Abstract
The fate of hematopoietic stem cells (HSCs) is regulated through a combinatorial action of proteins that determine their self-renewal and/or their commitment to differentiation. Stem cell leukemia/T-cell acute lymphoblastic leukemia 1 (SCL/TAL1), a basic helix-loop-helix (bHLH) transcription factor, plays key roles in controlling the development of primitive and definitive hematopoiesis during mouse development but its function in adult HSCs is still a matter of debate. We report here that the lentiviral-mediated enforced expression of TAL1 in human CD34+ cells marginally affects in vitro the differentiation of committed progenitors, whereas in vivo the repopulation capacity of the long-term SCID (severe combined immunodeficient) mouse–repopulating cells (LT-SRCs) is enhanced. As a consequence, the production of SRC-derived multipotent progenitors as well as erythroid- and myeloid-differentiated cells is increased. Looking at the lymphoid compartment, constitutive TAL1-enforced expression impairs B- but not T-cell differentiation. Expression of a mutant TAL1 protein that cannot bind DNA specifically impairs human LT-SRC amplification, indicating a DNA-binding dependent effect of TAL1 on primitive cell populations. These results indicate that TAL1 expression level regulates immature human hematopoietic cell self-renewal and that this regulation requires TAL1 DNA-binding activity.
Introduction
The hematopoietic system is constituted by a hierarchy of cells that originates from a small population called the hematopoietic stem cells (HSCs). The HSCs can self-renew and are subjected to successive steps of differentiation, leading to the lymphoid, myeloid, and erythroid lineages. The self-renewal process allows the maintenance of an HSC pool, and thus a long-term production of blood cells during the entire life span of individuals.1 Only a limited number of factors are known to control HSC self-renewal, including transcription factors such as HOXB4 (homeobox B4), BMI1 (B-lymphoma Moloney murine leukemia virus insertion region 1), and GATA2 and receptors/ligands such as the NOTCH and WNT (Wingless-type) pathways.2-6
The T-cell acute lymphoblastic leukemia 1 (TAL1; also named TCL-5 [T-cell leukemia 5] or SCL [stem cell leukemia]) protein is a transcription factor that belongs to the basic helix-loop-helix (bHLH) family of proteins and has been characterized in a translocation occurring in T-cell leukemia in children.7 The bHLH transcription factors include 3 main classes of proteins that are different in terms of their structure, expression patterns, capacity to homodimerize or heterodimerize, and DNA binding capacity (see Massari and Murre8 for review). These bHLH factors determine cell fate and direct differentiation of many cell types, as shown for MYF5 (myogenic factor 5) or MYOD (myogenic determination factor) in muscles9 and for Id proteins in hematopoiesis.10 TAL1 is a class II bHLH factor, predominantly expressed, in the adult, in the vascular and hematopoietic systems, precisely in hematopoietic primitive progenitors and erythroid, megakaryocytic, and mast cell precursors.11,12 Tal1 is considered as a master gene for the establishment of primitive and definitive hematopoiesis. Tal1-null embryos fail to develop any hematopoietic cells, and this defect can be rescued by expressing TAL1.13-15 Tal1-null embryonic stem (ES) cells also fail to participate to hematopoiesis when injected into blastocysts but do so for any other tissue genesis.13,15 The role of TAL1 in adult hematopoiesis has been studied using human and murine hematopoietic cell lines, gene transfer strategy in human bone marrow (BM), or umbilical cord blood (UCB) cells and more recently a conditional knock-out strategy in mice.16-18 These studies clearly demonstrated that TAL1 regulates erythroid and megakaryocytic differentiations. However, the importance of TAL1 in the transplantability and self-renewal properties of mouse adult HSCs is unclear because repopulation levels differ widely depending on the transplantation protocol used.16,17,19
The biologic activity of TAL1 relies at least on 2 domains within the protein. Its HLH domain is required for heterodimerization with class I bHLH such as the E2A (early region 2A) proteins,18 and its basic domain is required for binding of the heterodimer to DNA on the E-box consensus sequence (CANNTG) and further transactivation of target genes. Studies of deletions or point mutations in these 2 domains of TAL1 indicated that the DNA-binding domain, but not the HLH domain, is dispensable for some TAL1 properties,20-22 such as the rescue of hematopoietic development from tal1-/- embryonic stem cells.
The role of TAL1 in primary human hematopoietic cells has been poorly evaluated, because it was difficult to genetically modify such cells in a reproducible way, and in vitro and in vivo assays for human immature cells were not easily available. In the course of studying the role of key proteins during human hematopoiesis, we have developed lentiviral vectors and culture conditions that allow high levels of gene transfer and homogeneous transgene expression into the whole human hematopoietic cell hierarchy, including very immature cells.23,24 Using this strategy, we have enforced TAL1 expression into UCB CD34+ cells and analyzed their hematopoietic properties in vitro24 and after transplantation into nonobese diabetic–severe combined immunodeficient (NOD-SCID) mice to study of the effect of TAL1 overexpression on different human hematopoietic cells, including lymphoid- and myeloid-committed progenitors and very immature cells. We show that TAL1 expression level regulates the self-renewal of long-term SCID-repopulating cells (LT-SRCs), the most immature human cells that can be tested,25 whereas committed progenitors are not extensively affected. To evaluate the importance of the DNA-binding property of TAL1 in expanding LT-SRCs, a DNA-binding domain deleted-TAL1 (called ΔbTAL1) was expressed into CD34+ UCB cells. We show that the mutant ΔbTAL1 protein has similar effects than the full-length TAL1 protein in vitro on committed progenitors but interferes with human hematopoiesis in NOD-SCID mice by acting on the proliferation/differentiation, but not on the migration, of LT-SRCs.
Materials and methods
Lentiviral vector
The lentiviral constructs TRIPΔU3-EF1α encoding enhanced green fluorescent protein (GFP), TAL-1, and ΔbTAL-1 have been described.24 The batches of the 3 concentrated vector particles were prepared together to ensure similar transduction efficiencies.
Purification of hematopoietic CD34+ cells
UCB samples were collected with the informed consent of the mothers. CD34+ cells (> 80% purity) were purified from mononuclear cells by immunomagnetic selection (Miltenyi Biotec, Paris, France).
Transduction protocol
Human CD34+ UCB cells were transduced as described.23 Briefly, thawed or freshly purified CD34+ UCB cells (106 cells/mL) were incubated with the lentiviral vectors (2500 ng/mL P24) in Iscoves medium containing 15% BIT serum-free medium (Stem Cell Technologies, Vancouver, Canada) plus stem cell factor (SCF; 100 ng/mL; Amgen, Neuilly-sur-Seine, France), Flt3-ligand (Flt3-L; 100 ng/mL; Immunex, Seattle, WA), thrombopoietin (TPO; 10 ng/mL; Genosys Biotechnologies, St Quentin en Yvelines, France), and interleukin 3 (IL-3; 60 ng/mL; Novartis, Rueil Malmaison, France) during 3 days. At the end of the transduction period, cells were washed and used for transplantation or cultures. Colonies (n = 122, 3 experiments) derived from TRIP-TAL1– and ΔbTAL1-transduced cells grown in colony-forming cell (CFC) assays were harvested, and transgene integration was tested by polymerase chain reaction (PCR). Forty-nine of 61 (82% ± 8%) and 44 of 61 (72% ± 8%) colonies were positive, respectively, for the TRIP-TAL1 and TRIP-ΔbTAL1 vector integration. Flow cytometry analysis of cells transduced with TRIP–GFP vector indicated that 74% ± 20% (n = 5) were GFP+.
Western blot analysis
Western blot was performed exactly as in Ravet et al.24
Lymphoid (B, natural killer [NK]) and myeloid differentiation
Transduced CD34+ cells and CD34+/CD19-CD14-CD15- or CD34+/CD38lo cells sorted from the BM of NOD-SCID mice (2-4 × 103/mL) were cultured during 3 weeks with a confluent layer of murine MS-5 stromal cells in Iscoves medium containing 10% human AB serum and 5% fetal calf serum with SCF (50 ng/mL), recombinant human (rhu)–IL-2 (5 ng/mL; Diaclone, Besançon, France) and rhu–IL-15 (1 ng/mL; Diaclone).26 Culture medium was half changed every week, and cells were harvested by vigorously pipetting the culture at the end of the incubation time. Cells were then subjected to fluorescence-activated cell sorting (FACS) analysis using human lineage-specific antibodies.
Clonogenic and LTC-IC progenitor assays
Assays for CFCs were carried out in methylcellulose cultures supplemented with SCF (50 ng/mL), IL-3 (8 ng/mL), GM-CSF (10 ng/mL; Schering Plough, Dardilly, France), granulocyte colony-stimulating factor (G-CSF; 25 ng/mL; Amgen), and erythropoietin (EPO; 2 U/mL; Kirin Brewery, Gunma, Japan).27 Long-term culture (LTC) assays were carried out on a confluent layer of the MS-5 cell line as in Issaad et al.28
NOD-SCID repopulating assay
NOD/LtSz-scid/scid mice were housed in the animal facility of the Institut A Lwoff (Villejuif, France). They were irradiated at 3.25 Gy and treated with anti-CD122/TMβ1 antibody29,30 24 hours prior to intravenous30 and intrabone31 injections of transduced human UCB CD34+ cells (1-2 × 105 CD34+ cells/mouse).30,31 At the indicated time points, human hematopoietic cells were harvested from the mouse hematopoietic organs and analyzed. In the case of the intrabone injection, the bone marrow cells were harvested separately from the injected femur and from the noninjected long bones. When specified, engrafted human CD45+ cells were isolated using the EasySep kit (StemCell Technologies).
Secondary transplantations were performed using intravenous injection of bone marrow cells pooled from part of the cells of up to 5 primary mice into NOD-SCID secondary recipients. Every secondary mouse received the equivalent of 2 to 2.5 × 106 human CD45+ cells or 2 × 105 CD34+ cells isolated from primary mice. In every secondary transplantation, a minimum of 6 mice was injected per group of primary mice. Cells from primary mice were cultured overnight in Iscoves medium containing 15% BIT with SCF (50 ng/mL), Flt3-L (50 ng/mL), and IL-6 (10 ng/mL) prior to secondary transplantation.
All experiments and procedures were performed in compliance with French Ministry of Agriculture regulations for animal experimentation.
Flow cytometry analysis and cell sorting
Lineage analysis of human hematopoietic cells was performed by flow cytometry.26 Mouse anti–human monoclonal antibodies (MoAbs) used were CD14 (clone RMO52), CD15 (80H5), CD19 (clone J4-119), CD34 (581 and Qbend10), CD45 (J33), and Glycophorin A (GPA; 11E4B-7-6) from Beckman-Coulter/Immunotech (Marseille, France). MoAbs were labeled with fluorescein isothiocyanate, phycoerythrin, phycoerythrincyanin5, or allophycocyanine. In all the FACS analyses, nonspecific staining was measured using isotypic controls such as immunoglobulin G1 (IgG1), IgG2a, and IgM MoAbs.
When specified, human CD34+ cells were purified from mouse bone marrow by immunomagnetic selection (Miltenyi Biotec). CD34+/CD19-CD14-CD15- and CD34+/CD38lo cells were further sorted with an ELITE cytofluorometer (Beckman-Coulter, Miami, FL) equipped with an argon ion laser (Innova 70-4-Coherent Radiation, Palo Alto, CA) tuned to 488 nm and operating at 500 mw. Purity of the cells was greater than 90%. The sorted populations were cultured in CFC and LTC–initiating cell (IC) conditions or processed for gene expression analysis.
Gene expression analysis
RNA was isolated from 104 to 105 sorted cells using 200 μL TRIzol reagent (Invitrogen, Groningen, Netherlands). Total RNA was reverse transcribed using random hexamers and the Superscript reverse transcriptase (RT) kit (Invitrogen) according to manufacturer's instructions. Synthesized cDNA concentration was normalized between samples by varying the number of cycle of PCR using S14 primers. PCR amplification was performed with AmpliTaqGOLD polymerase (Applied Biosystems, Courtaboeuf, France) using primers and conditions described. Semiquantitative analysis was confirmed by real-time quantification PCR running on a light cycler rapid thermal cycler system (Roche Diagnostics, Lewers, United Kingdom) used according to the manufacturer's instructions. cDNA reactions were diluted 1/10 in water and used as template in real-time PCR reactions using the light cycler-DNA Master SYBRGreen I mix (Roche Diagnostics) supplemented with 3.5 mM MgCl2 and 0.5 μM specific primer pairs. The GAPDH housekeeping gene was used as internal control to normalize cDNA input.
Quantitative results for each sample were obtained by the threshold cycle number, which was determined from the crossing point between Ft and the plotted curve. Each RNA value was expressed as the ratio to the GAPDH value. Results obtained with GFP+ control cells were considered as 100%. Each sample was tested at least 2 times.
Primers for semiquantitative PCR were as follows: S14: S, GGCAGACCGAGATGAATCCTCA, and AS, CAGGTCCAGGGGTCTGGTCC (annealing temperature 65°C, 29 cycles or 28, 30, 32 ×); CD19: S, TCACCGTGGCAACCTGACCATG, and AS, GAGACAGCACGTTCCCGTTACTG (65°C, 37 ×); EBF: S, CTCACTTTGAGAAGCAGCCGC, and AS, CATGTCACGTGGGTTTCCCGC (65°C, 32 ×); Pu.1: S, TGGAAGGGTTTCCCCTCGTC, and AS, TGCTGTCCTTCATGTCGCCG (60°C, 32 ×); GATA-1: S, CAGTAAACGGGCAGGTACTC, and AS, CATAAAGCCACCAGCTGGTC (60°C, 32 ×); GATA-2: S, AGCCGGCACCTGTTGTGCAA, and AS, TGACTTCTCCTGCATGCACT (60°C; 28, 30, 32 ×); GATA-3: S, ACTCCTACATGGACGCGGC, and AS, GGTGGATGGACGTCTTGGAG (60°C, 32 ×); HOXB4: S, GGAGCCCGGCCAGCGCTGCGAGG, and AS, ACCCGAGCGGATCTTGGTGTTGGGCAA (68°C; 36, 38, 40 ×); Bmi1: S, CCAGGGCTTTTCAAAAATGA, and AS, GCATCACAGTCATTGCTGCT (60°C; 28, 30, 32 ×).
Expressions of endogenous and transgenic TAL1 were tested using a primer (S), which sequence hybridized with the coding sequence of human TAL1 (“Common TAL1”) and 2 different AS primers. The “Transgenic TAL1” primer hybridizes with the vector sequence and the “Endogenous TAL1” hybridizes with the 3′ noncoding sequence of TAL1 that is missing in the TAL1 lentiviral vector; Common TAL1: S, TCTGAAGCAAGGCGGTGGAC; Endogenous TAL1: AS, GGAAGACCGTGCCGTCTTCA (60°C, 32 ×); Transgenic TAL1: AS, AATCAGCATTGGTAGCTGCTGT (60°C, 32 ×).
Primers for quantitative PCR were as follows: GAPDH: S, GGGAAACTGTGGCGTGAT, and AS, GGAGGAGTGGGTGTCGCTGTT; c-kit receptor tyrosine kinase (KIT): S, TTCTTACCAGGTGGCAAAGG, and AS, AAATGCTTTCAGGTGCCATC; GATA-1: S, CAGTAAACGGGCAGGTACTC, and AS, CATAAAGCCACCAGCTGGTC; GATA-2: S, AGCCGGCACCTGTTGTGCAA, and AS, TGACTTCTCCTGCATGCACT; GATA-3: S, GGATGCCAAGAAGTTTAAGGA, and AS, GGCAACTGGTGAACGGTAACA; Bmi1: S, AGATTGGATCGGAAAGT, and AS, TAGGCAATATCCATTAGTGTA; HOXB4: S, CCGATACCCAGCGAAAGC, and AS, TCAGTGAATGGGCACGAAAGA.
Vector integration
CFC-derived colonies were processed as described previously.24 DNA was purified from 105 purified engrafted CD45+ cells using classic phenolchloroform extraction and resuspended into a volume of 100 μL. Semiquantitative PCR was performed on 1 μL, using the following primers: TAL1 Vector: S, ATCCACTTTGGCTGATACGC, and AS, GGTCATCCTGGGGCATATTT (60°C; 28, 30, 32 ×); BCL6: S, CCGCTGCTCATGATCATTATTT, and AS, TAGACACGATACTTCATCTCAT (55°C; 31, 33, 35 ×).
Statistics
Statistical analysis was performed using the paired Student t test.
Results
In vitro cultures of CD34+ cells with enforced TAL1 expression
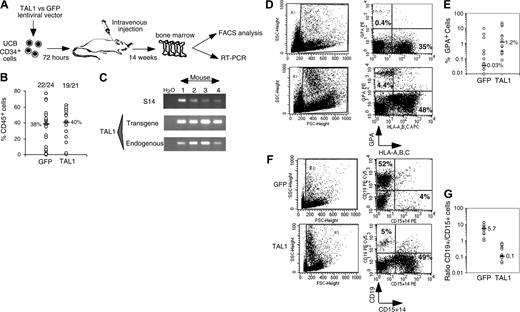
The human TAL1 cDNA was cloned into the TRIPΔU3-EF1α lentiviral vector (Figure 1A) and transduced into CD34+ UCB cells following a protocol that allows gene transfer and expression into greater than 70% of the cells (Figure 1B and as described previously23,24 ). Enforced gene expression resulted in a 6-fold increase of TAL1 expression over the endogenous level as measured by Western blot (Ravet et al24 and Figure 1C). To investigate the biologic function of TAL1 in the human committed progenitor compartment, cells transduced with the TAL1 recombinant vector were cultured during 3 weeks in conditions that allow lymphoid and myeloid differentiations.26 Enforced TAL1 expression decreased the differentiation of CD34+ cells toward B-cell but only mildly affected myeloid cell maturation, with a 40% decrease of CD19+ B cell and a 20% increase of granulomacrophagic (CD14+/CD15+ G/M) cell productions (Figure 1D-E). The mild effect of enforced TAL1 expression on myeloid cell differentiation was also observed when transduced cells were cultured in myeloid-restricted liquid cultures as well as on myeloid clonogenic progenitors (granulocyte/macrophage colony-forming unit [CFU-G/M]) (data not shown and Ravet et al24 ). Moreover, TAL1-expressing cells did not display any cytokine hypersensitivity, as measured in CFCs or in myeloid-restricted liquid cultures (data not shown). Altogether, these results suggested that, in vitro, enforced TAL1 had a limited action on the differentiation of myeloid-committed progenitors.
Effects of enforced TAL1 expression on the differentiation of SRCs
To analyze the effect of enforced TAL1 expression on the primitive cell compartment, transduced cells (equivalent of 1-2 × 105 fresh CD34+ cells/mouse) were intravenously transplanted into irradiated NOD-SCID mice. Human hematopoiesis was allowed to proceed for 14 weeks (Figure 2A) to study the differentiation of the LT-SRCs that are closely related to human HSCs.25
After 14 weeks, human cell engraftment, estimated by the percentage and absolute numbers of CD45+ cells, was similar in the BM of mice that received a transplant with TAL1+ cells and control GFP+ cells (Figure 2B). Transgene expression in engrafted human hematopoietic cells was assessed by FACS analysis for GFP (data not shown) and by RT-PCR for TAL1 (Figure 2C). The biologic activity of the transgenic TAL1 protein was evidenced by the increased proportion of GPA+ cells, because SCL/TAL1 is a positive regulator of erythropoiesis.16-18 GPA+ cells were 40 times increased in SRC-derived TAL1+ cells (median: 1.2%; n = 14 mice; 4 experiments) as compared with GFP+ cells (median: 0.03%; n = 18 mice; 4 experiments) (Figure 2D-E).32,33 Moreover, the proportion of B-lymphoid cells and myeloid cells was inverted in TAL1+ cells (B/myeloid ratio = 0.1) as compared with GFP+ cells (B/myeloid ratio = 5.7) (Figure 2F-G). These data are in agreement with reports showing impaired B-cell development as the result of titration of E2A by TAL1.34,35 The production of granulomonocytic (proportion and absolute cell number of CD14+ and/or CD15+) cells was also increased (D.R. and F.P., unpublished data, December 2003; and Figure 2F) similarly to the results described recently in the mouse.36
Effect of enforced TAL1 expression on the differentiation of CD34+ cells in lymphomyeloid cultures. (A) Lentiviral construct and localization of primers designed to evaluate the transduction efficiency. (B) Transduction efficiency revealed by flow cytometry (GFP+ cells) or by PCR (TAL1+ cells). GFP-transduced cells were cultured during 3 days in lymphoid/myeloid conditions, and GFP expression was detected by flow cytometry (indicated is the percentage of GFP-expressing cells, left). TRIP-TAL1–transduced cells were plated in CFC assay; colonies were harvested after 14 days and individually subjected to PCR analysis to detect the vector integration. Shown in the right panel is a typical analysis of 11 colonies of which 9 are positive for the vector integration. Controls are Jurkat-L4 cells transduced (+) or nontransduced (-) with TRIP-TAL1 vector. (C) Western blot analysis of TAL1 and ΔbTAL1 protein expression in human CD34+ cells transduced with the TRIP vectors. Blots were stripped and reprobed with antiactin antibody as a control of protein loading. (D-E) In vitro production of CD19+ B cells and of CD14+/CD15+ myeloid cells from transduced cells. Immediately after transduction, cells were cultured in lymphoid/myeloid conditions during 3 weeks, and the proportion of CD19+ B cells and CD14+/CD15+ granulomonocytic cells was evaluated by FACS. Shown is a typical experiment (D) and the overall 6 independent experiments performed (E). Numbers indicate the percentage of positive cells among gated cells. Results obtained with GFP+ cells and TAL1+ cells shown (E) are linked for every experiment. Indicated is the ratio (expressed as a mean percentage) obtained by comparing results from TAL1+ cells and GFP+ cells. Each symbol represents an independent experiment.
Effect of enforced TAL1 expression on the differentiation of CD34+ cells in lymphomyeloid cultures. (A) Lentiviral construct and localization of primers designed to evaluate the transduction efficiency. (B) Transduction efficiency revealed by flow cytometry (GFP+ cells) or by PCR (TAL1+ cells). GFP-transduced cells were cultured during 3 days in lymphoid/myeloid conditions, and GFP expression was detected by flow cytometry (indicated is the percentage of GFP-expressing cells, left). TRIP-TAL1–transduced cells were plated in CFC assay; colonies were harvested after 14 days and individually subjected to PCR analysis to detect the vector integration. Shown in the right panel is a typical analysis of 11 colonies of which 9 are positive for the vector integration. Controls are Jurkat-L4 cells transduced (+) or nontransduced (-) with TRIP-TAL1 vector. (C) Western blot analysis of TAL1 and ΔbTAL1 protein expression in human CD34+ cells transduced with the TRIP vectors. Blots were stripped and reprobed with antiactin antibody as a control of protein loading. (D-E) In vitro production of CD19+ B cells and of CD14+/CD15+ myeloid cells from transduced cells. Immediately after transduction, cells were cultured in lymphoid/myeloid conditions during 3 weeks, and the proportion of CD19+ B cells and CD14+/CD15+ granulomonocytic cells was evaluated by FACS. Shown is a typical experiment (D) and the overall 6 independent experiments performed (E). Numbers indicate the percentage of positive cells among gated cells. Results obtained with GFP+ cells and TAL1+ cells shown (E) are linked for every experiment. Indicated is the ratio (expressed as a mean percentage) obtained by comparing results from TAL1+ cells and GFP+ cells. Each symbol represents an independent experiment.
Because TAL1 is involved in human T-cell acute lymphoblastic leukemia (T-ALL), T-cell potential of transduced SRC-derived cells was also investigated in vivo in NOD-SCID thymuses. Indeed, the treatment of NOD-SCID mice with an anti-CD122 monoclonal antibody allows human T-cell development following long-term engraftment.30 Surprisingly, enforced TAL1 expression did not affect T-cell differentiation (data not shown). This result is compatible with in vitro data of xenogenic fetal thymic organ culture (FTOC) that also showed that T-cell differentiation is not altered in the presence of enforced TAL1 expression levels (data not shown).
Effects of enforced TAL1 expression on SRC-derived CD34+ cells
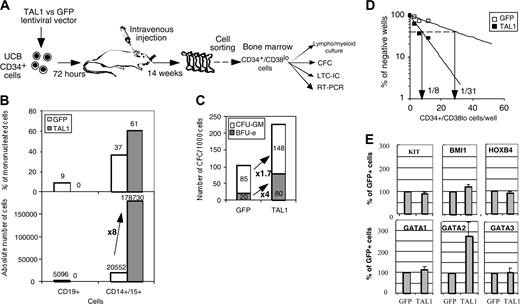
We next investigated whether the NOD-SCID–engrafted CD34+ progenitor compartment was modified by enhanced TAL1 expression. Although enforced TAL1 expression did not play a major role on myeloid cell differentiation in vitro (Figure 1),24 the myeloid cell compartment was expanded in NOD-SCID mice. To study the origin of this expansion, we studied the percentage of cells with an immature phenotype (CD34+/CD19-CD15-CD14- [lineage negative, Lin-]) and found that it was 4-fold higher in the TAL1+ cell population than in the GFP+ cell population (Figure 3A-B). RT-PCR analysis demonstrated that the primitive TAL1+/CD34+/Lin- cells had a myeloid/erythroid profiling (CD19-/EBF-/GATA1+/PU1high), whereas control GFP+/CD34+/Lin- cells were lymphoid restricted (CD19lo/EBF+ [early B-cell factor]/GATA1-/PU1low) (Figure 3C). Altogether, these results indicate that, in vivo, enforced TAL1 expression prevents B-cell differentiation, promotes myeloid and erythroid cell development, and increases the number of cells with a primitive CD34+/Lin- phenotype.
To evaluate whether the effects of TAL1 overexpression in NOD-SCID mice resulted from an increase of the progenitor cell compartment, lymphoid/myeloid, CFC, and LTC-IC potentials of sorted NOD-SCID–derived CD34+/Lin- cells were characterized (Figure S1; see the Supplemental Figure link at the top of the online article, at the Blood website). Similar assays were performed on CD34+/CD38lo cells (Figure 4A). TAL1+/SRC-derived CD34+/Lin- and CD34+/CD38lo cell populations exhibited similar functional characteristics. These include a reduced in vitro capacity to generate human CD19+ B cells, whereas their CD14+/CD15+ myeloid cell progeny was increased (Figure 4B and Figure S1A). The enforced TAL1 expression increased the SRC-derived CFC compartment by up to 6-fold, and this effect was obtained on both CFU-GMs and BFU-Es (erythroid burst-forming units) (Figure 4C and Figure S1B). As expected the increase in the number of BFU-Es was higher than that of CFU-GMs, probably resulting from an effect on immature progenitors and from a specific effect on erythroid-committed progenitors.24 Finally enforced TAL1 expression enhanced LTC-IC potentials from engrafted CD34+/Lin- and CD34+/CD38lo cells as measured by quantitative limiting dilution analysis. LTC-IC frequencies were 4- to 11-fold increased in TAL1+/CD34+/Lin- or /CD38lo cells as compared with phenotypically similar GFP+ control cells (Figure 4D and Figure S1C). Interestingly, the LTC-ICs derived from TAL1+/CD34+ cells generated larger and more diverse CFCs than the LTC-ICs derived from GFP+ controls (Figure S1C). These results clearly indicate that TAL1 regulates the differentiation of LTC-IC progeny and/or allows increased development of a class of LTC-ICs undetectable in GFP control cells. Quantitatively, taking into account mouse bone marrow cellularity, enforced TAL1 expression increases the number of CFCs by 3- to 22-fold and the number of LTC-ICs by 4- to 40-fold compared with GFP controls. Finally, assuming that the CFC and LTC-IC frequencies of CD34+ UCB cells are 20% and 2% to 5%, respectively,27,28 enforced TAL1 expression increases the number of CFCs and LTC-ICs during the period of engraftment by at least 2- and 11-fold, respectively.
Effect of enforced TAL1 expression on the differentiation of SRCs in NOD-SCID mice. (A) Experimental strategy. (B) Engraftment levels measured by the percentage of human CD45+ cells in the mouse bone marrow; indicated are median values of 4 independent experiments; the numbers of engrafted mice over the total injected mice are also given. (C) Endogenous and transgenic tal1 mRNA levels measured in the human cells engrafted in 4 NOD-SCID recipients from a representative experiment. Semiquantitative RT-PCR was performed with primers that do not amplify the mouse tal1 cDNA. Human S14 levels are used to control the human engraftment levels in every mouse. (D-E) Human erythroid cell engraftment in the BM of NOD-SCID mice. Cells were labeled with anti-GPA antibody and subjected to FACS analysis. Percentages of GPA+ cells in 2 representative mice (D) and in all tested mice (E) are shown (P = .01). (F-G) FACS analysis of the mature human CD19+ B cells and CD14+/CD15+ myeloid cells present among nucleated cells in the BM of NOD-SCID mice. Results from two representative mice are shown (F), and the ratio between CD19+ B cells and CD14+/CD15+ myeloid cells is given for each tested mouse (G). The numbers indicated in panels E and G are median values. Numbers in each FACS quadrant indicate the percentage of positive cells among gated (R1) cells.
Effect of enforced TAL1 expression on the differentiation of SRCs in NOD-SCID mice. (A) Experimental strategy. (B) Engraftment levels measured by the percentage of human CD45+ cells in the mouse bone marrow; indicated are median values of 4 independent experiments; the numbers of engrafted mice over the total injected mice are also given. (C) Endogenous and transgenic tal1 mRNA levels measured in the human cells engrafted in 4 NOD-SCID recipients from a representative experiment. Semiquantitative RT-PCR was performed with primers that do not amplify the mouse tal1 cDNA. Human S14 levels are used to control the human engraftment levels in every mouse. (D-E) Human erythroid cell engraftment in the BM of NOD-SCID mice. Cells were labeled with anti-GPA antibody and subjected to FACS analysis. Percentages of GPA+ cells in 2 representative mice (D) and in all tested mice (E) are shown (P = .01). (F-G) FACS analysis of the mature human CD19+ B cells and CD14+/CD15+ myeloid cells present among nucleated cells in the BM of NOD-SCID mice. Results from two representative mice are shown (F), and the ratio between CD19+ B cells and CD14+/CD15+ myeloid cells is given for each tested mouse (G). The numbers indicated in panels E and G are median values. Numbers in each FACS quadrant indicate the percentage of positive cells among gated (R1) cells.
Finally, we measured mRNA levels of transacting factors known to promote primitive cell expansion,2-4 HOXB4, BMI1, and GATA2 (Figure 4E and Figure S1D). Levels of KIT mRNAs were also tested since the KIT gene is expressed on primitive progenitor cells and is regulated by TAL1.22 GATA1 and GATA3 mRNAs were measured because they participate to protein complexes with TAL1. Quantitative RT-PCR analysis demonstrated a 2.8- ± 0.6-fold increase in GATA2 mRNA levels in the TAL1+ cells as compared with GFP+ controls (P < .05). KIT, HOXB4, and BMI1 but also GATA1 and GATA3 mRNA levels were similar in TAL1+ and GFP+ populations (Figure 4E and Figure S1D). The increase in GATA2 mRNAs suggested that GATA2 could participate to the progenitor amplification promoted by enforced TAL1 expression.
Enhanced TAL1 expression increases SRC activity
We then assessed whether the effect of enforced TAL1 expression on primitive SRC-derived CFC and LTC-IC progenitor cells are linked to an enhancement of the activity of SRCs. We performed secondary transplantations with either total BM cells (n = 3 experiments, 39 mice, injection of an equivalent of 1-2 × 106 human CD45+ cells) or CD34+ purified cells (n = 1 experiments, 6 mice, 2 × 105 cells/mouse) from primary mice. Five to 6 weeks after transplantation, human cells were detected in the bone marrow of 8 of 21 (38%) secondary mice that received a transplant with GFP+ control cells as compared with 18 of 24 (75%) of secondary mice that received a transplant with SRC-derived TAL1+ cells (Figure 5A, 5-6 weeks). The engraftment levels in CD45+, GFP+, and TAL1+ mice were comparable in both groups of secondary mice (respective median value, 1.7% and 3%) as in primary recipients (Figure 2B). The engrafted human cells included lymphoid, myeloid, and CD34+ cells, showing that they derived from multipotential cells. Interestingly, the myeloid cells were predominant in most of the TAL1+ secondary-engrafted mice as in primary recipients (Figures 5B and 2F). This suggests that, in agreement with persistent TAL1 transgene expression in the human CD45+ cells recovered from the BM of secondary mice (Figure 5C), the effect of TAL1 transgene persists throughout serial transplantations. These results show that enforced TAL1 expression enhances the activity of multipotential SRCs.
Effect of enforced TAL1 expression on the CD34+ cells engrafted into the BM of NOD-SCID mice. (A-B) Measurement of the percentage of CD34+/CD14-CD15-CD19- (Lin-) cells by FACS. CD34+ cells were first gated for their lack of CD14 and CD15 expression. Proportion of CD34+/Lin- was obtained by measuring the percentage of CD34+/CD19- in the CD34+/CD14-CD15- population. Shown are the results of 2 representative mice (A) and the summary of the percentage of CD34+/Lin- cells in the nucleated cells of the BM of engrafted individual NOD-SCID mice (B). Median values are indicated in panel B. (C) Gene expression analysis in CD34+/Lin- cells. RT-PCR were performed on RNA extracted from sorted CD34+/Lin- cells using primers that amplify only human cDNA. Results shown are representative of 2 independent experiments. Each measurement was done twice to ensure reproducibility. Numbers in FACS quadrants indicate the percentage of positive cells in a mononucleated cell gate (panel A).
Effect of enforced TAL1 expression on the CD34+ cells engrafted into the BM of NOD-SCID mice. (A-B) Measurement of the percentage of CD34+/CD14-CD15-CD19- (Lin-) cells by FACS. CD34+ cells were first gated for their lack of CD14 and CD15 expression. Proportion of CD34+/Lin- was obtained by measuring the percentage of CD34+/CD19- in the CD34+/CD14-CD15- population. Shown are the results of 2 representative mice (A) and the summary of the percentage of CD34+/Lin- cells in the nucleated cells of the BM of engrafted individual NOD-SCID mice (B). Median values are indicated in panel B. (C) Gene expression analysis in CD34+/Lin- cells. RT-PCR were performed on RNA extracted from sorted CD34+/Lin- cells using primers that amplify only human cDNA. Results shown are representative of 2 independent experiments. Each measurement was done twice to ensure reproducibility. Numbers in FACS quadrants indicate the percentage of positive cells in a mononucleated cell gate (panel A).
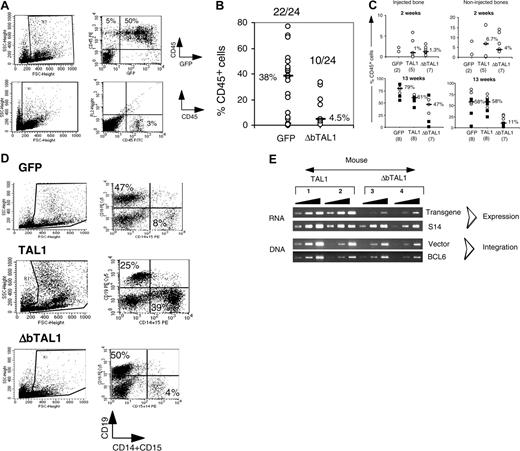
In NOD-SCID mice, a hierarchy of SRCs has been described to repopulate the mouse BM and to give rise to differentiated human cells. This hierarchy includes rapid-, short-term (ST)–, and LT-SRC, and their detection depends on the immunodepletion state of the NOD-SCID mice, the transplantation route, and the time of human cell engraftment analysis.25,37,38 Because (1) secondary NOD-SCID mice were treated before transplantation with anti-CD122 antibodies and (2) human cell engraftment levels were tested 5 to 6 weeks after transplantation, enhanced TAL1 expression could enhance ST-SRC and not LT-SRC activity.19 To address this issue, groups of secondary mice were analyzed 12 weeks after transplantation, that is the hallmark of LT-SRC detection. Similar to the mice analyzed at 5 to 6 weeks, enforced TAL1 expression improved human cell engraftment into secondary mice as 7 of 7 mice (100%, n = 2 experiments) TAL1+/12 week/secondary mice were positive for human cells as compared with 3 of 7 (42%) of GFP controls (Figure 5A, 12 weeks). The engraftment level was enhanced at 12 weeks in both groups, although TAL1+ mice had higher engraftment level (median CD45+ cells, 8%; range, 0.1%-24%) than GFP controls (median CD45+ cells, 1.3%; range, 0.17%-6.9%). The phenotype of the human cells detected in the 12-week engrafted mice was similar to the one of the 5- to 6-week engrafted animals and included CD34+/CD38lo cells and GPA+ cells for several TAL1+ mice (not shown). These results clearly indicated that enforced TAL1 expression enhances immature human hematopoietic cell activity.
Expression of a DNA-binding mutant of TAL1 (ΔbTAL1) abrogates LT-SRC activity
Finally, we investigated whether the activity of the enforced TAL1-dependent amplification of LT-SRC–derived immature progenitors relied on TAL1 DNA binding or a TAL1-directed titration of bHLH proteins that regulate the activity of immature human progenitors. We and others have previously described dependent and independent DNA-binding functions of TAL1 during hematopoiesis in humans and mice.20,21,24
To discriminate between the 2 possibilities, CD34+ UCB cells were transduced with a lentiviral construct containing a TAL1 cDNA deleted in the DNA-binding domain (named ΔbTAL1). We showed that this mutant has a dominant effect on the endogenous TAL1 DNA-binding activity (data not shown). Transduced cells were then transplanted into NOD-SCID mice. It is noteworthy that culture of ΔbTAL1+ cells in lymphoid/myeloid and in LTC conditions affected the differentiation of such progenitors in a limited fashion (data not shown and Ravet et al24 ). In contrast to these in vitro results and to the effect of enforced TAL1 expression in SRCs, ΔbTAL1 expression decreased human cell engraftment into NOD-SCID mice. Of 24 mice that received a transplant, only 10 (42%) mice exhibited human cell engraftment after 10 to 12 weeks, and the engraftment levels were very low (median, 4.5%; range, 0.7%-20.2% CD45+ cells) (Figure 6A-B). Among the repopulating cells, few transduced cells were detected with low transgene expression, and the engrafted cells were mainly CD19+ B cells similar to GFP+ cells (not shown). These data strongly suggest that DNA binding is necessary for the SRC-dependent hematopoietic activity.
The low engraftment of ΔbTAL1+ cells could result from an impaired migration of the ΔbTAL1-transduced SRCs to marrow niches or from their reduced capacity to establish human hematopoiesis. This latest phenomenon requires proliferation and differentiation and have been described to be altered in the presence of high levels of HLH Id protein that could explain the results obtained with ΔbTAL1.39,40 To distinguish between these hypothesis, transduced cells were directly injected into one femur of recipient mice.31,41 This experimental system allows the study of the proliferation and differentiation abilities of progenitor cells in a bone marrow environment avoiding the bias of migration when analyzing the engraftment levels in the injected bone.41 Human cell engraftment in the injected femur was consistently observed 2 and 13 weeks after transplantation in the 35 of 36 injected mice (n = 2 experiments), irrespective of the construct (GFP/TAL1/ΔbTAL1) used (Figure 6C). At 2 weeks, engraftment levels were similar in injected as well as in noninjected bones in all groups of mice (Figure 6C), suggesting that homing is not regulated by TAL1 or ΔbTAL1. Differentiated (CD15+, GPA+, CD19+) as well as primitive CD34+/CD38lo cells were detected (data not shown). These data further suggest that differentiation of rapid-SRCs, a class of mature SRCs,31 is not affected by TAL1 and ΔbTAL1 expression. These results are consistent with the data obtained in vitro (Figure 1) and further show that enforced TAL1 and ΔbTAL1 expression mildly affect the differentiation of committed progenitors. However, at 13 weeks after transplantation, the presence of ΔbTAL1 reduced the percentage of CD45+ human cells as compared with TAL1 and GFP controls in both the injected and noninjected bones (Figure 6C), and the engrafted cells mainly generated B cells as in the GFP controls (Figure 6D). Analysis of vector integration and transgene expression showed that integration and expression of transgenic ΔbTAL1 were low in human cells recovered from the injected bones compared with the ones measured in cells transduced with the TAL1 vector (Figure 6E).
Enforced TAL1 expression amplifies SRC-derived CFCs and LTC-ICs. (A) SRC-derived CD34+/CD38lo cells were purified by cell sorting from the NOD-SCID bone marrow, and their function was studied in different culture conditions. (B) The progeny of sorted CD34+/CD38lo cells was counted and analyzed by FACS after 3 weeks of culture in lymphomyeloid conditions. Results are expressed as the percentage (upper quadrant) and as the absolute number (lower quadrant) of CD19+ B cells and CD15+ myeloid cells. (C) Sorted CD34+/CD38lo cells were cultured in methylcellulose for the detection of human CFCs. Results are expressed in absolute numbers of CFCs generated for 1000 plated cells. Fold increase in absolute numbers of cells or of CFCs between GFP+ cells and TAL1+ cells is indicated (A-B). (D) CD34+/CD38lo cells were cultured at limiting dilutions in LTC conditions. Frequencies of LTC-ICs were calculated according to Poisson statistics and are indicated for GFP and TAL1 transgenes. (E) mRNA levels of KIT, Bmi1, HOXB4, GATA-1, GATA-2, and GATA-3 in CD34+/CD38lo cells were measured by quantitative RT-PCR analysis. After standardization over GAPDH mRNA levels, results were expressed as percentage of GFP+ cells. The mRNA level of every factor was tested at least twice. Error bars indicate standard deviations between measurements. P < .05 for GATA-2 expression.
Enforced TAL1 expression amplifies SRC-derived CFCs and LTC-ICs. (A) SRC-derived CD34+/CD38lo cells were purified by cell sorting from the NOD-SCID bone marrow, and their function was studied in different culture conditions. (B) The progeny of sorted CD34+/CD38lo cells was counted and analyzed by FACS after 3 weeks of culture in lymphomyeloid conditions. Results are expressed as the percentage (upper quadrant) and as the absolute number (lower quadrant) of CD19+ B cells and CD15+ myeloid cells. (C) Sorted CD34+/CD38lo cells were cultured in methylcellulose for the detection of human CFCs. Results are expressed in absolute numbers of CFCs generated for 1000 plated cells. Fold increase in absolute numbers of cells or of CFCs between GFP+ cells and TAL1+ cells is indicated (A-B). (D) CD34+/CD38lo cells were cultured at limiting dilutions in LTC conditions. Frequencies of LTC-ICs were calculated according to Poisson statistics and are indicated for GFP and TAL1 transgenes. (E) mRNA levels of KIT, Bmi1, HOXB4, GATA-1, GATA-2, and GATA-3 in CD34+/CD38lo cells were measured by quantitative RT-PCR analysis. After standardization over GAPDH mRNA levels, results were expressed as percentage of GFP+ cells. The mRNA level of every factor was tested at least twice. Error bars indicate standard deviations between measurements. P < .05 for GATA-2 expression.
Enforced TAL1 expression enhances SRC activity. (A) Secondary transplantations were performed by intravenous injection of total bone marrow cells containing 2 to 2.5 × 106 human CD45+ cells (39 mice, white symbols) or of purified human CD34+ cells (6 mice, black symbols) from primary mice. Secondary mice were analyzed at 5 to 6 weeks (45 mice, 3 experiments) and at 12 weeks after transplantation (14 mice, 1 experiment). Shown are the levels of engraftment measured by the percentage of CD45+ cells achieved in secondary recipients. Median values are indicated. A positive mouse contained at least 0.1% human cells in its BM. (B) Phenotype analysis of the human cells detected in 2 representative secondary recipients analyzed 5 weeks after transplantation. Percentage of positive cells is indicated in each quadrant for every surface marker tested. (C) TAL1 transgene expression in human CD45+ cells enriched from the bone marrow of 3 secondary recipients that received BM cells from primary mice that received a transplant 5 weeks before with TAL1+ cells.
Enforced TAL1 expression enhances SRC activity. (A) Secondary transplantations were performed by intravenous injection of total bone marrow cells containing 2 to 2.5 × 106 human CD45+ cells (39 mice, white symbols) or of purified human CD34+ cells (6 mice, black symbols) from primary mice. Secondary mice were analyzed at 5 to 6 weeks (45 mice, 3 experiments) and at 12 weeks after transplantation (14 mice, 1 experiment). Shown are the levels of engraftment measured by the percentage of CD45+ cells achieved in secondary recipients. Median values are indicated. A positive mouse contained at least 0.1% human cells in its BM. (B) Phenotype analysis of the human cells detected in 2 representative secondary recipients analyzed 5 weeks after transplantation. Percentage of positive cells is indicated in each quadrant for every surface marker tested. (C) TAL1 transgene expression in human CD45+ cells enriched from the bone marrow of 3 secondary recipients that received BM cells from primary mice that received a transplant 5 weeks before with TAL1+ cells.
Expression of ΔbTAL1 mutant protein interferes with the SRC activity. (A-B) Engraftment levels in the BM of NOD-SCID mice that received a transplant with GFP- and ΔbTAL1-transduced cells. (A) Percentages of human CD45+ cells in the bone marrow of 2 representative NOD-SCID mice that received a transplant intravenously 14 weeks before with GFP- and ΔbTAL1-transduced CD34+ cells. (B) Summary of the results of 4 independent experiments. The ratios of positive to total injected mice and the median value of the percentage of CD45+ cells for each group of mice are indicated. (C) Human cell engraftment levels obtained from mice that received a transplant by intrabone injection with GFP-, TAL1-, and ΔbTAL1-transduced cells. Shown are the percentages of human CD45+ cells detected in the injected bone (left) and in the noninjected side bones (right). Engraftment levels were analyzed 2 (1 experiment) and 13 weeks (2 experiments) after transplantation. Median percentages of CD45+ cells are indicated as well as the number of tested mice. (D) Phenotypic analysis of the human cells detected in the BM of intrabone injected NOD-SCID mice. GFP-, TAL1-, and ΔbTAL1-transduced cells were recovered from all the bones of individual NOD-SCID mice and analyzed by FACS for the presence of CD19+ cells and CD15+ cells. Shown are the results of 3 representative mice. Numbers indicate the percentage of positive cells in a gated population. (E) Transgenic TAL1 and ΔbTAL1 expression and vector integration measured by semiquantitative PCR analysis in human cells purified from the BM of 4 mice that received a transplant by intrabone injection with TAL1 (mice 1 and 2) and ΔbTAL1 (mice 3 and 4) transduced CD34+ cells. Primers used in RT-PCR were designed to amplify both TAL1 and ΔbTAL1.
Expression of ΔbTAL1 mutant protein interferes with the SRC activity. (A-B) Engraftment levels in the BM of NOD-SCID mice that received a transplant with GFP- and ΔbTAL1-transduced cells. (A) Percentages of human CD45+ cells in the bone marrow of 2 representative NOD-SCID mice that received a transplant intravenously 14 weeks before with GFP- and ΔbTAL1-transduced CD34+ cells. (B) Summary of the results of 4 independent experiments. The ratios of positive to total injected mice and the median value of the percentage of CD45+ cells for each group of mice are indicated. (C) Human cell engraftment levels obtained from mice that received a transplant by intrabone injection with GFP-, TAL1-, and ΔbTAL1-transduced cells. Shown are the percentages of human CD45+ cells detected in the injected bone (left) and in the noninjected side bones (right). Engraftment levels were analyzed 2 (1 experiment) and 13 weeks (2 experiments) after transplantation. Median percentages of CD45+ cells are indicated as well as the number of tested mice. (D) Phenotypic analysis of the human cells detected in the BM of intrabone injected NOD-SCID mice. GFP-, TAL1-, and ΔbTAL1-transduced cells were recovered from all the bones of individual NOD-SCID mice and analyzed by FACS for the presence of CD19+ cells and CD15+ cells. Shown are the results of 3 representative mice. Numbers indicate the percentage of positive cells in a gated population. (E) Transgenic TAL1 and ΔbTAL1 expression and vector integration measured by semiquantitative PCR analysis in human cells purified from the BM of 4 mice that received a transplant by intrabone injection with TAL1 (mice 1 and 2) and ΔbTAL1 (mice 3 and 4) transduced CD34+ cells. Primers used in RT-PCR were designed to amplify both TAL1 and ΔbTAL1.
Taken together, these data suggest that ΔbTAL1 expression impairs the function of a primitive progenitor cell population that has long-term hematopoiesis establishment capacities, independently of homing. These results favor a DNA-binding dependent action of enforced TAL1 in the enhancement of LT-SRC activity and also demonstrates the importance of bHLH protein function in human immature hematopoietic cells.
Discussion
Our results demonstrate that enforced TAL1 expression enhances the hematopoietic activity of human immature cells whereas in vitro–tested committed progenitors (LTC-ICs, CFCs, and myeloerythroid mature cells) are much less affected.
SCL/TAL1 has a major function in the establishment of primitive and definitive hematopoiesis (for review see Begley and Green18 ). Its conditional knockout alters erythroid and megakaryocytic differentiation programs, whereas the effect of such manipulation on HSC proliferation, differentiation, and transplantation is unclear because of discordant results generated in different transplantation protocols.16,19 The effect of enforced TAL1 expression we describe can be related to enhancing the survival or the self-renewal of primitive cells, which will be definitively proven using quantitative transplantation experiments. It is possible also, although unlikely from the length of the engraftment period, that enforced TAL1 expression enhances the engraftment potential and thus the survival of myeloid-restricted progenitors since the differentiated progeny of TAL1+/LT-SRCs was biased toward the myeloid lineage. Discordance between our results and the conditional knockout of TAL1 in mouse exists that can be due to several points, including the target cell population (adult BM versus UCB fetal cells), the mouse versus human system, or a possible redundancy by other bHLH class 2 proteins, such as lymphoblastic leukemia derived sequence 1 (LYL1), in the TAL1 knockout mouse experiments. Moreover, another study indicated that enforced TAL1 expression changes the differentiation but not the repopulation capability of mouse HSCs.36 However, in this study, no secondary transplantation was performed to assay the effect of long-time exposure of mouse HSCs to enforced TAL1 expression; thus, no real conclusion can be drawn on the effect of TAL1 on mouse HSC self-renewal.
As a consequence of the enhancement of LT-SRC activity, we show an amplification of SRC-derived myeloid/erythroid progenitors (LTC-ICs and CFCs), finally resulting into an increase of mature myeloid and erythroid cells. This latter phenomenon is however quantitatively less important than the amplification of their direct progenitors and maybe reminiscent of HSC amplification by HOXB4 whereby hematopoietic homeostasis limits the expansion of differentiated cells3 or because of the reduced crossreactivity of mouse cytokines implicated in late steps of human differentiation. This last hypothesis is supported by a recent report from Kunisato et al36 in which myeloid differentiation has been described to be highly enhanced in the presence of enforced TAL1 expression.
The relatively small expansion of SRC-derived differentiated cells following enforced TAL1 expression most probably results from the dramatic decrease of B-cell differentiation. In the NOD-SCID model, human hematopoiesis is constituted mainly of B cells and their progenitors and to a much lesser extent of granulocytic/monocytic cells.27,42,43 Enforced TAL1 expression during B-cell development is known to interfere with B-cell differentiation as a consequence of a titration of E2A proteins34,35 that are required for this differentiation pathway.45 A transitory TAL1 expression system into LT-SRCs will definitively show that the limited expansion of the differentiated cell compartment in our conditions is due to the depletion of B cells and is linked to the constitutive expression of TAL1 in our lentiviral construct. The development of such an experimental design is also crucial in the issue of determining the immature cell population (multipotential versus myeloid restricted) and the process enhanced by the enforced TAL1 expression.
Human T-cell development was not affected in the presence of enforced TAL1 expression in our experiments, and no leukemia developed during the time course of our experiments. Treatment of NOD-SCID mice prior to CD34+ transplantation with anti-CD122/interleukin 2 receptor β (IL2Rβ) chain antibodies allows human T-cell development in mouse thymuses.30 We obtained similar results and further observed that lentiviral transduction in our conditions decreased by 2-fold the proportion of human thymocyte-positive mice (data not shown) probably related to long-time exposure of CD34+ cells to cytokines.45 When TAL1+/CD34+ cells were transplanted into NOD-SCID mice, no changes in terms of the proportions of human thymocyte-positive mice and the quality of human T-cell differentiation were observed. These in vivo results were confirmed by data we obtained in FTOC (data not shown). Altogether these results are singular because, as for B-cell development, interference of E2A activity during human and mouse T-cell development results in a severe T-cell depletion and eventually to T-cell leukemia.39,46 They are different from the ones obtained in transgenic mouse.35 They may suggest that human T and B cells generated in NOD-SCID mice may not derive from a common progenitor and/or the level of TAL1 expression obtained from our construct is not elevated enough to completely block E2A activity in T-cell progenitors. This last point is however contradictory with the fact that TAL1 expression in TAL1+ T-ALL is highly variable. In some SIL–TAL1/T-ALL cells the TAL1 protein is not detectable,47 whereas TAL1 expression level in T-ALL–derived cells in which the TAL1 gene is regulated by translocated cis-acting regulatory elements can be very high. The levels of TAL1 expression in our experiments is probably halfway between SIL-TAL1/T-ALL and T-cell receptor-δ (TCRδ)–TAL1/T-ALL, suggesting that the mechanism of leukemogenesis is different from low TAL1 expressing to high TAL1 expressing cells possibly related to the involvement of TAL1 to different protein complexes.
In this study, the positive effect of enforced TAL1 expression on human primitive progenitors requires DNA binding. The mutant ΔbTAL1 not only did not mimic TAL1 effects on LT-SRCs but abrogated their reconstitution ability. This result contrasts with the rescue of hematopoietic development of mouse Tal1-/- ES cells by a TAL1 DNA-binding mutant21 and suggests that one of the molecular mechanisms of enforced TAL1 effect in human primitive cells is mediated by regulation of target genes. The effect of ΔbTAL1 expression further shows the importance of bHLH proteins in human hematopoiesis and parallels previous experiments using dominant-negative Id proteins that suggested that Id proteins are key factors for hematopoietic differentiation, especially for the lymphoid T and B programs.10,39 In our experimental system, expression of ΔbTAL1 impairs the proliferation or the differentiation of primitive but not committed progenitors, as shown by comparing our data from cultures, short-term, and long-term engraftment of ΔbTAL1+/CD34+ cells into NOD-SCID mice. The independence of the ΔbTAL1 effect from migration is shown by comparing short-term and long-term engraftment levels in experiments in which cells were transplanted directly into the mouse BM.31 A higher effect of ΔbTAL1 expression was detected in the noninjected bones, in which invasion by human primitive cells necessitates a homing process. Although one could argue that this last result indicates that the homing process is altered in the presence of ΔbTAL1, it is now established that homing is a property of primitive cells.48 It is thus difficult in this case to separate an effect on homing from an effect on primitive cells with homing properties.
New concepts have emerged in the field of leukemias in the past decade. In particular, it appears that certain leukemia can be generated from a phenotypically immature cell that is called leukemic stem cell (LSC).49 Two main hypotheses are proposed to explain the origin of LSCs. (1) They derive from true HSCs that are transformed and retain some abilities to self-renew and differentiate.50 (2) They derive from committed progenitors in which the transforming event allows the reacquisition of some stem cell properties, such as self-renewal.51 The fact that enforced TAL1 expression improves the hematopoietic capacities of human primitive progenitor cells is of potential relevance to understand the mechanism of TAL1-induced transformation in T-cell leukemia and could represent the reacquisition of an amplification/self-renewal/survival event for a given committed T-cell progenitor.
Prepublished online as Blood First Edition Paper, June 16, 2005; DOI 10.1182/blood-2005-02-0557.
Supported by the scientific review board of INSERM, Association Française contre les Myopathies (AFM/ATG 1999, AFM 9721), Association pour la Recherche sur le Cancer (ARC 7551), and Fondation de France (Engt 2002 004534). D.R. was supported by fellowships from ARC and Ligue Nationale Contre le Cancer (LNCC; RAB05013KKA). E.R. was supported by fellowships from the Ministère de la Recherche et des Technologies, Fondation pour la Recherche Médicale, and Société Française d'Hématologie.
D.R. and E.R. contributed equally to this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank N. Taylor, I. Dusanter, and C. Francastel for helpful discussions and critically reviewing the manuscript. We thank B. Izac for the excellent production of lentiviral vectors, M.-C. Rouillez who performed all the RQ-PCR analyses, and J.-C. Deschemin for the production of TM-β1 antibody. All the cord bloods were sampled in the Clinique des Noriets, Vitry-sur-Seine, France, and we are indebted to the midwives and to Dr Rouquet that were devoted to help us. We thank all the staff of the animal colony of the Institut A Lwoff, Villejuif, France, for their precious help in the mouse work. Cell sorting was performed in the common facilities of the Institut Cochin.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal