Abstract
NKG2D is an activation receptor that allows natural killer (NK) cells to detect diseased host cells. The engagement of NKG2D with corresponding ligand results in surface modulation of the receptor and reduced function upon subsequent receptor engagement. However, it is not clear whether in addition to modulation the NKG2D receptor complex and/or its signaling capacity is preserved. We show here that the prolonged encounter with tumor cell-bound, but not soluble, ligand can completely uncouple the NKG2D receptor from the intracellular mobilization of calcium and the exertion of cell-mediated cytolysis. However, cytolytic effector function is intact since NKG2D ligand-exposed NK cells can be activated via the Ly49D receptor. While NKG2D-dependent cytotoxicity is impaired, prolonged ligand exposure results in constitutive interferon γ (IFNγ) production, suggesting sustained signaling. The functional changes are associated with a reduced presence of the relevant signal transducing adaptors DNAX-activating protein of 10 kDa (DAP-10) and killer cell activating receptor-associated protein/DNAX-activating protein of 12 kDa (KARAP/DAP-12). That is likely the consequence of constitutive NKG2D engagement and signaling, since NKG2D function and adaptor expression is restored to normal when the stimulating tumor cells are removed. Thus, the chronic exposure to tumor cells expressing NKG2D ligand alters NKG2D signaling and may facilitate the evasion of tumor cells from NK cell reactions. (Blood. 2005;106:1711-1717)
Introduction
Numerous activation receptors can trigger natural killer (NK) cell-mediated cytotoxic and/or cytokine responses. The NKG2D receptor is specific for endogenous ligands (ie, histocompatibility 60 (H60), retinoic acid early inducible gene-1 (RAE-1), and mouse UL16-binding proteinlike transcript 1 (MULT1) in mice), which are usually not expressed on normal resting cells but which are readily induced upon different forms of cellular distress (for a review see Raulet1 ). Thus, the de novo expression of NKG2D ligands renders cells susceptible to NK cell-mediated lysis. On many tumor cell lines of distinct tissue origins, NKG2D ligands are constitutively expressed. The ectopic expression of NKG2D ligands induces the rejection of several transplantable tumor cell lines.2,3
Despite NKG2D's prominent role in NK cell activation and T-cell costimulation, it is noteworthy that many established tumor cell lines constitutively express NKG2D ligands, suggesting that tumors can escape NKG2D recognition. Indeed, T and NK cells, which were infiltrating major histocompatibility class I chain-related protein A (MICA)-positive tumors had reduced NKG2D cell surface levels.4,5 In vitro, soluble MICA or tumor cells expressing MICA induced the down-modulation and degradation of NKG2D expressed by CD8+ T cells, which led to reduced effector functions.4 In addition, NK cells derived from nonobese diabetic (NOD) mice (a strain of mice prone to autoimmune diabetes) displayed impaired NKG2D function.6 NKG2D dysfunction and modulation were explained by an interaction of NKG2D with its ligands, which are aberrantly induced on NOD NK cells upon culture.6
In general, reduced NKG2D function is thought to be the consequence of and directly related to the lower NKG2D cell surface levels. However, it has not been determined whether sustained NKG2D engagement leaves the NKG2D receptor complex and/or its signal transduction capacity intact. To this end we established a culture system in which NK cells are exposed to tumor cells expressing NKG2D ligand. We show that chronic engagement with tumor cell-bound NKG2D ligand can uncouple NKG2D recognition from the exertion of NK cell-mediated target cell lysis. NKG2D dysfunction is explained by striking reduction of the NKG2D signaling adaptor molecules DNAX-activating protein of 10 kDa (DAP-10) and killer cell activating receptor-associated protein/DNAX-activating protein of 12 kDa (KARAP/DAP-12). These findings suggest that tumor cells expressing NKG2D ligand can efficiently shut down and consequently evade NKG2D-mediated NK-cell reactions.
Materials and methods
Mice
C57BL/6 (B6) mice were purchased from Harlan (Horst, The Netherlands). KARAPki mice, B6 backcross 9, have been described.7 Animal experimentation followed protocols approved by the Service Vétérinaire de l'Etat de Vaud.
Cell lines
NK cell culture
B6 splenocytes were depleted of B and T cells with nylon wool and monoclonal antibody (mAb) 17A2 (CD3ϵ, Ludwig Institute for Cancer Research [LICR], Lausanne, Switzerland) plus anti-rat immunoglobulin G (IgG) M450 Dynabeads (Dynal, Oslo, Norway), respectively, resulting in 20% to 35% CD3-NK1.1+ cells. For kinetic experiments, NK cells were enriched further by negative selection using mAbs to F4.80, CD5, and B220 (at suboptimal concentration), resulting in 70% to 80% pure NK cells. Carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR)-labeled and irradiated (3000 rad) RMA or RMA H60 cells were added at a 1:1 ratio to NK cells (at 106/mL). After 3 days of coculture in the presence of interleukin 2 (IL-2; 500 ng/mL), remaining CFSE+ RMA cells were removed using mAbs to CD3ϵ, murine leukemia virus (MuLV) glycoprotein 70 (gp70) (RL2.5.6, LICR), CD5 (53-7.213.2, LICR), and Dynabeads, yielding 50% to 80% NK cells, less than 3% T cells, and less than 5% residual RMA cells.
Flow cytometry
Cells (1 × 106) were incubated with mAb 24G2 (anti-CD16/32) to reduce background and stained with the following mAbs: PK136 (NK1.1), 4E5 (Ly49D; BD PharMingen, San Diego, CA), 2.4G2 (anti-CD16/32, LICR), and NKG2D (R&D Systems, Oxon, United Kingdom). Staining with soluble NKG2D has been described.8
NK cells were loaded with indo-1/AM (5 μM; Sigma) at 37°C for 1 hour, washed, incubated with primary mAb (20 μg/mL) for 30 minutes at 4°C and washed. The Ca2+ flux was measured at 37°C using an LSR flow cytometer (Becton Dickinson, San Jose, CA). Data acquisition was interrupted for cross-linking with 25 μg/mL goat anti-rat IgG (Sigma) or to add ionomycin (1 μg/mL).
NK cells (1.5 × 105) were stimulated with 2 × 105 tumor cells (for 6 hours or 18 hours) or with plate-bound mAb (for 6 hours). Brefeldin A (10μg/mL; Sigma) was added for the last 3 hours of stimulation. IFNγ was determined by intracellular flow cytometry.
Cytotoxicity assays
Standard (4-hour) 51Cr release assays were performed as described elsewhere.8 For CHO killing, Ly49D was blocked with anti-Ly49D (4E5) mAb at 10 μg per 106 effector cells.
Immunoblot and immunoprecipitation
Total cell lysates (3 × 106 NK cells) were prepared in sample buffer (containing 1.5% sodium dodecyl sulfate [SDS] and dithiothreitol [DTT]). Lysates were boiled at 95°C for 5 minutes and loaded on 16% SDS-polyacrylamide gel electrophoresis (PAGE) gels. After transfer onto nitrocellulose, immunoblots were performed with Abs to FcRγ (Upstate Biotechnology, Lake Placid, NY), DAP-10,10 or DAP-12,10 CD3ζ (H146), and α-tubulin (B-5-1-2; Sigma) followed by horseradish peroxidase (HRP)-coupled secondary Abs (Southern Biotechnology, Birmingham, AL) and visualization by enhanced chemiluminescence (ECL; Amersham Bioscience, Buckinghamshire, United Kingdom).
NK cells (5 × 106) were incubated with anti-NKG2D or anti-Ly49D followed by goat anti-rat Ab. After washing, the cells were lysed in ice-cold 0.2% dodecyl maltoside lysis buffer (20 mM Tris-HCl pH 8.2, 100 mM NaCl, 10 mM EDTA, 50 mM NaF, 1 mM Na3VO4) and protease inhibitor cocktail (Roche, Mannheim, Germany) followed by immunoprecipitation with Protein G Sepharose (Amersham Bioscience) and anti-rat IgG agarose (Sigma). Immunoprecipitates were analyzed as described for cell lysates.
Production of recombinant protein
The production of soluble NKG2D has been described elsewhere.8 The extracellular portion including the endogenous leader peptide of H60 (amino acids 1-213) was amplified by polymerase chain reaction (PCR) from A20 B-cell cDNA, using the following primers (restriction sites are underlined): H60 5′ sense, actg AAGCTT tgagggaagacc ATG GCAAAG G; H60 3′ antisense, actg GTCGAC CTG GTT GTC AGA ATT ATG TCG GAA G.
The PCR product was directionally cloned into modified PCR-3 vectors (Invitrogen, San Diego, CA) to add a COOH-terminal FLAG epitope or to generate an Fc fusion protein (kindly provided by P. Schneider, Department of Biochemistry, University of Lausanne, Switzerland). The resulting H60-FLAG and H60-Fc constructs were sequenced to ensure their identity with the published sequences. Fusion proteins were expressed by transient transfection of 293T cells as before.8
RT-PCR
Total RNeasy (Qiagen, Hilden, Germany) and cDNA was synthesized using oligo(dT) priming (Amersham Bioscience) using avian myeloblastic virus (AMV) reverse transcriptase (RT; Roche). PCR primers were as follows: hypoxanthine guanine phosphoribosyl transferase (HPRT) 5′ AGT CCC AGC GTC GTG ATT AGC and 3′ GAC ATC TCG AGC AAG TCT TTC AGT C.
Results
Sustained engagement blunts NKG2D function in NK cells
To determine the impact of sustained engagement on NKG2D function we exposed B6 NK cells to tumor cells expressing NKG2D ligand. RMA cells, which do not express endogenous NKG2D ligands,8,12 were stably transfected with the high-affinity NKG2D ligand H60 (Figure 1A). Freshly isolated, T-cell-depleted NK cells from B6 mice were then activated with IL-2 in the presence of irradiated RMA-H60 or control RMA cells. Three days later, residual tumor cells and T cells were removed and NK cell reactivity was tested.
RMA-H60-exposed NK cells were almost completely unable to kill RMA-H60 target cells (Figure 1B). In contrast, NK cells exposed to control RMA cells efficiently killed RMA-H60 but, as expected, not the parental RMA cells (Figure 1B). On average, 14-fold ± 5-fold more RMA-H60-exposed than control NK cells were needed to obtain an equivalent specific lysis of RMA-H60 cells.
The deficient lysis of RMA-H60 cells was not due to cold target competition. Addition of unlabeled RMA-H60 cells to control NK cells (at a concentration equivalent to that remaining after depletion in RMA-H60 cocultures, usually less than 5%) had a negligible effect on the lysis of Cr51-labeled RMA-H60 cells (Figure 1B).
RMA-H60-exposed NK cells killed CHO cells, albeit at a reduced efficiency (Figure 1B). However, RMA-H60 coculture specifically reduced the Ly49D-independent part of CHO lysis (ie, the part that cannot be blocked with anti-Ly49D mAb). If Ly49D-independent lysis is subtracted, the Ly49D-dependent part of CHO killing remains essentially unchanged (Figure 1B). Thus, sustained RMA-H60 exposure does not exhaust the NK cell's lytic capacity as demonstrated by efficient NK cell activation via Ly49D.
We further determined the time span required to induce NKG2D dysfunction. A significant but incomplete inactivation of NKG2D function was already observed when RMA-H60 cells were present for 24 hours (Figure 1C). At 48 hours, the control RMA-exposed NK cells had increased activity while NKG2D inactivation remained incomplete in RMA-H60 cultures. At 72 hours, the NKG2D pathway of RMA-H60 exposed NK cells was completely inactive (Figure 1C).
Chronic engagement abrogates NKG2D function. (A) RMA cells stably transfected with the NKG2D ligand H60 bind soluble NKG2D. (B) The lytic capacity of NK cells exposed for 3 days to RMA-H60 or control RMA cells was determined. To exclude cold-target competition, unlabeled RMA-H60 cells were added to control NK cells during the lysis assay (at a percentage equivalent to that remaining in the RMA-H60 cultures [ie, < 5%]) (▪). These results are from one representative experiment of 14 performed. The lysis curves were shifted relative to the content of NK cells in the effector cell populations. To demonstrate the Ly49D dependence of CHO lysis, Ly49D was blocked with mAb 4E5 (▦ for controls and  for RMA-H60-exposed NK cells). (C) NK cells were cultured in the presence of IL-2 plus RMA-H60 (▦) or RMA cells (□). The lytic activity of NK cells against RMA-H60 was determined after 24, 48, and 72 hours of coculture. NK cell activity was tested in triplicate at effector-target (E/T) ratios of 50:1 and 10:1. The data are representative of 2 independent experiments. (D) NK cells were cultured (for 72 hours in IL-2) separate from RMA-H60 (▪) or control RMA cells (□) using transwell plates. The lytic activity of these NK cells against RMA-H60 was determined. (B-D) Graphs depict the mean (±SD) of triplicate determinations at various E/T ratios.
for RMA-H60-exposed NK cells). (C) NK cells were cultured in the presence of IL-2 plus RMA-H60 (▦) or RMA cells (□). The lytic activity of NK cells against RMA-H60 was determined after 24, 48, and 72 hours of coculture. NK cell activity was tested in triplicate at effector-target (E/T) ratios of 50:1 and 10:1. The data are representative of 2 independent experiments. (D) NK cells were cultured (for 72 hours in IL-2) separate from RMA-H60 (▪) or control RMA cells (□) using transwell plates. The lytic activity of these NK cells against RMA-H60 was determined. (B-D) Graphs depict the mean (±SD) of triplicate determinations at various E/T ratios.
Chronic engagement abrogates NKG2D function. (A) RMA cells stably transfected with the NKG2D ligand H60 bind soluble NKG2D. (B) The lytic capacity of NK cells exposed for 3 days to RMA-H60 or control RMA cells was determined. To exclude cold-target competition, unlabeled RMA-H60 cells were added to control NK cells during the lysis assay (at a percentage equivalent to that remaining in the RMA-H60 cultures [ie, < 5%]) (▪). These results are from one representative experiment of 14 performed. The lysis curves were shifted relative to the content of NK cells in the effector cell populations. To demonstrate the Ly49D dependence of CHO lysis, Ly49D was blocked with mAb 4E5 (▦ for controls and  for RMA-H60-exposed NK cells). (C) NK cells were cultured in the presence of IL-2 plus RMA-H60 (▦) or RMA cells (□). The lytic activity of NK cells against RMA-H60 was determined after 24, 48, and 72 hours of coculture. NK cell activity was tested in triplicate at effector-target (E/T) ratios of 50:1 and 10:1. The data are representative of 2 independent experiments. (D) NK cells were cultured (for 72 hours in IL-2) separate from RMA-H60 (▪) or control RMA cells (□) using transwell plates. The lytic activity of these NK cells against RMA-H60 was determined. (B-D) Graphs depict the mean (±SD) of triplicate determinations at various E/T ratios.
for RMA-H60-exposed NK cells). (C) NK cells were cultured in the presence of IL-2 plus RMA-H60 (▦) or RMA cells (□). The lytic activity of NK cells against RMA-H60 was determined after 24, 48, and 72 hours of coculture. NK cell activity was tested in triplicate at effector-target (E/T) ratios of 50:1 and 10:1. The data are representative of 2 independent experiments. (D) NK cells were cultured (for 72 hours in IL-2) separate from RMA-H60 (▪) or control RMA cells (□) using transwell plates. The lytic activity of these NK cells against RMA-H60 was determined. (B-D) Graphs depict the mean (±SD) of triplicate determinations at various E/T ratios.
The preactivation of NK cells before the addition of RMA-H60 in vitro (with IL-2 for 3 days) or in vivo (by poly I-C injection) did not prevent NKG2D inactivation (data not shown). Finally, NK-cell inactivation by RMA-H60 also occurred when NK cells were cultured in IL-12/IL-18 instead of IL-2 (data not shown). These data suggest that NKG2D inactivation requires prolonged exposure to RMA-H60 cells and that it is induced irrespective of the NK-cell activation status.
To determine whether direct cell contact was required, NK cells and RMA-H60 cells were separated during the coculture using transwell plates. This separation resulted in a minimal impact on NKG2D function (Figure 1D). Similarly, a marginal effect on NKG2D function was observed when NK cells were exposed to soluble or plastic-bound H60-Ig fusion protein, despite the fact that H60-Ig binds NKG2D based on flow cytometry (not shown). Thus, profound NKG2D dysfunction is dependent on prolonged encounter with ligand in a cellular context.
NK cells exposed to RMA-H60 for 72 hours, in which NKG2D was completely dysfunctional, retained 38% ± 12% of the NKG2D cell surface levels observed on NK cells exposed to RMA (Figure 2A-B). Lower NKG2D levels (10% and 22%, again as compared with day-3 RMA-exposed NK cells) were observed when NK cells were cocultured with RMA-H60 for 24 or 48 hours (Figure 2B), yet at these time points NKG2D was partially functional (Figure 2B). Finally, the separation of RMA-H60 and NK cells in transwell plates reduced NKG2D levels to 79% (not shown), while the effect on NKG2D function was marginal (Figure 1D). Thus, the extent of NKG2D down-modulation is not a very accurate predictive factor for NKG2D function.
In contrast to NKG2D, other NK-cell activation receptors, including Ly49D and NK1.1, were expressed at normal levels (Figure 2A). Notwithstanding, NK cells exposed to RMA-H60 cells expanded on average 2.3-fold less than those cocultured with RMA (not shown). In addition, their forward scatter (size) was below that of control NK cells, yet clearly increased as compared with nonstimulated NK cells (Figure 2A). Thus, besides NKG2D function, prolonged engagement of this receptor also impacts IL-2-driven NK cell growth and expansion.
Sustained NKG2D engagement induces constitutive IFNγ production
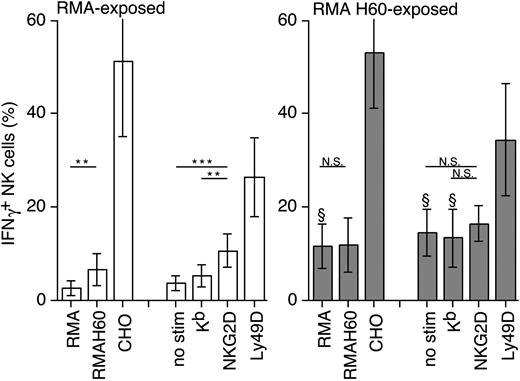
In addition to NKG2D-dependent target cell lysis, we tested whether sustained RMA-H60 exposure impacted IFNγ production. Surprisingly, after 3 days of coculture with RMA-H60, a substantial fraction (10%-15%) of NK cells were intracellular (ic) IFNγ+ even in the absence of any restimulation (Figure 3). Additionally, small but significant amounts of IFNγ were released into the culture medium (not shown). Thus, the prolonged exposure to RMA-H60 cells induces constitutive IFNγ production, indicating sustained NKG2D signaling.
Phenotype of NK cells exposed to RMA-H60. (A) Histograms show the expression of the indicated activation receptors or the forward size scatter (FSC) of control RMA- (open histogram) and RMA-H60-exposed (gray shaded histogram) NK cells. Numbers indicate the mean fluorescence intensity (MFI) of staining. The dotted line depicts the FSC of fresh splenic NK cells. These results are from one representative experiment of 6 performed. (B) Bar graphs show the mean MFI (±SD) of NKG2D staining normalized to that of NK cells cocultured with control RMA cells (□, RMA; ▦, RMA-H60) for 3 days. Data for ex vivo-isolated NK cells (▨; n = 8) and for the 72-hour time point (n = 12) are compiled from numerous independent experiments, whereas individual values (circles) and their means (bars) from 2 independent experiments are shown for 24 hours and 48 hours of coculture. Statistical significance of the difference at 72 hours of coculture was evaluated using the one-tailed unpaired Student t test; *P < .001.
Phenotype of NK cells exposed to RMA-H60. (A) Histograms show the expression of the indicated activation receptors or the forward size scatter (FSC) of control RMA- (open histogram) and RMA-H60-exposed (gray shaded histogram) NK cells. Numbers indicate the mean fluorescence intensity (MFI) of staining. The dotted line depicts the FSC of fresh splenic NK cells. These results are from one representative experiment of 6 performed. (B) Bar graphs show the mean MFI (±SD) of NKG2D staining normalized to that of NK cells cocultured with control RMA cells (□, RMA; ▦, RMA-H60) for 3 days. Data for ex vivo-isolated NK cells (▨; n = 8) and for the 72-hour time point (n = 12) are compiled from numerous independent experiments, whereas individual values (circles) and their means (bars) from 2 independent experiments are shown for 24 hours and 48 hours of coculture. Statistical significance of the difference at 72 hours of coculture was evaluated using the one-tailed unpaired Student t test; *P < .001.
The restimulation of NK cells that were previously exposed to control RMA cells increased the proportion of icIFNγ+ NK cells approximately 2-fold (Figure 3). In contrast, the restimulation of RMA-H60-exposed NK cells did not further enhance the proportion of icIFNγ+ NK cells (Figure 3). Nevertheless, a robust additional increase of the percentage of icIFNγ+ cells was induced when RMA-H60-exposed NK cells were restimulated with anti-Ly49D mAb or CHO cells (Figure 3). Thus, chronic exposure to ligand results in the sustained production of IFNγ production, whereas it blunts NKG2D-dependent target cell lysis.
Constitutive IFNγ production by RMA-H60-exposed NK cells. RMA- (□) and RMA-H60-exposed NK cells (▦) were restimulated with the indicated cell lines or plastic-bound mAbs. The bar graph depicts the mean percentage (± SD) of intracellular IFNγ+ NK cells obtained in 4 to 8 independent experiments. Statistical significance of differences between the indicated groups were determined using the 2-tailed Student t test. **P < .02; ***P < .01; NS, not significant; §P < .02 as compared with the corresponding values in the control RMA-exposed group.
Constitutive IFNγ production by RMA-H60-exposed NK cells. RMA- (□) and RMA-H60-exposed NK cells (▦) were restimulated with the indicated cell lines or plastic-bound mAbs. The bar graph depicts the mean percentage (± SD) of intracellular IFNγ+ NK cells obtained in 4 to 8 independent experiments. Statistical significance of differences between the indicated groups were determined using the 2-tailed Student t test. **P < .02; ***P < .01; NS, not significant; §P < .02 as compared with the corresponding values in the control RMA-exposed group.
Basis for defective NKG2D function
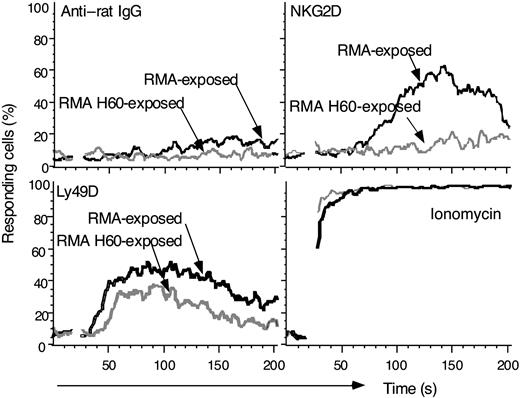
Granule exocytosis is dependent on intracellular mobilization of calcium ([Ca2+]i).13 Control, RMA-exposed NK cells showed a robust [Ca2+]i response upon NKG2D cross-linking. In contrast, [Ca2+]i mobilization was essentially absent in RMA-H60-exposed NK cells (Figure 4), suggesting that NKG2D on these NK cells is signaling deficient. In sharp contrast, [Ca2+]i mobilization was detected following Ly49D cross-linking. This is consistent with the induction of target cell lysis and IFNγ production upon Ly49D engagement. Stimulation with the Ca2+ ionophore ionomycin further indicated that inactivated NK cells have adequate intracellular Ca2+ stores and loading of Indo-1.
Sustained NKG2D engagement impacts NKG2D receptor complexes
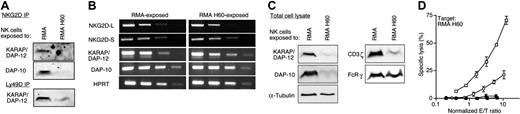
Since NKG2D cross-linking failed to induce [Ca2+]i mobilization, we next tested whether NKG2D was associated with its signaling adaptors KARAP/DAP-12 and DAP-10.10,11,14 Coimmunoprecipitation of KARAP/DAP-12 or DAP-10 with NKG2D from inactivated NK cells was below detection. In contrast, both adaptors were readily detected in association with NKG2D in control NK cells (Figure 5A). The absence of KARAP/DAP-12 in immunoprecipitates may be due to a failure to induce the expression of NKG2D-S, the NKG2D isoform, which can associate with KARAP/DAP-12. However, a semiquantitative RT-PCR assay ensured that mRNA for NKG2D-S is present in inactive NK cells (Figure 5B). Thus, the association of KARAP/DAP-12 and DAP-10 signaling adaptors with NKG2D is below detection, providing first insights into defective NKG2D function.
Significant but low amounts of KARAP/DAP-12 were found in association with Ly49D in RMA-H60-exposed NK cells (Figure 5A). The low amounts of adaptor protein are apparently not limiting for proper Ly49D function. Thus, chronic engagement and possibly signaling affects preferentially the NKG2D receptor complex.
Since low amounts of KARAP/DAP-12 were coimmunoprecipitated with Ly49D in silenced NK cells (Figure 5A), we further determined the DAP-10 and KARAP/DAP-12 protein contents in total NK-cell lysates. Remarkably, the abundance of both adaptors was severely reduced (Figure 5C). A semiquantitative RT-PCR assay showed that mRNA for DAP-10 and KARAP/DAP-12 was approximately equally abundant in inactive and control NK cells (Figure 5C). Thus, the small pools of cellular DAP-10 and KARAP/DAP-12 adaptor protein are the result of reduced translation and/or enhanced protein turnover.
In preliminary experiments, we tested whether DAP-10 and KARAP/DAP-12 are lost with similar kinetics upon the exposure of NK cells to RMA-H60 cells. DAP-10 levels were reduced after 24 hours of coculture, whereas KARAP/DAP-12 reduction was evident at 48 hours (data not shown). Whereas NKG2D-L is constitutively expressed, in IL-2-stimulated NK cells NKG2D-S mRNA is expressed at 24 hours.11 These data are consistent with the interpretation that due to the inducible expression of NKG2D-S, NK cell activation via KARAP/DAP-12, and its loss is delayed as compared with DAP-10.
Based on these findings we further determined the presence of FcRγ and CD3ζ, 2 additional signaling adapters used by NK cell activation receptors. Whereas CD3ζ was also severely diminished in inactive NK cells, FcRγ levels were unperturbed (Figure 5C). Thus, NKG2D engagement has an unexpectedly broad impact on the abundance of signaling adaptors in NK cells. Even though CD3ζ does not associate with NKG2D11 (data not shown), it is depleted in an NKG2D-induced fashion. The precise connection between CD3ζ and NKG2D will thus require further investigation.
NK cell inactivation in the absence of KARAP/DAP-12 function
To address whether KARAP/DAP-12 function was critical for NK cell inactivation, we used KARAP knock-in (KARAPki) mice. In these mice, the immunoreceptor tyrosine-based activation motif (ITAM) in KARAP/DAP-12 was rendered nonfunctional, while the mutant molecule is expressed.7 As compared with B6, KARAPki NK cells exposed to control RMA showed greatly reduced, yet still significant, lysis of RMA-H60 targets (Figure 5D).11 This residual lysis was NKG2D-dependent since RMA cells were not killed at all (not shown). Thus, short-term (3 days) IL-2-activated NK cells can inefficiently kill via NKG2D in the absence of functional KARAP/DAP-12, likely using DAP-10, consistent with other studies.10,11,14
Deficient Ca2+mobilization by RMA-H60-exposed NK cells. NK cells were loaded with Indo-1 before incubating with either anti-NKG2D antiserum or anti-Ly49D (4E5) mAb. Residual B and T cells were excluded by gating on CD19- and CD3- cells. The data acquisition was interrupted for cross-linking the bound mAb with goat anti-rat IgG. The results shown are from one experiment of 2 performed.
Deficient Ca2+mobilization by RMA-H60-exposed NK cells. NK cells were loaded with Indo-1 before incubating with either anti-NKG2D antiserum or anti-Ly49D (4E5) mAb. Residual B and T cells were excluded by gating on CD19- and CD3- cells. The data acquisition was interrupted for cross-linking the bound mAb with goat anti-rat IgG. The results shown are from one experiment of 2 performed.
KARAPki NK cells exposed to RMA-H60 cells completely failed to kill RMA-H60 targets (Figure 5D). Thus, NK-cell inactivation can occur in the absence of KARAP/DAP-12 function, suggesting that a single signaling adaptor is sufficient for NKG2D inactivation. Obviously, this does not exclude the possibility that KARAP/DAP-12 mediates NK cell inactivation.
Expression and function of signaling adaptors. (A) NKG2D (top) and Ly49D (bottom) were immunoprecipitated from RMA and RMA-H60-exposed NK cells. Immunoprecipitates (IP) were analyzed using DAP-10 or KARAP/DAP-12 Abs. The results shown are representative of 2 independent experiments performed. (B) Total cellular mRNA was extracted from RMA and RMA-H60-exposed NK cells. The relative abundance of specific transcripts was estimated by RT-PCR analysis on serial 10-fold cDNA dilutions. The results shown are representative of 5 independent experiments performed. (C) Immunoblot analysis of total cell lysates of day-3 cocultures using the indicated Abs. The results shown are representative of 4 independent experiments performed. (D) KARAPki NK cells were cocultured with RMA (○) or RMA-H60 (•) for 3 days. The lytic activity of KARAPki NK cells toward RMA-H60 cells was compared with that of B6 NK cells cocultured with RMA (□) or RMA-H60 (▪). The graph depicts the mean (±SD) of triplicate determinations at various E/T ratios. The results shown are representative of 2 independent experiments performed.
Expression and function of signaling adaptors. (A) NKG2D (top) and Ly49D (bottom) were immunoprecipitated from RMA and RMA-H60-exposed NK cells. Immunoprecipitates (IP) were analyzed using DAP-10 or KARAP/DAP-12 Abs. The results shown are representative of 2 independent experiments performed. (B) Total cellular mRNA was extracted from RMA and RMA-H60-exposed NK cells. The relative abundance of specific transcripts was estimated by RT-PCR analysis on serial 10-fold cDNA dilutions. The results shown are representative of 5 independent experiments performed. (C) Immunoblot analysis of total cell lysates of day-3 cocultures using the indicated Abs. The results shown are representative of 4 independent experiments performed. (D) KARAPki NK cells were cocultured with RMA (○) or RMA-H60 (•) for 3 days. The lytic activity of KARAPki NK cells toward RMA-H60 cells was compared with that of B6 NK cells cocultured with RMA (□) or RMA-H60 (▪). The graph depicts the mean (±SD) of triplicate determinations at various E/T ratios. The results shown are representative of 2 independent experiments performed.
Restoration of NKG2D function
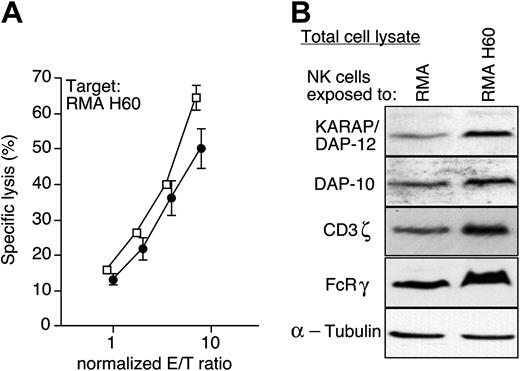
Finally, we addressed whether or not the deficient NKG2D function was permanent. Following the separation of NK cells from RMA-H60 cells and overnight culture in medium containing IL-2, NK cells readily regained a functional NKG2D pathway as RMA-H60 cells were now efficiently killed (Figure 6A).
The functional recovery was associated with a restoration of a normal cell size and normal NKG2D cell surface levels (not shown). Moreover, the pools of cellular DAP-10, KARAP/DAP-12, and CD3ζ protein returned to normal (Figure 6B). Thus, NKG2D dysfunction is not permanent, and continuous presence of NKG2D ligand maintains the inactive state.
Discussion
Here we show that sustained stimulation with tumor cell-bound ligand uncouples the NKG2D receptor from the intracellular mobilization of calcium and the exertion of NK cell-mediated cytolysis, while it induces the continuous production of IFNγ. These functional changes are associated with a low abundance of the NKG2D signaling adaptors DAP-10 and KARAP/DAP-12. Since the removal of the stimulating tumor cells restores both NKG2D function and adaptor expression, the findings suggest that sustained NKG2D signaling drastically alters NKG2D function.
The partial inactivation of human NK cells using a brief exposure (6 hours-18 hours) to K562 tumor target cells has previously been reported.15,16 However, K562 cells engage multiple NK cell activation receptors, including Nkp30, Nkp46, and NKG2D.17 It is thus difficult to draw conclusions regarding the involvement and importance of specific activation pathways for NK cell inactivation.
The functionality of the NKG2D pathway following ligand exposure has recently been studied using a coculture system6 similar to the one shown here. However, the authors observed a minor (2-fold) negative effect on NKG2D function, while we observed more than 10-fold effects, despite a comparable 2- to 3-fold reduction of NKG2D cell surface levels. While the affinities of NKG2D for H60 and RAE1ϵ ligands are comparable,18 B16 melanoma cells were used before, whereas we used RMA lymphoma cells. The type of NKG2D ligand-expressing tumor cells may thus be of importance to determine the extent of NKG2D dysfunction. Consistent with this possibility, the lysis of NKG2D ligand-transduced B16 cells is more dependent on DAP-12 than that of ligand-transduced RMA cells,1 indicating qualitative and/or quantitative differences in how NK cells are activated by these 2 cell lines.
The NKG2D receptor complex
Experiments using KARAPki NK cells demonstrate that KARAP/DAP-12 is not necessary for NK cell inactivation via NKG2D. Since DAP-10 is the only known remaining adaptor normally associated with NKG2D, the experiments suggest that DAP-10 is sufficient to inactivate NK cells. In this context it is interesting to note that human NKG2D associates exclusively with DAP-10,19,20 raising the possibility that chronic exposure to NKG2D ligands may also be sufficient for the inactivation of human NK cells. Obviously, the above data do not exclude the possibility that NK cell inactivation can be induced via KARAP/DAP-12 signaling.
Restoration of NKG2D function. (A) After 3 days of coculture with RMA (□) or RMA-H60 cells (•), residual tumor cells were removed and NK cells were cultured for an additional 18 hours with IL-2. The cytolytic activity of the NK cells was determined toward RMA-H60 targets. The graph depicts the mean (±SD) of triplicate determinations at various E/T ratios. The results shown are representative of 5 independent experiments performed. (B) Immunoblots of total cell lysates from “recovered” NK cells using the indicated Abs.
Restoration of NKG2D function. (A) After 3 days of coculture with RMA (□) or RMA-H60 cells (•), residual tumor cells were removed and NK cells were cultured for an additional 18 hours with IL-2. The cytolytic activity of the NK cells was determined toward RMA-H60 targets. The graph depicts the mean (±SD) of triplicate determinations at various E/T ratios. The results shown are representative of 5 independent experiments performed. (B) Immunoblots of total cell lysates from “recovered” NK cells using the indicated Abs.
The transport of NKG2D to the cell surface requires the presence of KARAP/DAP-12 or DAP-10 adaptors11 (data not shown). However, in inactivated NK cells, KARAP/DAP-12 or DAP-10 association with NKG2D was not detected even though significant NKG2D cell surface expression is detectable. To reconcile these findings it is possible that NKG2D is transported to the cell surface as NKG2D/adaptor complexes. However, chronic NKG2D engagement and signaling, which is in line with the constitutive IFNγ production, may disrupt NKG2D complexes and result in the continuous loss of the adaptors. Indeed, the dissociation of transmembrane adapter molecules from multichain receptor complexes has been observed in ligand-stimulated T-cell receptors (TCRs).21 In preliminary experiments, we have attempted to prevent the loss of adaptor molecules by the addition of inhibitors of lysosomal function. However, the exposure of primary NK cells to inhibitors resulted in nonspecific negative effects on the expression of signaling adaptors. Heterologous systems may thus be needed to address this issue.
Alternatively, it is possible that an unknown, signaling-incompetent adaptor may replace KARAP/DAP-12 and DAP-10 in inactivated NK cells. Along these lines, it has been shown that FcRγ can replace CD3ζ in the TCR complex of chronically stimulated T cells.22 However, FcRγ did not become part of the NKG2D complex in inactivated NK cells (data not shown).
Irrespective of the precise basis, constitutive NKG2D signaling seems to limit KARAP/DAP-12 and DAP-10 levels. This view is supported by the observation that the removal of the stimulating tumor cells restores KARAP/DAP-12 and DAP-10 protein expression and NKG2D-dependent target cell lysis.
Reaction of NK cells to chronic NKG2D stimulation
Besides KARAP/DAP-12 and DAP-10, we have also noted the loss of CD3ζ as a consequence of sustained NKG2D engagement by tumor cell-bound ligand. However, CD3ζ does not directly associate with NKG2D11 (data not shown). Therefore, the precise connection between CD3ζ loss and chronic NKG2D engagement will require further investigation. However, it is interesting to note that the loss of CD3ζ has also been observed in chronically activated NK cells and T cells in cancer patients, infectious diseases, and autoimmune states (for review see Baniyash23 ). In essentially all of these instances, up-regulation of NKG2D ligands has since then also been documented.4,6,12,24 Since NKG2D is constitutively expressed on human CD8+ T cells25 and upon activation on murine CD8+ T cells,12 it will be important to test whether chronic NKG2D engagement results in DAP-10 loss and/or is involved in CD3ζ loss in T cells. It is tempting to speculate that CD3ζ loss and NK-cell (and/or T-cell) dysfunction reflects in part or entirely the chronic exposure to cells expressing NKG2D ligand.
Modulation of NK-cell activation receptors
Reduced cell surface expression of specific NK-cell activation receptors has first been reported in humans.26 NK cells with high levels of NCR expression (natural cytotoxicity receptors; ie, NKp30, NKp44, and NKp46 in humans) display high NK-cell activity toward a variety of target cells, whereas those with low levels showed low NK activity. While NK cells from occasional healthy donors have an NCRlow phenotype, the majority of patients with acute myeloid leukemia (AML) display this NCRlow phenotype, which correlates with low NK activity.27 Similar findings were reported in patients with HIV.28 To our knowledge the basis for the low NCR levels is in most cases unknown. Thus, in situations where the levels of NK-cell activation receptors are reduced, it will be important to determine whether or not their signaling capacity and/or the association with adaptors is preserved.
For NKG2D, down-modulation is induced upon ligand encounter. In this instance, lower NKG2D expression is observed despite normal expression of the genes coding for NKG2D and its adaptor genes, suggesting that postranscriptional mechanisms are responsible to reduce cell surface expression6 (shown here). Lower NKG2D cell surface expression is also observed upon the exposure of NK cells to transforming growth factor β (TGFβ).29,30 In this case, the reduced NKG2D levels are due to reduced NKG2D gene expression.29
Activation versus inactivation
Recent studies have highlighted an important role of NKG2D ligands on murine tumor models for the induction of antitumor immunity.2,3,31,32 Despite this, many NKG2D ligand-positive tumors progress, providing circumstantial evidence that in vivo NKG2D function may be impaired at some stage during tumor progression. Indeed, we provide in vitro evidence that profound NK-cell dysfunction can result from the direct and prolonged interaction with tumor cell-bound NKG2D ligand. In addition to tumors, chronic virus infections may generate a comparable situation. It will be crucial to precisely define the factors that determine the consequences of NKG2D engagement (ie, reactivity versus inactivity). Answering this question will be of great value to fully exploit the fact that distressed host cells have the capacity to alert the immune system.
Prepublished online as Blood First Edition Paper, May 10, 2005; DOI 10.1182/blood-2005-03-0918.
Supported in part by a grant from the Swiss Cancer League (Oncosuisse) (W.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Pascal Schneider (Department of Biochemistry, University of Lausanne, Switzerland) for providing expression vectors; Wayne Yokoyama (Washington University, St Louis, MO) for CHO Pro5 cells; and Jean-Charles Cerottini (LICR, Lausanne, Switzerland) for critical reading of the manuscript.

![Figure 1. Chronic engagement abrogates NKG2D function. (A) RMA cells stably transfected with the NKG2D ligand H60 bind soluble NKG2D. (B) The lytic capacity of NK cells exposed for 3 days to RMA-H60 or control RMA cells was determined. To exclude cold-target competition, unlabeled RMA-H60 cells were added to control NK cells during the lysis assay (at a percentage equivalent to that remaining in the RMA-H60 cultures [ie, < 5%]) (▪). These results are from one representative experiment of 14 performed. The lysis curves were shifted relative to the content of NK cells in the effector cell populations. To demonstrate the Ly49D dependence of CHO lysis, Ly49D was blocked with mAb 4E5 (▦ for controls and for RMA-H60-exposed NK cells). (C) NK cells were cultured in the presence of IL-2 plus RMA-H60 (▦) or RMA cells (□). The lytic activity of NK cells against RMA-H60 was determined after 24, 48, and 72 hours of coculture. NK cell activity was tested in triplicate at effector-target (E/T) ratios of 50:1 and 10:1. The data are representative of 2 independent experiments. (D) NK cells were cultured (for 72 hours in IL-2) separate from RMA-H60 (▪) or control RMA cells (□) using transwell plates. The lytic activity of these NK cells against RMA-H60 was determined. (B-D) Graphs depict the mean (±SD) of triplicate determinations at various E/T ratios.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/5/10.1182_blood-2005-03-0918/6/m_zh80170583280001.jpeg?Expires=1765895546&Signature=cdvOxXpCVte-E8zixaNHxUO8ZP8B2Kw75~s7B8qMiQ~kbDUNmEQ~B1YA2snHPJSaLNFEqy34NAvZygbFsbo2V3FFPHLTBJKISgdwcaesKsfaKFNUnqnFrw~4RJmLSA8rFhlP3QGLhznjZ3rixwtuj9qkm9lkA9OzQxzp6xW~DlDyaTxyfq63JxSL1qOejtlFxnx8Jvx26YnOU7xksTSFK18RIFDJod7lt2XBw5q7H44J-IBimrt-i0H9XzHZHxid0W5uUU-1mrmL-eGB5orshg3v7~XsnMo-LL1a5XH94kMwlXdUwlw1SbQdLIGi17gqpUAPbw12EA16C5wFpEhRsw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal