Abstract
The Notch signaling pathway is involved in several lineage commitment and differentiation events. One of these is fate determination of the common lymphoid progenitor, promoting T-cell development at the expense of B-cell differentiation. It has been suggested that this process relies on Notch's ability to inhibit E proteins, which are crucial for early B-cell development. Here, we report that Notch signaling also modulates the function of the transcription factor, early B-cell factor (EBF). Transient transfection of intracellular Notch1 (Notch1-IC) into a pre-B cell line resulted in the down-regulation of EBF-regulated promoters and diminished the capacity of EBF to activate these promoters in an epithelial cell line. This correlated with a reduction in the ability of EBF to bind DNA. Ligand-induced stimulation of endogenous Notch receptors with Delta4 mimicked the activity of Notch1-IC toward EBF. These data suggest that Notch signaling may affect B-versus T-lineage commitment by the targeting of both EBF and E2A.
Introduction
The development of B- and T-lymphoid cells from hematopoietic stem cells involves a complex multistep differentiation pathway proceeding through common lymphoid progenitors (CLPs).1 The CLPs receive instructing signals from the surrounding cells that appear to promote a developmental branch point, where the choice of lineage, B cells versus T cells, takes place.2 One such signaling pathway with a crucial role in this process is mediated through the Notch family receptors.3,4 Transgenic mice expressing a constitutively active form of intracellular Notch1 (Notch1-IC) within the hematopoietic compartment, display gross abnormalities in lymphoid development.5 These are manifested as an induction of T-cell development apparently at the cost of B-lymphoid differentiation.5 Targeted disruption of the Notch1 gene has the opposite effect, resulting in a dramatic reduction of T-cell number and the development of B-lymphoid cells in the thymus.6,7 The signal transduction cascade, initiated by the interaction of a Notch receptor with any of its ligands, Jagged1, Jagged2, Delta1, Delta3, or Delta4, involves a γ-secretase-mediated cleavage of the Notch protein.3,4 This releases the Notch intracellular domain (Notch-IC), which enters the nucleus, forms a complex with CSL (CBF1/RBP-Jk), and activates transcription.3,4 The potential of Notch signaling to impair B-cell development has been attributed to a functional inhibition of the E2A protein E47.8 E2A proteins are crucial in the development of B lymphocytes in a dose-dependent manner,9,10 supporting this notion. However, several lines of evidence are indicating that the function of E2A proteins in B-cell development is linked to the function of another transcription factor, early B-cell factor (EBF).11,12 This protein is of crucial importance for B-cell development because mice carrying a homologous disruption of the gene also display a block in early B-cell differentiation.13 EBF and E2A share several target genes, such as λ5, VpreB,12 and Cd79a,14 and have also been shown to act in a coordinated manner to support B-cell development in vivo.11 Furthermore, it has been shown that B-cell development in E2A-deficient mice can be rescued by ectopic expression of EBF in hematopoietic progenitors, suggesting a partially redundant function of the E2A gene products in B-cell development.15 A direct role for EBF in B-versus T-lineage commitment per se has been suggested from studies in which the protein was ectopically expressed by retroviral transduction in hematopoietic progenitor cells.16 This resulted in an impairment of T lymphopoiesis, suggesting a key role for EBF in the initiation and commitment into the B-lymphoid pathway.
The crucial function of both Notch signaling and EBF in early B-cell development prompted us to ask if EBF is affected by Notch signaling. We here present evidence that suggests that Notch signaling inhibits the activity of EBF as well as the coordinated activity of EBF and E2A. Thus, our results provide a novel insight into the orchestration of gene expression and lineage commitment in early lymphopoiesis.
Materials and methods
Tissue culture conditions and cell purification
HeLa and 230-238 cells lines were grown in RPMI medium supplemented with 10% fetal calf serum (FCS), 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 2 mM pyruvate, 50 μM 2-mercaptoethanol, and 50 μg gentamicin per milliliter (complete RPMI media). BaF/3 and the EBF-expressing BaF/3-EBF17 cell lines were kept in complete RPMI supplemented with 10% interleukin 3 (IL-3) containing WEHI3-conditioned media. 293T cells were grown in Dulbecco modified Eagle medium (DMEM) with 10% FCS and 50 μM 2-mercaptoethanol added (all purchased from Gibco Invitrogen, Paisley, United Kingdom). To block Notch signaling, growth medium was supplemented with 10 μM N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester (DAPT; Merck Biosciences, Darmstadt, Germany). CD19+ cells from murine bone marrow were enriched by positive selection by using magnetic-activated cell sorting (MACS) CD19 microbeads (Miltenyi Biotec, Auburn, CA), and kept in Optimem containing 10% FCS, 50 μM 2-mercaptoethanol, and 50 μg/mL gentamicin. All cells were cultured at 37°C and 5% CO2.
Transient transfections and luciferase assays
On transfection, approximately 2.5 × 106 230-238 lymphoid cells were incubated for 30 minutes at 20°C in 1 mL diethylaminoethyl (DEAE)-dextran (0.8 mg/mL Tris-buffered saline [TBS] containing 140 mM NaCl, 5 mM KCl, 25 mM Tris [tris(hydroxymethyl)aminomethane], pH 7.4, 0.6 mM Na2HPO4, 0.7 mM CaCl2); Pharmacia, Uppsala, Sweden) containing 200 μg cytomegalovirus (CMV)-controlled Renilla luciferase reporter gene (Promega, Madison, WI), used as internal control, 2 μg reporter construct, controlled by the λ5, B29, or Cd79a promoter, and additional plasmids; 3 μg empty vector, or 1 μg Notch1-IC plus 2 μg empty vector, or 3 μg Notch1-IC. The transfected cells were incubated in 5 mL complete RPMI medium in 6-well plates for 36 to 48 hours. Protein extracts were prepared by adding 80 μL cell lysis buffer (Promega).
HeLa cells, seeded into a 24-well tissue culture plate, were transfected according to the Invitrogen transfection protocol: “HeLa cells with LIPOFECTIN reagent and PLUS reagent.” Briefly, DNA expression plasmids were mixed in 75 μL serum-free medium Optimem (Gibco, Invitrogen), 7 μL PLUS reagent and 0.75 μL LIPOFECTIN reagent (Invitrogen Life Technologies, Carlsbad, CA). After replacement of cell growth medium with 300 μL of serum-free medium, the DNA-PLUS-LIPOFECTIN mixture was added to the cells. Plates were then incubated at 37°C in 5% CO2 for 3 hours, after which each well was supplemented with 1 mL growth medium. Cells were harvested 48 hours after transfection and protein extracts were prepared by adding 80 μL cell lysis buffer to each well. In each transfection, 40 ng of the λ5, B29, Cd79a, or x3 Cd79a reporter construct was cotransfected with 320 ng empty vector as control for basic promoter activity, or with 200 ng EBF expression plasmid18 where also 120 ng empty vector, or 40 ng Notch1-IC plus 80 ng empty vector, or 120 ng Notch1-IC was added. To verify the specificity of Notch1-induced repression, 120 ng Notch1-IC was cotransfected with an SV40 reporter construct. The CD19 reporter construct was activated by 200 ng Pax5, kindly provided by Dr Meinrad Busslinger. In the experiment in which EBF and E47 were cotransfected, 120 ng of each construct was used. The λ5 promoter controlled reporter was also activated by 200 ng of the fusion protein EBFVp16.18 To control the effect of Notch1-IC exerted on the Vp16 part of the EBFVp16, we cotransfected a G5 reporter construct together with the fusion protein Gal-4Vp16 (Promega) and 120 ng Notch1-IC. As internal control and for normalization of the luciferase activities, all transfections included 20 ng pRL-0, received from Dr Björn Olde (Lund, Sweden).
To generate a soluble form of a Notch1 ligand, 293T cells grown in a 10-cm Petri dish were transfected in the same way as described for HeLa cells, with either 5 μg Fc TRAIL-R4 or 5 μg Fc-Delta4 expression plasmid.19 After 48 hours, the growth medium was collected and filtered through a 0.45-μm syringe filter. Fc-TRAIL-R4 or Fc-Delta4 fusion protein was immobilized by incubating nontissue culture plates overnight in 4°C with 10 μg/mL rabbit anti-human IgG Fcγ antibody (Jackson ImmunoResearch Laboratories, West Grove, PA), and then incubated for 2 hours with respective filtered supernatant.
BaF/3 cells were transfected with DEAE, as described for 230-238 cells. Then, 0.5 μg λ5 reporter construct was cotransfected with 5 μg empty vector, or 3.5 μg EBF together with 1.5 μg empty vector or 1.5 μg Notch1-IC. The CD19 reporter and Pax5 were transfected in the same way as described. CMV controlled Renilla luciferase reporter gene was again used as internal control. The cells were grown for 48 hours on plates coated with Fc-TRAIL-R4 as control or the fused Notch ligand Fc-Delta4.
The luciferase assays for all transfections were performed with a Dual-Luciferase Reporter Assay System (Promega) using 20 μL of the total protein extract. All samples were made in triplicate.
RT-PCR and real-time quantitative PCR
RNA was prepared from cells using RNeasy Mini Kit (QIAGEN, VWR International, Stockholm, Sweden), and cDNA was generated by annealing 1 μg total RNA to 0.5 μg random hexamers in 10 μL diethyl pyrocarbonate-treated water. Reverse transcription (RT) reactions were performed with 200 U SuperScript II (Invitrogen Life Technologies, Carlsbad, CA) in accordance with the manufacturer's recommendations. One-twentieth of the RT reaction was used in the polymerase chain reaction (PCR) assays. PCR was performed with 1 U Taq polymerase (Invitrogen Life Technologies) in the manufacturer's buffer supplemented with 0.2 mM deoxynucleotide triphosphate (dNTP), in a total volume of 25 μL. β-actin was amplified by 25 cycles, and 40 cycles were used to amplify Notch1 cDNA (94°C, 30 seconds, 60°C, 30 seconds, and 72°C, 30 seconds). The following primers were added to a final concentration of 1 mM: β-actin sense: 5′-GTTTGAGACCTTCAACACC; β-actin antisense: 5′-GTGGCCATCCCTGCTCGAAGTC; Notch1 sense: 5′-GTGCCGTGGCCTCCAACACCGC; and Notch1 antisense: 5′-CACCAGGGTGCCGGCTGCCAGCC.
Each quantitative (Q)-PCR (ABI PRISM 7700 Sequence Detector; PE Applied Biosystems, Foster City, CA) involved cDNA extracted from approximately 500 cells. Reactions were conducted by mixing 2 × TaqMan Universal PCR Master Mix, 20 × Gene Expression Assay Mix from Assays-on-Demand or Assays-by-Design service, RNase-free H2O and 5 μL cDNA to a final volume of 25 μL (all reagents purchased from Applied Biosystems). The reaction was initiated by a hold for 10 minutes at 95°C followed by 45 cycles 15 seconds at 95°C and 1 minute at 60°C. The following Assays-on-Demand probes were used: HPRT: Mm00446968_m1; EBF: Mm22395519_m1; CD79a: Mm00432423_m1; CD79b: Mm00434143_m1; CD19: Mm00515420_m1; and Pax5: Mm00435501_m1.
The λ5 primer/MGB-sequences were constructed using Assays-by-Design: sense 5′-GGAACAACAGGCCTAGCTATGG; antisense 5′-CTCCCCGTGGGATGATCTG; MGB-probe: 5′-CCGGCAGCTCCTGTTC. Each reaction was performed in triplicate and was normalized to the housekeeping gene HPRT.
Protein extracts and electrophoretic mobility shift assay
Nuclear extracts were prepared according to Schreiber et al.22 DNA probes were labeled with γ[32P] adenosine triphosphate (ATP; Amersham Biosciences, Buckinghamshire, United Kingdom) by incubation with T4 polynucleotide kinase (Roche Diagnostics, Mannheim, Germany), annealed and purified on a mini-Quick Spin Oligo Column (Roche Diagnostics, Mannheim, Germany). Nuclear extracts were incubated with labeled probe (20 000 cpm, 3 fmol) for 30 minutes at room temperature in binding buffer (10 mM HEPES, pH 7.9, 70 mM KCl, 1 mM dithiothreitol, 1 mM EDTA [ethylenediaminetetraacetic acid], 2.5 mM MgCl2, 4% glycerol) with 0.75 μg Poly(dI/dC) (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). When EBF binding was investigated, 1 mM ZnCl2 was supplemented in the mixture. The samples were separated on a 6% polyacrylamide Tris boric acid EDTA (TBE) gel, which was dried and subjected to autoradiography.
Oligonucleotides used for electrophoretic mobility shift assays (EMSAs) were as follows: Mb-1 EBF sense: 5′-GAGAGAGACTCAAGGGAATTGTGG; Mb-1 EBF antisense: 5′-CCACAATTCCCTTGAGTCTCTCTC; Oct sense: 5′-TTCATTGATTTGCATCGCATGAGACGCTAACATCGTACGTTC; Oct antisense: 5′-GAACGTACGATGTTAGCGTCTCATGCGATGCAAATCAATGAA; λ5 EBF sense: 5′-CCAGGGGCCCTCAGGGACTGG; and λ5 EBF antisense: 5′-CCAGTCCCTGAGGGCCCCTGG.
Western blotting
For Western blotting, the HeLa cell nuclear extracts used for EMSA and the BaF/3 extracts used for luciferase assay were separated on a 7.5% PAGEr Gold Precast gel (Cambrex, In vitro Sweden AB, Stockholm, Sweden) and blotted onto a nylon membrane (anti-myc; Amersham Pharmacia Biotech) or immobilon polyvinylidene difluoride (PVDF; Millipore, Billerica, MA; anti-HES-1) by semidry electroblotting. For EBF detection, a primary mouse monoclonal anti-c-myc antibody (9E10) diluted 1:1000 in phosphate-buffered saline (PBS; Gibco Invitrogen) supplemented with 0.5% Tween 20 (PBS-T: Amersham Life Science, Ohio), and a 1:2000 dilution of a secondary horseradish peroxidase (HRP)-conjugated anti-mouse immunoglobulin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used. The primary anti-HES-1 antibody was diluted 1:8000 (kindly provided by Dr Tetsuo Sudo, Kamakura, Japan) in PBS-T, and HRP-conjugated donkey anti-rabbit antibody was here used as secondary antibody. Detection of the secondary antibody was obtained using an enhanced chemoluminescence (ECL) system (Amersham Pharmacia Biotech). All the steps were performed at room temperature.
Results
Ectopic expression of Notch1-IC reduces the functional activity of B-lineage promoters in a pre-B cell line
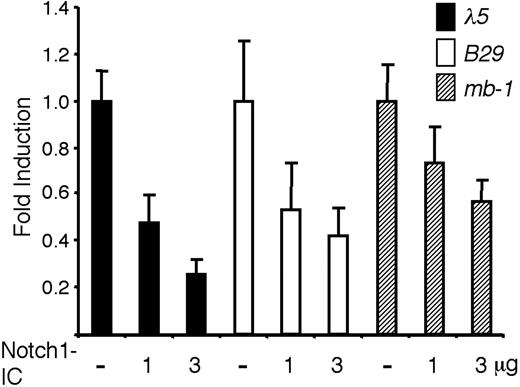
Even though it has been shown that active Notch signaling alters the expression of B cell-restricted genes,23 it is unclear if this effect is due to a general influence on lymphoid progenitors or a direct activity on pre-B cell-restricted promoters. The study of these phenomena in primary cells is complicated by the fact that functional alterations can be related to a lineage switch rather than the direct action of Notch signaling on target promoters. Thus, to investigate if Notch signaling has a direct impact on the activity of promoters active in a pre-B cell, we transiently transfected the pre-B cell line 230-238 with luciferase reporter constructs controlled by either the B29, Cd79a, or λ5 promoter together with an expression plasmid encoding the intracellular constitutively active domain of Notch-1, Notch1-IC (Figure 1). Inclusion of 1 μg of the Notch1-IC plasmid reduced the expression of the λ5 reporter to less than 48%, whereas 3 μg of the expression plasmid reduced the functional activity to 25% (Figure 1). Similar results were obtained with the B29 promoter, which was down-regulated to 53% and 42%, respectively (Figure 1). The effect was less pronounced on the Cd79a promoter with reductions to 73% and 57% by the inclusion of 1 and 3 μg Notch1-IC (Figure 1). Notch1-IC did not render any significant alternations in the activity of the cotransfected CMV-Renilla luciferase plasmid (data not shown), suggesting that there were no general pleiotropic effects exerted by Notch1-IC on these cells under the investigated time span of 36 hours. These data suggest that constitutively active Notch1 has the ability to directly reduce the functional activity of B-lineage restricted promoter elements in a pre-B cell line.
Notch1-IC expression reduces the functional activity of pre-B cell-restricted promoter elements in a pre-B cell environment. The resulting relative luciferase activity when 2 μg λ5, B29, or Cd79a promoter-controlled reporter constructs were transiently transfected into the 230-238 pre-B cell line along with 1 or 3 μg Notch1-IC, as indicated. Reporter activities were normalized to the internal control CMV-Renilla and are presented relative to the value obtained after cotransfection of each respective reporter construct along with empty expression plasmid. The data are based on 6 transfection experiments and error bars indicate SD.
Notch1-IC expression reduces the functional activity of pre-B cell-restricted promoter elements in a pre-B cell environment. The resulting relative luciferase activity when 2 μg λ5, B29, or Cd79a promoter-controlled reporter constructs were transiently transfected into the 230-238 pre-B cell line along with 1 or 3 μg Notch1-IC, as indicated. Reporter activities were normalized to the internal control CMV-Renilla and are presented relative to the value obtained after cotransfection of each respective reporter construct along with empty expression plasmid. The data are based on 6 transfection experiments and error bars indicate SD.
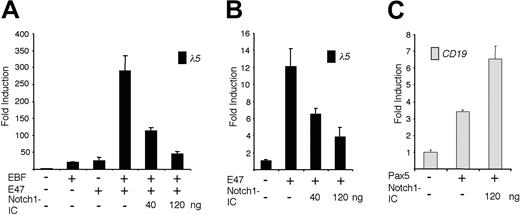
The ability of EBF and E47 to act synergistically on the λ5 promoter is abolished by expression of Notch1-IC. (A) The diagram shows the relative luciferase activity obtained after transiently transfecting HeLa cells with 40 ng of the λ5 reporter plasmid and 120 ng of the EBF and/or 120 ng of the E47-encoding expression plasmid, along with 40 or 120 ng of the Notch1-IC expression plasmid. (B) Graphs display the resulting relative activity of a λ5 reporter plasmid in HeLa cells after activation by ectopic expression of a forced dimer of E47 in the presence of increasing amounts of cotransfected Notch1-IC plasmid as indicated. (C) Columns representing the relative luciferase activity observed after cotransfecting 40 ng CD19 reporter construct and 200 ng Pax-5 alone, or together with 120 ng Notch1-IC, into HeLa cells. Reporter activities were normalized to the internal control pRL-0, and values obtained with 320 ng empty expression plasmid were set to 1. Data were calculated from a single representative of 3 experiments with 3 transfections. Error bars indicate SD.
The ability of EBF and E47 to act synergistically on the λ5 promoter is abolished by expression of Notch1-IC. (A) The diagram shows the relative luciferase activity obtained after transiently transfecting HeLa cells with 40 ng of the λ5 reporter plasmid and 120 ng of the EBF and/or 120 ng of the E47-encoding expression plasmid, along with 40 or 120 ng of the Notch1-IC expression plasmid. (B) Graphs display the resulting relative activity of a λ5 reporter plasmid in HeLa cells after activation by ectopic expression of a forced dimer of E47 in the presence of increasing amounts of cotransfected Notch1-IC plasmid as indicated. (C) Columns representing the relative luciferase activity observed after cotransfecting 40 ng CD19 reporter construct and 200 ng Pax-5 alone, or together with 120 ng Notch1-IC, into HeLa cells. Reporter activities were normalized to the internal control pRL-0, and values obtained with 320 ng empty expression plasmid were set to 1. Data were calculated from a single representative of 3 experiments with 3 transfections. Error bars indicate SD.
Ectopic expression of a constitutively active Notch1 reduces the functional activity of EBF
The λ5 and Cd79a promoters are both targets of EBF and E47.12,24 Because Notch signaling has been shown to inhibit E47 activity,8 we wanted to investigate how ectopic expression of Notch1-IC acts on the coordinated activity of EBF and E47. To this end we transfected a λ5 promoter reporter plasmid together with an EBF expression plasmid and a plasmid expressing a forced dimer of E47 (E47FD)12 into HeLa cells (Figure 2A). Relative to the λ5 promoter, EBF or E47FD alone gave rise to a 20-fold induction, whereas together they activated the reporter nearly 300-fold. Cotransfection of 40 ng or 120 ng Notch1-IC gradually diminished this to 110- and 45-fold, respectively (Figure 2A). A reduction in functional activity mediated by Notch1-IC could also be observed when the λ5 promoter was activated by E47FD alone (Figure 2B). The 12-fold induction of promoter activity was reduced to 7- and 3-fold on cotransfection with 40 or 120 ng Nothc1-IC-expressing plasmid. This indicates that the functional synergy between EBF and E47 is reduced by Notch signaling and suggests that EBF function is directly or indirectly modulated by active Notch signaling.
Whereas both the λ5 and the Cd79a promoter contain E-boxes with the ability to bind E47,12,14 the B29 promoter does not,20 indicating that Notch1 signaling acts on additional factors crucial for early B-cell development. To investigate the effect of Notch1-IC on the transcriptional activation exerted by Pax-5, we transfected HeLa cells as described in “Materials and methods.” Pax-5 activated the human CD19 promoter 3.5-fold, whereas the combination of Pax-5 and Notch1-IC further induced activity (Figure 2C). We therefore conclude that Pax-5 is not repressed by Notch1-IC and that the reduced function of early B-cell promoters cannot be explained by a loss of Pax-5 activity.
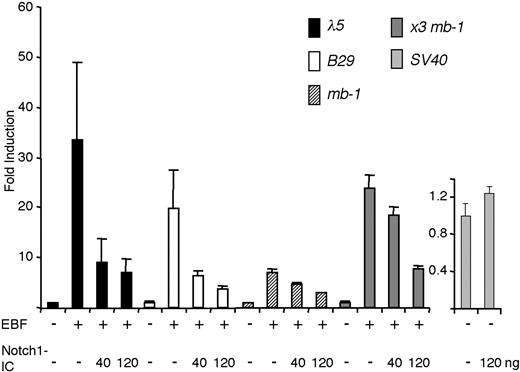
We next transfected a set of luciferase reporters containing EBF-responsive promoters into HeLa cells together with expression plasmids encoding EBF and Notch1-IC (Figure 3). The λ5 promoter was activated 33-fold by the inclusion of EBF-encoding plasmid, and this was reduced to 9-fold by the inclusion of 40 ng Notch1-IC, and further to 6-fold by the inclusion of 120 ng of the Notch1-IC encoding plasmid. The same amounts of Notch1-IC caused activity of the B29 promoter to decline from 20-fold to 6- and less than 4-fold, whereas the 10-fold induction of the Cd79a promoter was reduced to 7 and 3 times that of the basal activity of the promoter. To further link the reduced induction of the EBF target genes to EBF function, we cloned a synthetic promoter composed of 3 Cd79a EBF-binding sites (x3 Cd79a) in front of an Igκ promoter TATA box. Cotransfection of this reporter and EBF resulted in a 24-fold induction that was reduced to 18- and 8-fold in the presence of increasing amounts of Notch1-IC plasmid. The function of an SV40 promoter-controlled reporter plasmid was not significantly altered by cotransfection of Notch1-IC, supporting the idea that the repression was restricted to EBF and not a general effect. These data suggest that EBF function can be modulated by Notch1-IC.
Notch1-IC reduces the DNA-binding ability of EBF
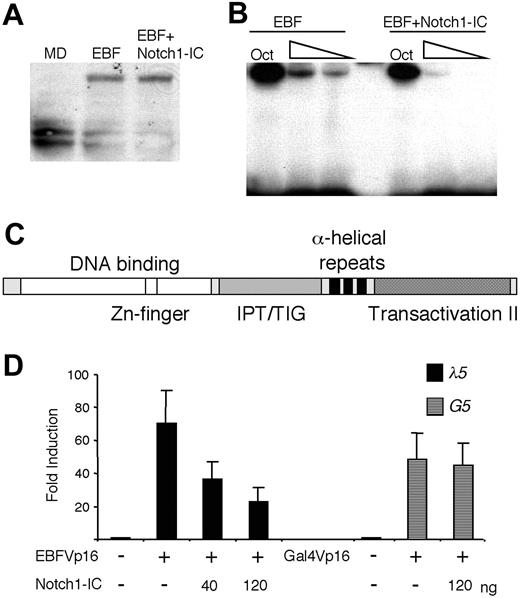
Notch signaling has been shown to induce expression of hairy/enhancer of split homologues (HES) proteins,3,4,25 potential repressors of E-protein activity, but also to stimulate ubiquitination and degradation of E47.26,27 To investigate the influence of Notch1-IC on EBF protein levels, we transfected HeLa cells with a myctagged EBF protein and assayed the level of EBF in nuclear extracts by Western blot using anti-myc antibody (Figure 4A). The vector-transduced cells did not generate any band of the expected size, whereas myc-EBF-transduced cells expressed the expected protein of roughly 55 to 60 kDa. The same protein (comparable intensity and size) could be detected in nuclear extracts from cells transduced with myc-EBF and Notch1-IC. Thus, we were unable to detect any signs that EBF is modified or degraded as a consequence of Notch1 signaling. To investigate the ability of the protein to bind DNA, we performed an EMSA, using the same nuclear extracts and an Cd79a promoter EBF-binding site as probe (Figure 4B). Even though all the extracts contained comparable amounts of Oct1 DNA-binding activity as assayed by binding of protein to a consensus octamer site, DNA binding by EBF to the Cd79a promoter site was reduced in the presence of Notch1-IC. To verify that the activity of Notch1 on EBF was directed to the DNA-binding part of the protein, we transiently transfected HeLa cells with an EBF fusion protein (EBFVp16), where the transactivation domain of EBF (Figure 4C) was replaced with that of the Vp16.18 This fusion protein activated the λ5 reporter 70-fold, which was reduced to 36- and 24-fold by the inclusion of 40 and 120 ng Notch1-IC, respectively (Figure 4D). By contrast, Notch1-IC had no effect on a Gal-4Vp16 fusion protein (Figure 4D). Taken together, these data argue that Notch1 signaling inhibits EBF function by reducing the DNA-binding activity of the protein.
Ectopic expression of Notch1-IC affects the ability of EBF to activate target promoters. The luciferase data were attained by transiently transfecting HeLa cells with 40 ng of respective indicated reporter construct and either 200 ng EBF alone or EBF in combination with 40 or 120 ng Notch1-IC-encoding expression plasmid. The data are based on a single representative of 3 experiments including 3 transfections normalized to the internal control pRL-0. The reporter activities obtained with empty expression plasmid were set to 1 and error bars indicate SD.
Ectopic expression of Notch1-IC affects the ability of EBF to activate target promoters. The luciferase data were attained by transiently transfecting HeLa cells with 40 ng of respective indicated reporter construct and either 200 ng EBF alone or EBF in combination with 40 or 120 ng Notch1-IC-encoding expression plasmid. The data are based on a single representative of 3 experiments including 3 transfections normalized to the internal control pRL-0. The reporter activities obtained with empty expression plasmid were set to 1 and error bars indicate SD.
Ectopic expression of Notch1-IC impairs the DNA-binding ability of EBF. (A) EBF protein levels displayed on a Western blot obtained with an anti-c-myc antibody and 10 μL of the nuclear extracts also used for the EMSA (B) or nuclear extract from HeLa cells transfected with empty vector (MD). (B) An EMSA analysis where an end-labeled Cd79a promoter EBF-binding site was incubated with 6, 4, or 2 μL nuclear extract from HeLa cells transfected either solely with EBF or with EBF and Notch1-IC-encoding plasmids collectively. For nuclear extract quality control, 1 μL of the respective nuclear extract was incubated with an Oct1 protein-binding probe. (C) A schematic drawing of the EBF protein28,29 with the DNA-binding domain indicated in white, an IPT/TIG (immunoglobulin-like, plexins, transcription factors/transcription factor immunoglobulin domain) domain by stripes, the dimerization domain by black, and the major transactivation domain substituted with that of the Vp16 protein by dots. (D) Diagrams display the relative luciferase activity of the λ5 reporter or a Gal-4 (G5) responsive reporter plasmid after transfection into HeLa cells in the presence or absence of EBFVp16 or Gal-4Vp16 protein and Notch1-IC-expressing plasmids. The data are based on a single representative of 3 experiments including 3 transfections normalized to the internal control pRL-0. The reporter activities obtained with empty expression plasmid were set to 1 and error bars indicate SD.
Ectopic expression of Notch1-IC impairs the DNA-binding ability of EBF. (A) EBF protein levels displayed on a Western blot obtained with an anti-c-myc antibody and 10 μL of the nuclear extracts also used for the EMSA (B) or nuclear extract from HeLa cells transfected with empty vector (MD). (B) An EMSA analysis where an end-labeled Cd79a promoter EBF-binding site was incubated with 6, 4, or 2 μL nuclear extract from HeLa cells transfected either solely with EBF or with EBF and Notch1-IC-encoding plasmids collectively. For nuclear extract quality control, 1 μL of the respective nuclear extract was incubated with an Oct1 protein-binding probe. (C) A schematic drawing of the EBF protein28,29 with the DNA-binding domain indicated in white, an IPT/TIG (immunoglobulin-like, plexins, transcription factors/transcription factor immunoglobulin domain) domain by stripes, the dimerization domain by black, and the major transactivation domain substituted with that of the Vp16 protein by dots. (D) Diagrams display the relative luciferase activity of the λ5 reporter or a Gal-4 (G5) responsive reporter plasmid after transfection into HeLa cells in the presence or absence of EBFVp16 or Gal-4Vp16 protein and Notch1-IC-expressing plasmids. The data are based on a single representative of 3 experiments including 3 transfections normalized to the internal control pRL-0. The reporter activities obtained with empty expression plasmid were set to 1 and error bars indicate SD.
EBF function is modulated by receptor-mediated Notch signaling in hematopoietic progenitor cells
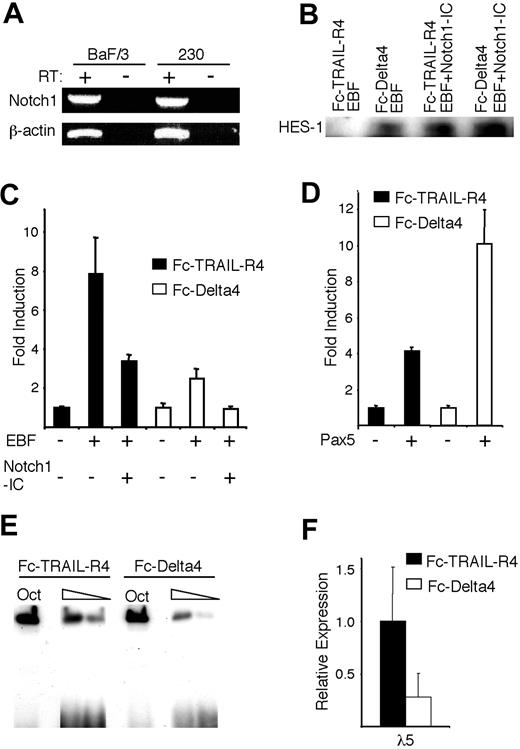
Although constitutively active Notch proteins (Notch-IC) are commonly used to study Notch activity, they may not fully mimic the Notch signaling pathway. To verify that EBF function is modulated by the interaction of the endogenous Notch1 receptor with its ligand Delta4, we transfected a Notch1-expressing hematopoietic progenitor cell line, BaF/3 (Figure 5A), with EBF and the λ5 promoter reporter construct, and then incubated these transfected cells for 36 hours in plates coated with either an Fc-Delta4 fusion protein, known to induce a robust Notch signal, or an Fc-TRAIL-R4 fusion protein as control.19,30 To confirm that Delta4 induced a Notch response in BaF/3 cells, we performed a Western blot against the Notch target HES-1 (Figure 5B). HES-1 protein was undetectable in the cells incubated on Fc-TRAIL-R4, but was clearly evident after incubation on Fc-Delta4 or after cotransfection with a Notch1-IC expression plasmid, supporting the idea that Fc-Delta4 is able to stimulate Notch activation in these cells. Incubation of the cells on Fc-TRAIL-R4 resulted in an 8-fold activation of the λ5 promoter after inclusion of EBF (Figure 5C). This activity was reduced by 60% after cotransfection with the Notch1-IC plasmid (Figure 5C). A reduced ability of EBF to activate the reporter gene was observed even when cells transduced with only EBF were incubated in wells coated with the Fc-Delta4 protein, because this resulted in a 2.5-fold rather than 8-fold activation of the λ5 reporter (Figure 5C). The inclusion of the Notch1-IC plasmid under these conditions resulted in a complete abrogation of the response of the reporter to EBF (Figure 5C). The opposite pattern could be observed when the CD19 promoter was cotransfected with a Pax-5-encoding expression plasmid, because this gave rise to a 5-fold induction of the reporter gene when the cells were incubated on Fc-TRAIL-R4 and a 10-fold induction when incubated on Fc-Delta4 (Figure 5D). No significant changes in expression of the cotransfected CMV-Renilla reporter were observed, arguing that reduced induction of the reporter was not a result of general effects, or a reduced function of the CMV promoter driving the expression of EBF. To investigate the effect of Notch signaling on the EBF-induced expression of the endogenous λ5 gene, we transduced the BaF/3 cells, normally not expressing either EBF or λ5, with an EBF-encoding retrovirus.12,17 EMSA analysis using the Cd79a promoter EBF site revealed that the DNA-binding activity of the ectopically expressed EBF was reduced (Figure 5E), and when the expression of the endogenous λ5 gene was investigated by real-time PCR, the level was lowered by 70% as compared with the control (Figure 5F). Together, these data suggest that the capacity of EBF to induce transcription from a target promoter in a hematopoietic progenitor cell is reduced by Notch1 signaling through the endogenous receptor.
Notch signaling induced by immobilized Delta4 ligand modulates EBF function. (A) RT-PCR analysis, visualized on an ethidium bromide-stained agarose gel, displaying the expression of Notch1 in the Baf/3 progenitor cell line and the 230-238 pre-B cell line. (B) Display of a Western blot analysis of HES-1 expression in BaF/3 cells after 36 hours of incubation on either the Fc-TRAIL-R4 or the Fc-Delta4 fusion protein, and after transfection of the cells with Notch1-IC as indicated. (C-D) Diagrams show the resulting relative luciferase activity obtained after DEAE-dextran-mediated transient transfections of 0.5 μg λ5 or CD19 promoter reporter constructs, 3.5 μg EBF, and 1.5 μg Notch1-IC, or 3.5 μg Pax-5 expression plasmids as indicated, into Baf/3 pro-B cells that then were incubated on plates coated with either the control protein Fc-TRAIL-R4 (▪) or Fc-Delta4 (□). The data are based on 3 transfections from a single representative of 3 experiments normalized to the internal control CMV Renilla. The reporter activities obtained with empty expression plasmid were set to 1 and error bars indicate SD. (E) Display of an EMSA where the DNA binding of ectopically expressed EBF to the Cd79a promoter EBF sites were assayed following incubation of the stably transduced cells on the control Fc-TRAIL-R4 or the Fc-Delta4 protein for 40 hours. (F) Diagrams show the expression of the endogenous λ5 gene in the cells stimulated in panel E. The data represent an average of 6 PCRs from 2 stimulation experiments and the error bars indicate the SD.
Notch signaling induced by immobilized Delta4 ligand modulates EBF function. (A) RT-PCR analysis, visualized on an ethidium bromide-stained agarose gel, displaying the expression of Notch1 in the Baf/3 progenitor cell line and the 230-238 pre-B cell line. (B) Display of a Western blot analysis of HES-1 expression in BaF/3 cells after 36 hours of incubation on either the Fc-TRAIL-R4 or the Fc-Delta4 fusion protein, and after transfection of the cells with Notch1-IC as indicated. (C-D) Diagrams show the resulting relative luciferase activity obtained after DEAE-dextran-mediated transient transfections of 0.5 μg λ5 or CD19 promoter reporter constructs, 3.5 μg EBF, and 1.5 μg Notch1-IC, or 3.5 μg Pax-5 expression plasmids as indicated, into Baf/3 pro-B cells that then were incubated on plates coated with either the control protein Fc-TRAIL-R4 (▪) or Fc-Delta4 (□). The data are based on 3 transfections from a single representative of 3 experiments normalized to the internal control CMV Renilla. The reporter activities obtained with empty expression plasmid were set to 1 and error bars indicate SD. (E) Display of an EMSA where the DNA binding of ectopically expressed EBF to the Cd79a promoter EBF sites were assayed following incubation of the stably transduced cells on the control Fc-TRAIL-R4 or the Fc-Delta4 protein for 40 hours. (F) Diagrams show the expression of the endogenous λ5 gene in the cells stimulated in panel E. The data represent an average of 6 PCRs from 2 stimulation experiments and the error bars indicate the SD.
Active Notch1 signaling reduces EBF DNA-binding capacity in pre-B cells
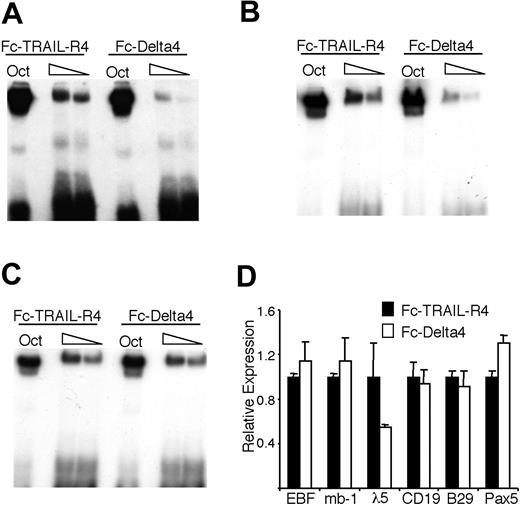
To investigate if EBF DNA-binding ability could be modulated by Notch signaling mediated through the endogenous receptors, we incubated 230-238 pre-B cells (which express Notch1 as judged by RT-PCR; Figure 5A) on plates coated either with the control Fc-TRAIL-R4 or the Fc-Delta4 fusion protein. In contrast to the BaF/3 cells, the 230-238 cells contained a background expression of HES-1, as judged by both real-time PCR and Western blot, but incubation on Fc-Delta4 still resulted in an about a 2-fold increase in HES-1 expression (data not shown). EMSA revealed that even though nuclear extracts from 230-238 cells grown on Fc-Delta4 or Fc-TRAIL-R4 gave rise to the same amounts of Oct1 DNA binding, there was a reduction in EBF binding to the Cd79a promoter in cells grown on Fc-Delta4 (Figure 6A). This effect was most dramatic after 48 hours of ligand exposure, whereas both earlier and later time points gave less definitive results (data not shown). Similar results were obtained when using an EBF site from the λ5 promoter (Figure 6B), arguing against a specific effect on EBF binding to the Cd79a promoter. However, on inclusion of the γ-secretase inhibitor DAPT in the cell cultures, no reduction of EBF DNA binding on the Cd79a promoter could be detected, providing further support for the idea that the observed effect is dependent on a functional Notch signaling pathway (Figure 6C). Q-RT-PCR analysis of the 230-238 cells incubated on Fc-TRAIL-R4 or Fc-Delta4 suggested that the EBF mRNA levels were comparable (Figure 6D), arguing against a direct effect on EBF transcription. Fc-Delta4 did not significantly affect the endogenous expression of Pax-5, B29, Cd79a, or CD19 either, but the expression of λ5 message was reduced by 50% (Figure 6D).
Notch signaling induced by immobilized Delta4 ligand modulates the DNA-binding activity of EBF in 230-238 pre-B cells. (A-B) The resulting autoradiogram of an EMSA experiment where radiolabeled Oct1 protein-binding sites, (A) Cd79a promoter EBF-binding site or (B) λ5 promoter EBF-binding site, were incubated with nuclear extracts from the 230-238 pre-B cell line cultured on either Fc-TRAIL-R4- or Fc-Delta4-coated plates for 48 hours. (C) Display and EMSA results as in panel A, but here the cells were incubated in the presence of the γ-secretase inhibitor DAPT. (D) Diagrams display Q-PCR analysis from the 230-238 cells used in the EMSA. The relative mRNA expression levels of EBF, Cd79a, Pax-5, CD19, B29, and λ5. The data are based on 6 PCR experiments from 2 independent stimulation experiments. Error bars indicate SD.
Notch signaling induced by immobilized Delta4 ligand modulates the DNA-binding activity of EBF in 230-238 pre-B cells. (A-B) The resulting autoradiogram of an EMSA experiment where radiolabeled Oct1 protein-binding sites, (A) Cd79a promoter EBF-binding site or (B) λ5 promoter EBF-binding site, were incubated with nuclear extracts from the 230-238 pre-B cell line cultured on either Fc-TRAIL-R4- or Fc-Delta4-coated plates for 48 hours. (C) Display and EMSA results as in panel A, but here the cells were incubated in the presence of the γ-secretase inhibitor DAPT. (D) Diagrams display Q-PCR analysis from the 230-238 cells used in the EMSA. The relative mRNA expression levels of EBF, Cd79a, Pax-5, CD19, B29, and λ5. The data are based on 6 PCR experiments from 2 independent stimulation experiments. Error bars indicate SD.
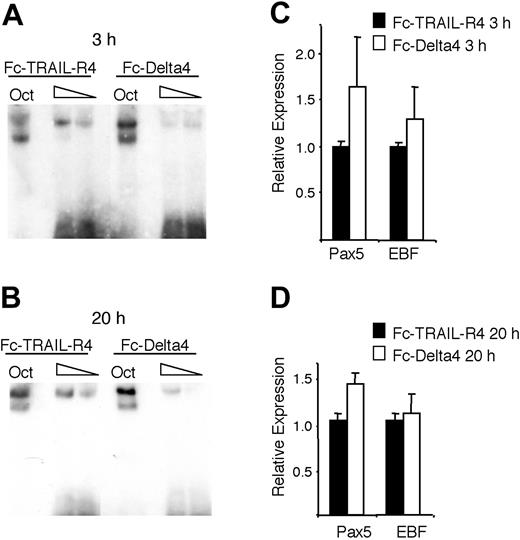
Notch signaling reduces the DNA-binding activity of EBF in primary CD19+ bone marrow cells. (A-B) The resulting autoradiogram of an EMSA experiment where either Oct1 protein-binding site or Cd79a promoter EBF-binding site was incubated with nuclear extracts from CD19+ bone marrow cultured on either Fc-TRAIL-R4- or Fc-Delta4-coated plates for 3 or 20 hours as indicated. (C-D) Diagrams display Q-PCR analysis from the CD19+ cells used in the EMSA. The relative mRNA expression levels of EBF and Pax-5 are shown. The data are based on 6 PCR experiments and 2 separate stimulations. Error bars indicate SD.
Notch signaling reduces the DNA-binding activity of EBF in primary CD19+ bone marrow cells. (A-B) The resulting autoradiogram of an EMSA experiment where either Oct1 protein-binding site or Cd79a promoter EBF-binding site was incubated with nuclear extracts from CD19+ bone marrow cultured on either Fc-TRAIL-R4- or Fc-Delta4-coated plates for 3 or 20 hours as indicated. (C-D) Diagrams display Q-PCR analysis from the CD19+ cells used in the EMSA. The relative mRNA expression levels of EBF and Pax-5 are shown. The data are based on 6 PCR experiments and 2 separate stimulations. Error bars indicate SD.
To investigate if Notch signaling could modulate EBF DNA binding in a normal B-lymphoid cell, we purified CD19+ cells from mouse BM and incubated these on plates coated with Fc-TRAIL-R4 or Fc-Delta4. This resulted in a down-regulation of DNA-binding activity after 3 hours (Figure 7A) and 24 hours (Figure 7B) of incubation without any dramatic effect on EBF or Pax-5 expression at these time points (Figure 7C-D). Although we may have expected a more widespread effect on gene expression, we conclude that bona fide Notch signaling reduces EBF DNA-binding activity in B-lineage cells.
Discussion
We provide data suggesting that the capacity of EBF to activate several target promoters is reduced by active Notch signaling and that this is associated with an impaired ability of EBF to bind DNA. This has important implications in the understanding of the mechanisms underlying the role of Notch signaling in B-cell versus T-cell commitment. The potential of Notch to inhibit both EBF and E47 function, while possibly enhancing the function of Pax-5, may provide an extended insight into definitive B-cell commitment. It has been shown that despite the fact that both EBF and E47, as well as several of their target genes, are expressed at high levels in B-cell progenitors from Pax-5-deficient mice,31,32 these cells are able to differentiate into other lineages, including T cells, in vivo.33 In addition, Pax-5-deficient cells are able to develop along the T-lineage pathway when cultured on Delta-ligand-expressing stromal cells.23 This strongly suggests that the definitive choice to proceed into the B-lymphoid pathway is incomplete even in the presence of EBF and E47 in the progenitor cell and proposes an absolute requirement for Pax-5 expression in the establishment of B lymphopoiesis. One mechanism that has been suggested to be of significant importance in the Pax-5-mediated lineage decision is the ability to suppress expression of Notch-1 in the progenitor cells.34 Thus, our current findings, in combination with previous data showing a functional repression of E47 by Notch signaling,8 provide a possible molecular explanation for the inability of EBF and E47 to lock the cells into the B-lymphoid developmental pathway in the absence of Pax-5 expression. Expression of Notch-1 in the absence of Pax-5 makes the cells susceptible to Delta or Jagged stimulation, and the corresponding drop in the functional activity of EBF and E47. Thus, the apparent role of EBF and E47 to induce but not lock B-cell development could be a result of susceptibility to the action of Notch signaling. Moreover, EBF is noted to be involved in the regulation of Pax-5 expression, suggesting an intricate interplay between transcription factors and Notch signaling in lymphoid lineage commitment.
These findings further strengthen the idea that the functions of EBF and E47 are closely linked in the earliest stages of B-cell development. These proteins have been shown to cooperate in the direct activation of pre-B cell-restricted target promoters such as Cd79a,14 λ5,12 V-preB,12 and B29.20 They are also likely to collaborate in the induction of immunoglobulin recombination events,35,36 making them key regulators of genes encoding critical components of the pre-B cell receptor. The exact mechanism of the functional synergy between EBF and E47 remains unclear but several lines of investigation suggest a cooperative binding to target promoters.14,18 In vivo studies of compound heterozygous mice lacking one allele of EBF and one of E2A revealed that these mice display a more pronounced impairment of B-cell development than mice lacking one allele of either EBF or E2A.11 This could indicate that the proteins act in a dose-dependent coordinated manner, and the ability of Notch to modulate the activity of either of the factors is likely to have an even greater impact on the functional synergy between these proteins. Both EBF and E47/E2A are independently crucial for B-cell development,9,13 but because B-cell development in E47-deficient mice can be rescued by ectopic expression of EBF,15 EBF may be a key factor to be modulated by Notch signaling in early lymphopoiesis. In contrast to the effects observed on target promoters after cotransfection of Notch1-IC (Figure 1), the PCR analysis of EBF target genes in the 230-238 pre-B cells suggested that the direct effect of receptor-mediated signaling on the endogenous gene expression was rather modest. The only significant decrease in mRNA levels was seen for λ5, possibly indicating that once the genetic program of the pre-B cell is established, the need of EBF and E47 for a set of target genes is reduced. It could also be a reflection of maintained or increased Pax-5 activity because this protein has been shown to share target genes with EBF and E47.17,37 However, the effect on λ5 transcription in both established pre-B cells and in the EBF-expressing BaF/3 cells (Figures 5, 6) shows that the reduced EBF activity is reflected in a reduction of target gene expression and this could be of crucial importance in a lineage decision event where the dose of an EBF target could be essential. This should also be considered in light of the fact that we probably have not yet identified the critical target genes for EBF in the earliest stages of B-cell development.
It has been suggested that the ability of Notch signaling to modulate E47 activity results from increased protein degradation26,27 and from direct inhibition of E2A function by HES proteins.25 We have not observed any apparent Notch1-induced EBF degradation, and the modest induction of HES-1 in 230-238 cells (Figure 6A and data not shown) was in contrast to the dramatic effect on EBF DNA binding, arguing against a direct influence of HES-1 on EBF DNA binding. To further investigate this we cotransfected EBF and HES-1 into HeLa cells and analyzed protein expression by Western blotting and DNA binding by EMSA. We were unable to observe any alternations in either EBF protein expression or DNA binding (data not shown), providing further support for the idea that the inability of EBF to bind DNA is not a direct effect of HES-1 action. These findings suggest that Notch signaling reduces the function of EBF by modulation of the DNA-binding activity of the protein in the context of a pre-B cell. Thus, we have not yet been able to unravel the mechanisms by which Notch inhibits EBF DNA binding, demanding additional biochemical analyses of the interaction between EBF and the Notch signaling pathway.
In addition to its important role in lymphoid development, Notch signal modulation of EBF activity may also be of critical importance in differentiation systems outside of the developing immune system. EBF stimulates neurogenesis in Xenopus38,39 as well as adipogenesis in mice,40 2 processes that are modulated by active Notch signaling.30,38,39 Key roles for the Notch/EBF link may also be revealed from further studies of these model systems for cellular differentiation.
Prepublished online as Blood First Edition Paper, May 26, 2005; DOI 10.1182/blood-2004-12-4744.
Supported by the Swedish Cancer Society (Cancerfonden), The Swedish Research Council (VR) [Vetenskaprådet], the Barn Cancer, Kocks, Österlunds, and Crafoord foundations, and the Medical Faculty at Lund University. The Lund Strategic Center for Stem Cell Biology and Cell Therapy is sponsored by a center grant from Stiftelsen för Strategisk Forskning (SSF).
E.M.K.S. conducted the large majority of the experiments with some help from P.Å., H.A., and M.S. T.K. provided crucial reagents as well as important ideas about experimental design. All authors contributed to the writing of the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Drs Warren Pear, Björn Olde, and Meinrad Busslinger for the kind gifts of reagents; Gerd Sten and Lillian Wittman for technical assistance; and Sten Eirik Jacobsen and Jalal Taneera for stimulating discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal