Abstract
Drug resistance in childhood acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) is associated with impaired ability to induce apoptosis. To elucidate causes of apoptotic defects, we studied the protein expression of Apaf-1, procaspases-2, -3, -6, -7, -8, -10, and poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) in cells from children with acute lymphoblastic leukemia (ALL; n = 43) and acute myeloid leukemia (AML; n = 10). PARP expression was present in all B-lineage samples, but absent in 4 of 15 T-lineage ALL samples and 3 of 10 AML cases, which was not caused by genomic deletions. PARP expression was a median 7-fold lower in T-lineage ALL (P < .001) and 10-fold lower in AML (P < .001) compared with B-lineage ALL. PARP expression was 4-fold lower in prednisolone, vincristine and L-asparaginase (PVA)-resistant compared with PVA-sensitive ALL patients (P < .001). Procaspase-2 expression was 3-fold lower in T-lineage ALL (P = .022) and AML (P = .014) compared with B-lineage ALL. In addition, procaspase-2 expression was 2-fold lower in PVA-resistant compared to PVA-sensitive ALL patients (P = .042). No relation between apoptotic protease-activating factor 1 (Apaf-1), procaspases-3, -6, -7, -8, -10, and drug resistance was found. In conclusion, low baseline expression of PARP and procaspase-2 is related to cellular drug resistance in childhood acute lymphoblastic leukemia. (Blood. 2005;106:1817-1823)
Introduction
The treatment of pediatric acute leukemia has greatly improved in the past 4 decades, resulting in long-term disease-free survival of approximately 80%1 for acute lymphoblastic leukemia (ALL) and 60% for acute myeloid leukemia (AML).2 Despite this progress, therapy resistance in a significant number of children still forms a major obstacle to successful treatment. We and others have previously shown that ALL patients whose leukemia cells exhibit in vitro resistance to antileukemic agents have a significantly worse prognosis than patients whose leukemic cells are sensitive.3-6 Compared with ALL, leukemic cells of children with AML have a less favorable prognosis and are in vitro more resistant to several antileukemic agents.7
Apoptosis is the predominant form of cell death triggered in vivo and in vitro by chemotherapeutic agents in hematologic malignancies.8 There are 2 major routes by which apoptosis can be induced: the intrinsic and the extrinsic apoptosis pathways. The intrinsic pathway is initiated by mitochondrial damage that leads to release of cytochrome c from the mitochondrial intermembrane space.9 Upon entry in the cytoplasm, cytochrome c interacts with apoptotic protease-activating factor 1 (Apaf-1), deoxyadenosine triphosphate (dATP), and procaspase-9 to form a complex known as the apoptosome.10 In the apoptosome, caspase-9 is activated, which in turn induces processing and activation of downstream effector caspases such as caspases-2, -3, -6, and -7.11,12 Activation of the effector caspases results in the cleavage of a number of structural and regulatory cellular proteins (eg, poly(adenosine diphosphate [ADP]-ribose) polymerase [PARP; also known as PARP1 and ADPRT]) and lamins.13,14 Initiation of the extrinsic apoptosis pathway involves ligand-induced aggregation of membrane receptors of the tumor necrosis factor receptor superfamily and subsequent cytoplasmic recruitment of Fas-associated protein with death domain (FADD), and procaspase-8 or procaspase-10 to form the death-inducing signaling complex (DISC).15,16 Within the DISC procaspase-8 or procaspase-10 is activated and released back into the cytoplasm, where it induces activation of downstream effector caspases. Both pathways converge at the level of caspase-3 activation and therefore have several downstream effector caspases and substrates (eg, PARP) in common.
Decreased apoptosis may be an important event in the acquisition of cellular drug resistance in pediatric acute leukemia.17 Studies evaluating the expression levels of apoptotic proteins in clinical samples are limited in leukemia and mainly restricted to adult acute leukemia.18-22 In the present study, we examined the expression of Apaf-1, procaspases-2, -3, -6, -7, -8, -10, and PARP in diagnostic samples containing at least 85% leukemic blasts of 43 children with ALL and 10 children with AML using quantitative Western blotting. The protein expression was compared between B-lineage ALL, T-lineage ALL, and AML, and between acute leukemia cases that are in vitro-sensitive and resistant to prednisolone (PRED; Bufa Pharmaceutical Products, Uitgeest, The Netherlands), vincristine (VCR; TEVA Pharma, Mijdrecht, The Netherlands), l-asparaginase (ASP; Paronal; Christiaens, Breda, The Netherlands), and daunorubicin (DNR; Cerubidine; Rhône-Poulenc Rorer, Amstelveen, The Netherlands).
Patients, materials, and methods
Leukemia samples
Bone marrow and peripheral blood samples were obtained at initial diagnosis after informed consent from children with newly diagnosed ALL who were enrolled on protocol Dutch Childhood Oncology Group (DCOG) ALL-9 at the Erasmus MC/Sophia Children's Hospital or on COALL-97 treatment protocol at one of the hospitals participating in the German COALL study group. Approval was obtained from the Erasmus MC/Sophia Children's Hospital institutional review board for these studies. Mononuclear cells were isolated by sucrose density gradient centrifugation (Lymphoprep, density 1.077 g/mL; Nycomed Pharma, Oslo, Norway), within 24 hours after sampling at the research laboratory of Pediatric Oncology, Erasmus MC/Sophia Children's Hospital, Rotterdam, The Netherlands. Cells were resuspended in culture medium consisting of RPMI 1640 (Dutch modification without l-glutamine; Gibco BRL, Life Technologies, Breda, The Netherlands) supplemented with 20% fetal calf serum (FCS; Integro, Zaandam, the Netherlands), 2 mM l-glutamine, 200 μg/mL gentamycin (Gibco BRL) 100 IU/mL penicillin, 100 μg/mL streptomycin, 0.125 μg/mL fungizone (Gibco BRL), and 5 μg/mL insulin, 5 μg/mL transferring, and 5 ng/mL sodium selenite (bovine insulin, human transferin, and sodium selenite [ITS] media supplement; Sigma-Aldrich Chemie B.V., Zwijndrecht, the Netherlands). If necessary, leukemic samples were further enriched to at least 85% leukemic blasts by removing nonmalignant cells with immunomagnetic beads (DynaBeads, Dynal Oslo, Norway).23 Cell pellets were immediately stored at -80°C until use.
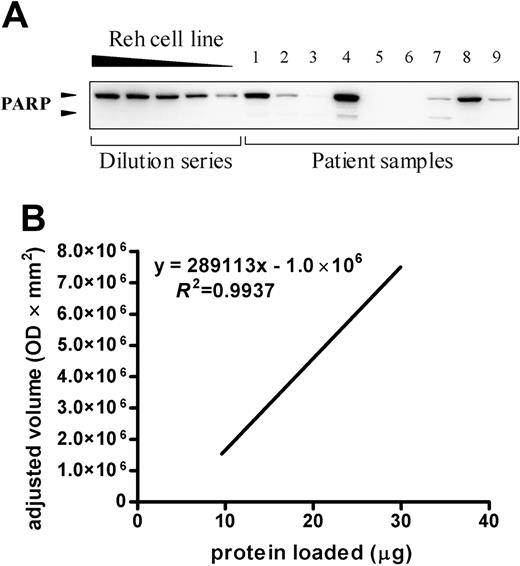
Quantification of apoptotic proteins in newly diagnosed childhood acute leukemia. A representative blot for cells of 9 acute leukemia samples incubated with monoclonal anti-PARP antibody is depicted in the top panel. The dilution series consists of 30, 25, 20, 15, and 10 μg Reh protein lysate. The presence of PARP was detected by enhanced chemiluminescent staining and the signal intensity was quantified by densitometry and plotted (bottom). This graph was used to estimate the relative quantity present in the patient's sample compared with the Reh cell line dilution series by linear regression. The 9 samples shown here are part of the group of 53 patients and include 3 B-lineage ALL samples, 3 T-lineage ALL samples, and 3 AML samples. OD indicates optical density.
Quantification of apoptotic proteins in newly diagnosed childhood acute leukemia. A representative blot for cells of 9 acute leukemia samples incubated with monoclonal anti-PARP antibody is depicted in the top panel. The dilution series consists of 30, 25, 20, 15, and 10 μg Reh protein lysate. The presence of PARP was detected by enhanced chemiluminescent staining and the signal intensity was quantified by densitometry and plotted (bottom). This graph was used to estimate the relative quantity present in the patient's sample compared with the Reh cell line dilution series by linear regression. The 9 samples shown here are part of the group of 53 patients and include 3 B-lineage ALL samples, 3 T-lineage ALL samples, and 3 AML samples. OD indicates optical density.
Antibodies
Monoclonal mouse antibodies to human procaspase-2 or PARP were purchased from PharMingen (San Diego, CA). Polyclonal rabbit antibodies to human procaspase-6, procaspase-10, and a monoclonal mouse antibody to human procaspase-8 were purchased from Cell Signaling Technology (Beverly, MA). In addition, we used monoclonal mouse antibodies to human procaspase-3 (Transduction Laboratories, Lexington, KY) and procaspase-7 (StressGen, Victoria, BC, Canada). Monoclonal antibodies tested for human procaspase-9 were purchased at Oncogene Research Products (Cambridge, MA), Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling Technology.
In vitro drug resistance assay
Responsiveness of leukemia cells to PRED, VCR, ASP, and DNR was determined by the 4-day in vitro (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) MTT drug resistance assay as described previously.3,4 The concentration ranges tested for these drugs were as follows: PRED, 0.008-250 μg/mL; VCR, 0.05-50 μg/mL; ASP, 0.003-10 IU/mL; and DNR, 0.002-2.0 μg/mL. The drug concentration lethal to 50% of the ALL cells (LC50 value) was used as the measure of cellular drug resistance. The PRED, VCR, and ASP (PVA) score was calculated as previously described (ie, each patient was given a score according to the LC50 value measured in the MTT assay).4 Patients were given a score of 1 if sensitive, 2 if intermediate, and 3 if resistant per agent. (The cut-off LC50 values used to assign these scores were those previously associated with treatment outcome in ALL4,24 and are provided in Table 1). The sum of the individual scores for PRED, VCR, and ASP resulted in the PVA score. Patients with a combined PVA score of 3 or 4 are relatively sensitive to PRED, VCR, and ASP, and have a favorable treatment outcome compared with patients with a combined PVA score of 7, 8, or 9, which are relatively resistant to these 3 drugs.
Cut-off LC50 values used to assign a patient as in vitro sensitive, intermediate, or resistant per drug
Drug . | In vitro sensitive . | In vitro resistant . |
|---|---|---|
| PRED, μg/mL | ≤0.100 | ≥150 |
| VCR, μg/mL | ≤0.391 | ≥1.758 |
| ASP, IU/mL | ≤0.033 | ≥0.912 |
| DNR, μg/mL | ≤0.075 | ≥0.114 |
Drug . | In vitro sensitive . | In vitro resistant . |
|---|---|---|
| PRED, μg/mL | ≤0.100 | ≥150 |
| VCR, μg/mL | ≤0.391 | ≥1.758 |
| ASP, IU/mL | ≤0.033 | ≥0.912 |
| DNR, μg/mL | ≤0.075 | ≥0.114 |
Western blotting
Leukemic cells were lysed in 50 mM Tris (tris(hydroxymethyl)-aminomethane; pH 7.6) containing 150 mM NaCl, 10 mM EDTA (ethylenediaminetetraacetic acid), and 1% Triton X-100. Protease inhibitors (200 μg/mL pepstatin A, 200 μg/mL leupeptin and 2 mM PMSF [phenylmethylsulfonyl fluoride]) were freshly added prior to use. Protein concentration was determined using the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). All samples were kept on ice during the protein isolation and lysates were either used directly or stored immediately at -80°C until use. Twenty micrograms of protein in Laemmli buffer (4% [wt/vol] sodium dodecyl sulfate [SDS], 100 mM Tris-HCl [pH 6.8], and 20% glycerol) was heated to 100°C for 3 minutes and loaded onto a 5% to 15% gradient polyacrylamide gel. After electrophoresis, samples were transferred to nitrocellulose blots, blocked with skim milk, and incubated for 1 hour with primary antibodies. Blots were washed and incubated for 1 hour with peroxidase-conjugated secondary antibody (DAKO, Carpentaria, CA). Presence of proteins was detected by enhanced chemiluminescent staining using the SuperSignal West Femto kit (Pierce). Signals were directly scanned by the ChemiGenius imaging system (SynGene, Cambridge, United Kingdom) and signal intensity was quantified using GeneTools 3.1 image analysis software (SynGene).
Quantification of apoptotic proteins
A dilution series of 30, 25, 20, 15, and 10 μg protein lysate of the human B-cell leukemia cell line Reh (N CRL-8286; ATCC, Rockville, MD) was included in each gel. Expression levels of protein were expressed in arbitrary units (AU) and were based on the relative quantity present in the patient's sample compared to the Reh cell line dilution series (Figure 1). Reproducibility of our method was assessed by subjecting 27 samples to duplicate Western blot analysis and is illustrated in Figure 2 for 16 representative samples.
Real time quantitative PCR
Total cellular mRNA was extracted using Trizol reagent (Gibco BRL) and cDNA was synthesized using random hexamers and oligo dT primers. mRNA expression levels of PARP and GAPDH as a reference, were quantified using real-time polymerase chain reaction (PCR) analysis on an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) as previously described.25 PCR reactions were performed and optimized to an amplification efficiency of more than 95%. The comparative cycle time (Ct) value of the target PCR was normalized by subtracting the Ct value of GAPDH (ΔCt). The ΔCt value was used to calculate the relative expression level to GAPDH for each target PCR using the following formula: relative mRNA expression = 2-ΔCt × 100%.25 PARP primer sequences used were: sense, 5′-AGGCTGCTTTGTCAAGAA-3′; and antisense, 5′-CTTGCTGCTTGTTGAAGAT-3′ and the probe sequence: 5′-ATGAGGTGGATGGAGTGGATGA-3′. GAPDH primers and probe sequences have been described elsewhere.26
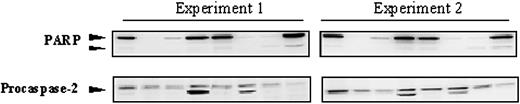
Reproducibility of PARP and procaspase-2 protein detection in childhood acute leukemia samples. Representative Western blots from 2 independent experiments incubated with monoclonal anti-PARP or anti-procaspase-2 antibodies are depicted.
Reproducibility of PARP and procaspase-2 protein detection in childhood acute leukemia samples. Representative Western blots from 2 independent experiments incubated with monoclonal anti-PARP or anti-procaspase-2 antibodies are depicted.
Fluorescence in situ hybridization analysis
To investigate the PARP gene copy number, dual colored fluorescence in situ hybridization (FISH) was applied using 2 combinations of probes: (1) Two 1q42-specific bacterial artificial chromosome (BAC) clones (Roswell Park Cancer Institute, Buffalo, NY), RP11-118H4 labeled with biotin-16-deoxyuridine triphosphate (dUTP), and RP11-125A15 labeled with digoxigenin-12-dUTP, with a 50-kilobase (kb) overlap in the PARP gene; and (2) a combination of the PARP-specific BAC clone RP11-118H4 (labeled with biotin-16-dUTP) and 2 P1-derived artificial chromosome (PAC) clones 203H23 and 213H16 (RCPI-6 Human PAC Library, Roswell Park Cancer Institute) labeled with digoxigenin-12-dUTP specific for the AF1q gene on 1q21. Cytospins of leukemic samples stored at -20°C were used for FISH analysis. The FISH protocol was based on that described previously.27 Briefly, slides were pretreated with RNase and pepsin and subsequently fixed with formaldehyde. Hybridization of 100 ng of each probe to the slides was performed overnight at 37°C. Biotinylated probes were detected via subsequent incubation with fluorescein isothiocyanate (FITC)-labeled avidin-d, biotinylated goat-anti-avidin and avidin-d FITC (Vector, Burlingame, CA), whereas digoxigenin-labeled probes were detected via sheep-anti-digoxigenin tetramethylrhodamine isothiocyanate (TRITC)-labeled antibodies (Boehringer Mannheim, Mannheim, Germany), followed by Texas-red-conjugated donkey antisheep antibodies (Jackson ImmunoResearch, Westgrove, PA). Slides were counterstained with DAPI (4′,6-diamidino-2-phenylindole) and embedded in Vectashield (Vector Laboratories, Burlingame, CA)/DABCO (1,4-diazabicyclo[2,2,2]octane). Hybridization results were examined using a Zeiss Axioplan 2 fluorescence microscope (Zeiss, Oberkochen, Germany).
Variation in expression of 8 apoptotic proteins in PRED, VCR, and ASP (PVA)-sensitive and -resistant childhood ALL. Protein levels of Apaf-1, procaspases-2, -3, -6, -7, -8, -10, and PARP of leukemic cells from 10 patients compared by Western blot analysis. Arrows indicate the positions of full-length procaspase-2 (48 kDa), procaspase-3 (32 kDa), procaspase-6 (33 kDa), procaspase-7 (35 kDa), procaspase-8 (50/55 kDa), procaspase-10 (58 kDa), full-length and cleaved PARP (116/89 kDa), and Apaf-1 (140 kDa). The in vitro drug responsiveness of each patient is indicated below each lane; R indicates in vitro resistant and S indicates in vitro sensitive toward PRED, VCR, and ASP (PVA), as explained in “Materials and methods.”
Variation in expression of 8 apoptotic proteins in PRED, VCR, and ASP (PVA)-sensitive and -resistant childhood ALL. Protein levels of Apaf-1, procaspases-2, -3, -6, -7, -8, -10, and PARP of leukemic cells from 10 patients compared by Western blot analysis. Arrows indicate the positions of full-length procaspase-2 (48 kDa), procaspase-3 (32 kDa), procaspase-6 (33 kDa), procaspase-7 (35 kDa), procaspase-8 (50/55 kDa), procaspase-10 (58 kDa), full-length and cleaved PARP (116/89 kDa), and Apaf-1 (140 kDa). The in vitro drug responsiveness of each patient is indicated below each lane; R indicates in vitro resistant and S indicates in vitro sensitive toward PRED, VCR, and ASP (PVA), as explained in “Materials and methods.”
Statistics
Protein expression in different leukemic subgroups (B-lineage ALL, T-lineage ALL, and AML) was compared using the Mann-Whitney U test. Spearman rank (rs) correlation test was used to correlate protein expression to several variables on the study. Statistical tests were performed at a 2-tailed significance level of .05. Whenever applicable, a Bonferroni correction was applied to correct for multiple comparisons.
Results
Expression of various apoptotic proteins in ALL
The expression of Apaf-1, procaspases-2, -3, -6, -7, -8, -10, and PARP was first determined in a test group of 20 children with ALL. These 20 patients were selected by having a PVA score of 3 or 4 (ie, relatively in vitro-sensitive to PRED, VCR, and ASP; n = 10) or 7, 8, or 9 (ie, relatively in vitro-resistant to PRED, VCR, and ASP; n = 10). The expression of procaspase-9 could not be analyzed in these samples since all procaspase-9 antibodies tested revealed abundant nonspecific staining, whereas staining of procaspase-9 itself was low. Most other apoptotic proteins were ubiquitously expressed in the ALL samples and varied markedly between patients (Figure 3). Exceptions were procaspase-10 and PARP, which were not expressed in 4 (2 B-lineage ALL, 2 T-lineage ALL) and 1 (T-lineage ALL) out of the tested 20 samples, respectively. Expression values in arbitrary units (AU) for each apoptotic protein are provided in Table 2. The variation in expression ranged between 5-fold for procaspase-3 and 44-fold for Apaf-1. In vitro cross-resistance to PVA is an important independent predictor of treatment failure and long-term outcome in childhood ALL.4,5,28 Therefore, the expression of the 8 apoptotic proteins was subsequently correlated to in vitro PVA resistance. Only procaspase-2 and PARP protein expression were significantly associated with in vitro PVA resistance in ALL. Procaspase-2 expression was 2.2-fold lower in cells of ALL patients resistant to PRED, VCR, and ASP, hereafter called PVA resistant (median: 6.2 AU) compared with cells of ALL patients sensitive to these 3 drugs, hereafter called PVA sensitive (median, 13.4 AU; P = .019). PARP expression was 2.4-fold lower in PVA-resistant (median, 15.1 AU) compared with PVA-sensitive children with ALL (median, 35.8 AU; P = .042). Based on these data, we examined the expression of PARP and procaspase-2 in a larger group of children with ALL (n = 43) and AML (n = 10).
Apoptosis-associated protein expression in PRED, VCR, and ASP (PVA)-sensitive and -resistant childhood ALL patients
Apoptotic protein . | PVA-sensitive patients, median AU (range) . | PVA-resistant patients, median AU (range) . | P . |
|---|---|---|---|
| Apaf-1TBLFN1 | 21.6 (12.7-124.4) | 17.4 (4.0-175.3) | .529TBLFN2 |
| Procaspase-2 | 13.4 (1.1-27.2) | 6.2 (1.2-10.1) | .019 |
| Procaspase-3 | 18.5 (9.9-41.0) | 20.8 (8.0-35.7) | .912 |
| Procaspase-6 | 17.5 (11.0-78.8) | 25.2 (16.3-49.0) | .280 |
| Procaspase-7 | 10.8 (6.1-32.4) | 8.6 (1.0-25.2) | .247 |
| Procaspase-8 | 32.4 (21.5-75.2) | 42.3 (17.0-110.1) | .393 |
| Procaspase-10 | 5.4 (0.0-40.8) | 6.4 (0.0-47.0) | .912 |
| PARP | 35.8 (6.5-48.9) | 15.1 (0.0-38.5) | .042 |
Apoptotic protein . | PVA-sensitive patients, median AU (range) . | PVA-resistant patients, median AU (range) . | P . |
|---|---|---|---|
| Apaf-1TBLFN1 | 21.6 (12.7-124.4) | 17.4 (4.0-175.3) | .529TBLFN2 |
| Procaspase-2 | 13.4 (1.1-27.2) | 6.2 (1.2-10.1) | .019 |
| Procaspase-3 | 18.5 (9.9-41.0) | 20.8 (8.0-35.7) | .912 |
| Procaspase-6 | 17.5 (11.0-78.8) | 25.2 (16.3-49.0) | .280 |
| Procaspase-7 | 10.8 (6.1-32.4) | 8.6 (1.0-25.2) | .247 |
| Procaspase-8 | 32.4 (21.5-75.2) | 42.3 (17.0-110.1) | .393 |
| Procaspase-10 | 5.4 (0.0-40.8) | 6.4 (0.0-47.0) | .912 |
| PARP | 35.8 (6.5-48.9) | 15.1 (0.0-38.5) | .042 |
Expression of 8 apoptotic proteins was compared in 10 PVA-sensitive (8 B-lineage ALL, 2 T-lineage ALL) and 10 PVA-resistant (5 B-lineage, 5 T-lineage) patients. For both groups, n = 10.
The expression of the predominantly expressed isoform was examined for all proteins.
Expression of an apoptotic protein in patient samples related to the expression of protein in a dilution series of the Reh leukemic cell line; see “Materials and methods.”
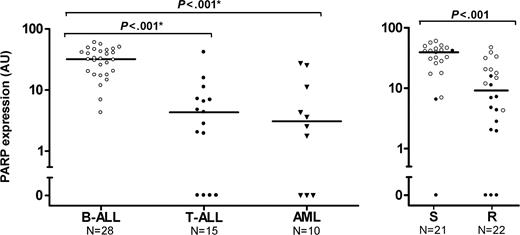
PARP expression levels in different types of childhood acute leukemia and in drug-resistant versus -sensitive childhood ALL. (A) PARP expression in different types of childhood acute leukemia. Each dot represents an individual patient and the horizontal bar represents the median protein level. Protein expression level of PARP was quantitated in 28 B-lineage ALL samples (○), 15 T-lineage ALL samples (•) and 10 AML samples (▾). (B) PARP expression versus in vitro PRED, VCR, and ASP (PVA) resistance in childhood acute lymphoblastic leukemia. The protein expression level of PARP quantitated in 21 PVA-sensitive (S) and 22 PVA-resistant (R) patients with B-lineage ALL (○) or T-lineage ALL (•). *Statistically significant after Bonferroni correction for multiple comparisons. AU indicates arbitrary units.
PARP expression levels in different types of childhood acute leukemia and in drug-resistant versus -sensitive childhood ALL. (A) PARP expression in different types of childhood acute leukemia. Each dot represents an individual patient and the horizontal bar represents the median protein level. Protein expression level of PARP was quantitated in 28 B-lineage ALL samples (○), 15 T-lineage ALL samples (•) and 10 AML samples (▾). (B) PARP expression versus in vitro PRED, VCR, and ASP (PVA) resistance in childhood acute lymphoblastic leukemia. The protein expression level of PARP quantitated in 21 PVA-sensitive (S) and 22 PVA-resistant (R) patients with B-lineage ALL (○) or T-lineage ALL (•). *Statistically significant after Bonferroni correction for multiple comparisons. AU indicates arbitrary units.
Expression of PARP in acute leukemia
Twenty-eight patients with B-lineage ALL, 15 patients with T-lineage ALL, and 10 patients with AML were studied. PARP expression varied between 0 and 59.8 AU in these patients with the relatively highest expression level found in B-lineage ALL (Figure 4A). The expression of PARP was significantly lower in T-lineage ALL (median, 4.3 AU; P < .001) and in AML (median, 3.1 AU; P < .001) compared to B-lineage ALL (median, 32.0 AU). PARP expression did not differ significantly between AML and T-lineage ALL. Whereas all patients with B-lineage ALL showed PARP expression, we identified 4 T-ALL and 3 AML patients without detectable PARP expression. PARP mRNA levels correlated with PARP protein levels (rs = .58, P = .018). FISH analysis of the 4 patients with T-lineage ALL and 3 patients with AML with absent PARP protein expression revealed that the lack of PARP protein and mRNA expression was not associated with genomic deletions in the PARP gene region of these patients (data not shown). Epigenetic changes such as DNA methylation of promoter regions may result in decreased mRNA levels; however, no cytosine-phosphate-guanine (CpG) islands have been reported within the promoter region of PARP, nor could we identify these using the CpGPlot program (http://www.ebi.ac.uk/emboss/cpgplot/).29
Next, we compared PARP expression in sensitive and resistant patients with acute leukemia to PRED, VCR, ASP, or DNR. After correction for multiple comparisons (Bonferroni model), we observed a significantly decreased PARP expression in samples resistant to PRED, VCR, and ASP (Table 3). PVA resistance data were available for 8 AML samples, 15 T-lineage ALL samples, and 28 B-lineage ALL samples; of these, 5 AML samples (63%) and 12 T-lineage ALL samples (80%) were PVA resistant, whereas only 10 B-lineage ALL samples (36%) were PVA resistant. This is in line with our previous findings that children with AML and T-lineage ALL are relatively in vitro-resistant to various drugs including PRED, VCR, and ASP.7,30 Since PRED, VCR, and ASP are an integral part of ALL therapy, PARP expression was compared between PVA-sensitive and PVA-resistant ALL samples. Within the total group of ALL cases, PVA-resistant cases (median, 39.2 AU) had a 4-fold lower expression of PARP compared with sensitive cases (median, 9.2 AU, P < .001). Our observation that PARP expression is lower in T-ALL and that this immunophenotype is associated with cellular drug resistance partly explains our finding that low PARP expression is associated with PVA resistance in pediatric ALL (Figure 4B). Within the B-lineage group, PVA-resistant patients (n = 10; median, 20.2 AU) also had a 2-fold lower expression of PARP compared with sensitive patients (n = 18, median, 39.8, P = .040; Figure 4B, open symbols). No difference was found in PARP expression between ALL patients with an M1 bone marrow response at day 15 and those having an M2 or M3 bone marrow response (P = .11). Due to limited follow-up of patients no relevant comparisons with long-term outcome could be made.
PARP and procaspase-2 expression in childhood acute leukemia patients sensitive and resistant to PRED, VCR, ASP, and DNR
. | PARP . | . | . | Procaspase-2 . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | No. studied . | Median, AU . | P . | No. studied . | Mdeian, AU . | P . | ||||
| PRED sensitive | 20 | 39.8 | < .001* | 15 | 24.7 | .012* | ||||
| PRED resistant | 21 | 6.9 | 19 | 8.8 | ||||||
| VCR sensitive | 24 | 31.0 | .002* | 19 | 22.0 | .142 | ||||
| VCR resistant | 21 | 4.7 | 18 | 9.3 | ||||||
| ASP sensitive | 21 | 31.7 | .004* | 18 | 23.3 | .070 | ||||
| ASP resistant | 22 | 7.0 | 19 | 9.5 | ||||||
| DNR sensitive | 30 | 31.0 | .017 | 24 | 12.8 | .188 | ||||
| DNR resistant | 9 | 4.7 | 8 | 7.3 | ||||||
. | PARP . | . | . | Procaspase-2 . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | No. studied . | Median, AU . | P . | No. studied . | Mdeian, AU . | P . | ||||
| PRED sensitive | 20 | 39.8 | < .001* | 15 | 24.7 | .012* | ||||
| PRED resistant | 21 | 6.9 | 19 | 8.8 | ||||||
| VCR sensitive | 24 | 31.0 | .002* | 19 | 22.0 | .142 | ||||
| VCR resistant | 21 | 4.7 | 18 | 9.3 | ||||||
| ASP sensitive | 21 | 31.7 | .004* | 18 | 23.3 | .070 | ||||
| ASP resistant | 22 | 7.0 | 19 | 9.5 | ||||||
| DNR sensitive | 30 | 31.0 | .017 | 24 | 12.8 | .188 | ||||
| DNR resistant | 9 | 4.7 | 8 | 7.3 | ||||||
Statistically significant after Bonferroni correction for multiple comparisons.
Expression of procaspase-2 in acute leukemia
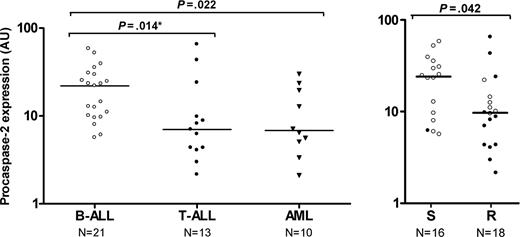
A slightly smaller group of 34 ALL cases (besides 10 AML) were analyzed for procaspase-2 expression due to limitations in availability of material. Procaspase-2 expression was detectable in all leukemic samples. As shown in Figure 5A, procaspase-2 expression varied between 2.1 and 65.4 AU in these patients, with the relatively highest expression level found in B-lineage ALL. The expression of procaspase-2 was 3-fold lower in T-lineage ALL (median, 7.0 AU; P = .014) and in AML (median, 6.80; P = .022) compared with B-lineage ALL (median, 22.0 AU). Procaspase-2 expression did not differ significantly between AML and T-lineage ALL (P = .976).
Procaspase-2 expression levels in different types of childhood acute leukemia and in drug-resistant versus -sensitive childhood ALL. (A) Procaspase-2 expression in different types of childhood acute leukemia. Each dot represents an individual patient and the horizontal bar represents the median protein level. Protein expression level of procaspase-2 quantitated in 21 B-lineage ALL samples (○), 13 T-lineage ALL samples (•), and 10 AML samples (▾). (B) Procaspase-2 expression versus in vitro PRED, VCR, and ASP (PVA) resistance in childhood acute lymphoblastic leukemia. The protein expression level of PARP quantitated in 16 PVA-sensitive (S) and 18 PVA-resistant (R) patients with B-lineage ALL (○) or T-lineage ALL (•). *Statistically significant after Bonferroni correction for multiple comparisons.
Procaspase-2 expression levels in different types of childhood acute leukemia and in drug-resistant versus -sensitive childhood ALL. (A) Procaspase-2 expression in different types of childhood acute leukemia. Each dot represents an individual patient and the horizontal bar represents the median protein level. Protein expression level of procaspase-2 quantitated in 21 B-lineage ALL samples (○), 13 T-lineage ALL samples (•), and 10 AML samples (▾). (B) Procaspase-2 expression versus in vitro PRED, VCR, and ASP (PVA) resistance in childhood acute lymphoblastic leukemia. The protein expression level of PARP quantitated in 16 PVA-sensitive (S) and 18 PVA-resistant (R) patients with B-lineage ALL (○) or T-lineage ALL (•). *Statistically significant after Bonferroni correction for multiple comparisons.
Procaspase-2 expression was compared between in vitro-sensitive and -resistant patients for PRED, VCR, ASP, or DNR as single drugs. After correction for multiple comparisons, we observed that decreased procaspase-2 expression was related to in vitro PRED resistance only, although a trend with resistance to the other drugs was found (Table 3). Within the total group of ALL patients, PVA-resistant cases (median, 9.7 AU) had a 2-fold lower expression of procaspase-2 compared with sensitive cases (median, 24.0 AU, P = .042; Figure 5B). Within the B-lineage group, PVA-resistant patients (n = 6; median, 11.8 AU) had a 2.1-fold lower expression of procaspase-2 compared with sensitive patients, although this difference was not statistically significant (n = 15, median, 24.7 AU, P = .132; Figure 5B, open symbols).
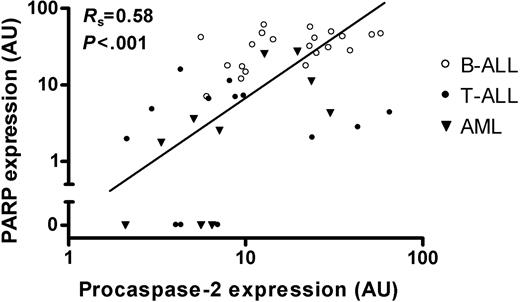
Since the expression of procaspase-2 and PARP showed similar expression patterns, we examined the possibility whether the expression of both proteins was correlated. As shown in Figure 6 there is a strong positive correlation between the expression of PARP and procaspase-2 (rs = .58, P < .001). Procaspase-2 expression did not differ between ALL patients with an M1 bone marrow response at day 15 and those having an M2 or M3 bone marrow response (P = .30). Due to limited follow-up of patients no relevant comparisons with long-term outcome could be made.
Discussion
The prognosis of childhood acute lymphoblastic leukemia (ALL) has improved remarkably over the past 4 decades due to the introduction of effective combination risk-adapted therapies. Conventional factors used to stratify patients are clinical and biological parameters such as age and white blood cell (WBC) count at diagnosis, immunophenotype, the presence of specific genetic abnormalities,1 and initial response to PRED treatment.31 Newer approaches are measurement of minimal residual disease after induction of initial remission32 and in vitro drug resistance profiles.4 We and others showed that children whose leukemic cells exhibit in vitro resistance to antileukemic agents, especially to PRED, VCR, and ASP, have a significantly worse prognosis than patients whose ALL cells are sensitive.3-6 Children with T-lineage ALL and AML are, in vitro, more resistant to a variety of drugs.7,30 The use of more intensive risk-adapted treatment regiments, however, has resulted in a prognosis for children with T-lineage ALL similar to that of children with B-lineage ALL.33 In the present study, the expression of Apaf-1, procaspases-2, -3, -6, -7, -8, -10, and PARP was studied in childhood ALL and AML and was related to immunophenotype and in vitro resistance to PRED, VCR, and ASP (PVA) as well as to resistance to PRED, VCR, and ASP and DNR individually.
Correlation between PARP and procaspase-2 protein levels in newly diagnosed childhood acute leukemia. Comparison between PARP and procaspase-2 protein expression levels in 21 B-lineage ALL samples (○), 13 T-lineage ALL samples (•), and 10 AML samples (▾). Diagonal line indicates line of best fit.
Correlation between PARP and procaspase-2 protein levels in newly diagnosed childhood acute leukemia. Comparison between PARP and procaspase-2 protein expression levels in 21 B-lineage ALL samples (○), 13 T-lineage ALL samples (•), and 10 AML samples (▾). Diagonal line indicates line of best fit.
We observed no relation between the expression of Apaf-1, procaspases-3, -6, -7, -8, and -10, and PVA resistance in childhood ALL in the present study. The absence of a relation with drug resistance may explain why no prognostic value was found for these proteins in adult ALL and AML,22 although data in children and adults are not necessarily interchangeable. In a previous study we demonstrated that the degree of caspase-3 activation (ie, cleavage of procaspase-3 into caspase-3) correlates with the degree of resistance to PRED and ASP in childhood ALL.17 In our present study no correlation was found between the baseline expression levels of procaspase-3 and resistance to these chemotherapeutic drugs. Taken together, these studies suggest that resistance to these 2 drugs may not be caused by decreased availability of the active enzyme due to decreased expression of procaspase-3, but rather may be caused by defects upstream of caspase-3 that inhibit caspase-3 activation upon PRED or ASP exposure.
PARP (Enzyme Commission [EC] 2.4.2.30) is an abundantly expressed nuclear enzyme, which binds to single- or double-stranded DNA breaks in response to DNA damage. At the breakage site, PARP catalyzes the transfer of the ADP-ribose polymers from the respiratory coenzyme NAD+ to nuclear acceptor proteins involved in chromatin structure, DNA repair, and DNA metabolism.34 Its poly(ADP-ribosylation) activity as well as its association with components of base-excision repair35 contribute to the role of PARP in DNA repair and the maintenance of genomic integrity. In the present study, we observed that in vitro drug-resistant childhood leukemia subtypes (ie, T-lineage ALL and AML), have a decreased and occasionally even absent expression of PARP. Complete absence of PARP expression was observed in 4 out of 15 patients with T-lineage ALL and 3 out of 10 patients with AML, but in none of the patients with B-lineage ALL. The variation in PARP expression between leukemia subtypes suggests that caution should be taken when interpreting studies that examining protein expression in a mixed group of B-lineage ALL, T-lineage ALL, and AML samples without correcting for leukemia subtypes. The absence of PARP was not caused by deletions of the PARP gene. Given the absence of CpG islands within the PARP promoter, it seems unlikely that the absence of PARP is caused by methylation-induced PARP silencing in these patients. The presence of putative binding sites for several transcription factors, including simian virus 40 protein 1 (Sp-1),36,37 activator protein-2 (AP-2),37 v-ets erythroblastosis virus E26 oncogene homolog 1 (Ets-1),38 and yin-yang 1 (YY1)39 within the PARP promoter region, suggests that PARP expression is regulated at the transcriptional level. In addition, we observed that decreased or absent expression of intact PARP is related to in vitro resistance to PRED, VCR, and ASP in childhood ALL. Other studies also reported that depletion of PARP, either by gene disruption,40,41 antisense RNA,40,42 or pharmacologic inhibitors,40,43-45 resulted in decreased drug-induced apoptosis. Various explanations have been proposed for the requirement of PARP during apoptosis, including depletion of cellular nicotinamide-adenine nucleotide (NAD)+ and adenosine triphosphate (ATP) pools,46 modification of proteins involved in apoptosis like p53,47,48 facilitation of oligonucleosomal DNA fragmentation,49 and up-regulation of P-glycoprotein in PARP-depleted cells.41 We previously observed that resistance to PRED and ASP, but not to VCR and DNR, is linked to decreased drug-induced PARP cleavage (ie inactivation of PARP).17 Our current observation that resistant cells express lower baseline levels of this protein suggests that the suitability to use cleaved PARP as read-out for functional apoptosis is drug/stimuli dependent.
Caspase-2 activity is implicated in the initiation50,51 as well as the effector phase52,53 of apoptosis induced by various stimuli. The present study shows a decreased procaspase-2 expression in in vitro drug-resistant childhood leukemia subtypes (ie, T-ALL and AML). In addition, we observed a relation between decreased procaspase-2 expression and in vitro PVA resistance in childhood ALL. This suggests that sufficient intracellular amounts of procaspase-2 are required to respond to these drugs, and that relative deficits in procaspase-2 expression levels may contribute to cellular PVA resistance in childhood acute leukemia. In adult acute leukemia, high expression of procaspase-2 was not linked to prognosis.19-22 The lack of prognostic value for the response to combination chemotherapy in these studies does not rule out a role for procaspase-2 in cellular responses to PRED and/or other drugs in adult leukemias. Since this relationship has not been addressed in adults so far, no comparison between childhood and adult studies can be made. In cell lines the requirement of caspase-2 for drug-induced apoptosis was shown to be highly cell specific.51,54,55
Decreased PARP and procaspase-2 expression did correlate with in vitro PVA resistance, whereas these expression levels did not correlate with an early bone marrow response at day 15. One may argue that this is exactly the time period when the leukemia is treated with PRED, VCR, and ASP in most protocols. However, in the COALL-97 protocol, patients are first treated with a DNR window (up-front combination chemotherapy) for 7 days, followed by PRED, VCR, and DNR for 4 weeks. After this period of 5 weeks, ASP is given for the first time. Since PARP and procaspase-2 expression are not linked to DNR resistance, and this drug is used up-front of combination chemotherapy as well as in the first 4 weeks, this may explain the lack of correlation between both proteins and day-15 bone marrow response of COALL-97 patients.
In our previous study we observed that resistance to each of the 4 studied drugs (PRED, VCR, ASP, and DNR) was associated with a decreased ability to expose phosphatidyl serine on the outer membrane and decreased collapse of the mitochondrial membrane potential.17 Caspase-3 activation and PARP inactivation correlated to PRED and ASP resistance, but not to VCR and DNR resistance. This finding suggested that cleaved caspase-3 and PARP are not important determinants of the VCR- and DNR-triggered apoptotic route. Both phosphatidyl serine exposure and the collapse of the mitochondrial membrane potential occurred earlier than caspase-3 activation and PARP inactivation, implying that resistance to these 4 drugs is caused by a blockade at the level of or upstream of mitochondrial function. The identity of these upstream causes of resistance are part of extensive research in the past few years by different groups (reviewed by Pieters and Den Boer56 ).
Recently, new insights were obtained by comparing gene expression profiles of drug-resistant and -sensitive cells of children with ALL.24 In this latter study, drug-specific genes rather than more general apoptosis-associated genes (such as caspases, Bcl-2 family members, and inhibitors of apoptosis [IAP] factors) were among the most discriminative genes that were associated with resistance to PRED, VCR, ASP, and DNR in ALL cells. These findings do not rule out a role for altered expression and/or activity of apoptosis genes, but suggest that, at the mRNA level, genes upstream of the apoptosis-execution pathway are more important for resistance to these drugs. At the protein level, we here show that the basal expression levels of Apaf-1 and procaspase-3, -6, -7, -8, and -10 are not differentially expressed between drug-resistant and -sensitive patients with acute leukemia. Expression of procaspase-2 and PARP were decreased in children with drug-resistant ALL. The causal aspect of this association should be further determined with functional studies showing the sensitizing effect of ectopic expression of PARP and procaspase-2 on drug resistance in leukemic cells of children with ALL.
Prepublished online as Blood First Edition Paper, May 17, 2005; DOI 10.1182/blood-2004-11-4296.
Supported in part by the Pediatric Oncology Foundation Rotterdam (A.H.) and the Dutch Cancer Society (EMCR 2003-2871; A.R.M.v.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all the hospitals participating in the German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL) and the staff of the Erasmus MC/Sophia Children's Hospital for providing the leukemia samples. In addition, we thank Renée X. Menezes (biostatistician) for statistical advice.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal