinderkrebszentrum Hamburg.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Monique Passier, Karin Kazemier, Anne von Bergh, and Anja Voigt for skillful technical assistance. We also thank Thomas Meyer, Pamela Cohen, Alfredo Romano, Debra Resta, and Doriano Fabbro from the Novartis Pharma PKC412 developing team. We further wish to express our gratitude to the members and participating hospitals of the INTERFANT-99 (Coordinated by R. Pieters, Dutch Childhood Oncology Study Group, The Netherlands) and the German COALL (coordinated by G. Janka-Schaub, Children's Hospital Eppendorf, Hamburg, Germany) study groups for supporting this study by providing leukemic samples.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal