Abstract
Glioblastoma 3 (Gli3) is a transcription factor involved in patterning and oncogenesis. Here, we demonstrate a role for Gli3 in thymocyte development. Gli3 is differentially expressed in fetal CD4–CD8– double-negative (DN) thymocytes and is most highly expressed at the CD44+ CD25– DN (DN1) and CD44–CD25– (DN4) stages of development but was not detected in adult thymocytes. Analysis of null mutants showed that Gli3 is involved at the transitions from DN1 to CD44+ CD25+ DN (DN2) cell and from DN to CD4+CD8+ double-positive (DP) cell. Gli3 is required for differentiation from DN to DP thymocyte, after pre–T-cell receptor (TCR) signaling but is not necessary for pre-TCR–induced proliferation or survival. The effect of Gli3 was dose dependent, suggesting its direct involvement in the transcriptional regulation of genes controlling T-cell differentiation during fetal development.
Introduction
Most αβT cells are produced in the thymus, where lymphocyte precursors pass through a series of developmental stages that depend on interactions with the thymic stroma. Although the stages of thymocyte development have been well characterized in terms of cell-surface protein expression, the molecular mechanisms and regulation of transcription underlying the control of T-cell maturation are less well understood (reviewed by Rothenberg1 and Michie and Zuniga-Pflucke2 ). Here, we show that the transcription factor glioblastoma 3 (Gli3) is differentially expressed in fetal CD4–CD8– double-negative (DN) thymocyte populations and that it is involved in the control of differentiation of fetal DN thymocytes.
Gli3 is a member of the Gli family of transcription factors, mammalian orthologues of the Drosophila Cubitus interruptus (Ci) protein. The Gli protein family (Gli1, Gli2, and Gli3) are large transcription factors that bind DNA in a sequence-specific manner through the last 3 fingers of their 5-zinc finger domain (reviewed by Matise and Joyner3 and Koebernick and Pieler4 ). In Drosophila, the single Ci protein mediates all signaling by a single Hedgehog (Hh) protein, while in mammals the functions of the 3 Gli proteins have diverged (reviewed by Koebernick and Pieler4 and Ingham and McMahon5 ).
The Gli proteins have evolved specialized functions with distinct temporal and tissue-specific expression patterns, acting as either positive or negative regulators of transcription (reviewed by Matise and Joyner3 and Kobernick and Pieler4 ). Gli1 acts exclusively as an activator of transcription and is not essential for mouse development or Hh signaling.6,7 Gli2 and Gli3 undergo processing to function as positive or negative regulators of transcription, depending on the presence or absence of Hh signaling, respectively.8-11 They are each essential for mouse development and have distinct, although partially overlapping, functions.12-14 Although both Gli2 and Gli3 can act as transcriptional activators, Gli2 has been shown in numerous in vivo studies to function primarily as a transcriptional activator downstream of Hh signaling.6,13,15,16 In contrast, Gli3 has been found to function primarily as a negative regulator of transcription in vivo (reviewed by Koebernick and Pieler4 and Ruiz i Altaba17 ), acting independently of Hh signaling.18 In fact, in most tissues studied, the phenotype of Gli3–/– is opposed to that of sonic hedgehog–/– (Shh–/–), confirming that Gli3 acts mainly as a repressor in the absence of Hh signaling.
The mouse mutant extra-toes (gli3Xt-J) carries a large spontaneous deletion of the 3′ end of the Gli3 gene.19 Heterozygous gli3Xt-J mice produce 50% of the Gli3 protein found in wild-type mice, and homozygous mutants do not express detectable Gli3 protein and die before birth with dorsal brain defects and severe polydactyly.20,21 The phenotype of the heterozygous gli3Xt-J mice is characterized by the formation of extra digits on the anterior side of the limb (polydactyly),22 but the mice are fertile and seem healthy. Human heterozygous mutations of the GLI3 locus are associated with dominant, autosomally inherited abnormalities, including the Pallister-Hall syndrome23 and Greig cephalopolysyndactyly syndrome.24
The thymus is formed during fetal development by seeding of the thymic rudiment with blood-borne lymphocyte progenitor cells from the fetal liver.25,26 The thymus continues to produce T cells throughout life and after birth is seeded by blood-borne progenitor cells from the bone marrow27,28 (reviewed by Shortman and Wu29 ). The precise phenotypic characteristics and potential of the population of progenitor cells that seed the thymus may differ between the fetal liver and the bone marrow.30,31 Both adult and fetal thymocytes then pass through a series of developmental stages, defined in terms of cell-surface expression of developmentally regulated markers: CD4–CD8– DN thymocytes progress to the CD4+CD8+ double-positive (DP) stage and then to mature CD4 or CD8 single-positive (SP) T cells. The DN population can be further subdivided by expression of c-kit, CD44, and CD25. The earliest c-kit+CD44+CD25– cells (DN1) migrate in the thymus, acquire CD25 expression (DN2), then lose CD44 and c-kit expression (DN3) and finally become c-kit–CD44–CD25– DN (DN4) cells before differentiating to DP cells, often via a CD8+ immature single-positive (ISP) intermediate (reviewed by Michie and Zuniga-Pflucker2 ). The transition from DN3 cell to DP cell depends on the expression of a functional pre–T-cell receptor (TCR) complex, allowing β-selection. The pre-TCR complex provides signals for proliferation, survival, and differentiation, but the dissection of molecular interactions that lead to these multiple outcomes has proved complicated (reviewed by Borowski et al32 ).
This sequence of thymocyte maturation is widely assumed to be the same in the fetal and adult thymus, but there are, however, likely to be differences in gene expression and the transcriptional regulation of differentiation, reflecting differences in function, kinetics, and microenvironment. For example, in the fetus, thymus organogenesis and patterning must occur, and this depends on stage-specific interactions between developing thymocytes and the developing thymic epithelium (reviewed by Manley26 ), whereas adult thymocytes mature in an organ that is already formed. Likewise, the kinetics of proliferation of DN thymocytes is different between the adult and fetal thymus.33
We have previously shown that Gli3 is expressed in the thymus.34 Here, we show that Gli3 is expressed in fetal but not adult thymocytes and, by analysis of null mutants, that Gli3 is involved in differentiation from fetal DN1 to DN2 cells, and from fetal DN to DP cells downstream of pre-TCR signaling.
Materials and methods
Real-time reverse transcription–polymerase chain reaction (RT-PCR) analysis
C57BL/6J embryonic day (E) 16.5 fetal and adult thymocytes were sorted on a Modular Flow Cytometer (MoFlo; Cytomation, Fort Collins, CO) at the Cancer Research UK FACS (fluorescence-activated cell sorting) laboratory. For purification of DN1 to DN4 populations, cells falling within the forward scatter/side scatter (FSC/SSC) live gate, greater than 98% of which were CD45.2+, were sorted using antibodies directed against CD25FITC (fluorescein isothiocyanate), CD44PE (phycoerythrin), and CD3/CD4/CD8Cychrome. For the DP and SP populations thymocytes were stained with CD4PE and CD8Cychrome. Purity of all populations was greater than 98%. RNA was extracted from the sorted thymocytes with the Absolutely RNA miniprep kit (Stratagene, La Jolla, CA), and cDNA was synthesized with Superscript II (Invitrogen, Carlsbad, CA). The cDNA samples were analyzed in triplicate by real-time PCR on an iCycler (Bio-Rad Laboratories, Hercules, CA) using the iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions. At least one primer for each amplification pair was designed to span exon-exon boundaries to avoid amplification of genomic DNA. For the amplification of gli3 transcripts we used primers Gli3F GCT CCA ACA TTT CCA ACA C and Gli3R TGT GGG CTT GCT CTG TGA GG and for the amplification of the HPRT transcripts we used primers F53333 TGA TTA TGG ACA GGA CTG AAA G and F53334 GGT CAG CAA AGA ACT TAT AGC C. For each sample gli3 transcript levels were normalized to HPRT levels prior to calculation of relative fold difference in transcription levels comparing with DN1 thymocytes.
Mice
C57BL/6 mice were purchased from B & K Universal (Hull, United Kingdom) and used to obtain embryos for the expression studies shown in Figure 1. Rag1–/–35 and C57BL/6J-Gli3Xt-J mice22 were purchased from Jackson Laboratories (Bar Harbor, ME). All mice were bred and maintained at the Central Biomedical Services unit at Imperial College London.
Timed mates were performed by mating a male with 2 females and monitoring the females for plugs. The day the plug was found was counted as E0.5.
PCR analysis
Although gli3Xt-J /Xt-J and gli3Xt-J/wt could be phenotypically distinguished from each other and from wild-type (wt) mice,22 phenotypic discrimination was difficult at day E13.5, and embryos were genotyped by PCR. The primers to amplify the mutated allele were XTJ580F TAC CCC AGC AGG AGA CTC AGA TT and XTJ580R AAA CCC GTG GCT CAG GAC AAG, and the primers to amplify the wt Gli3 allele were C3F GGC CCA AAC ATC TAC CAA CAC AT and C3R GTT GGC TGC TGC ATG AAG ACT GAC.19
Flow cytometry and antibodies
Thymocyte suspensions were prepared by crushing thymi between 2 pieces of ground glass. Cells were stained using combinations of directly conjugated antibodies obtained from BD Pharmingen (San Diego, CA): anti-CD44FITC, anti-CD44PE, anti-CD44Cychrome, anti-CD25PE, anti-CD25FITC, anti-CD4FITC, anti-CD4PE, anti-CD4Cychrome, anti-CD8αFITC, anti-CD8αPE, anti-CD8αCychrome, anti-CD3Cychrome, anti-CD45.2FITC, anti-CD2FITC, anti-CD2PE, anti-NK1.1FITC, CD117Cychrome, TCRβPE. The TCRγδ antibody used was biotin labeled, and an additional staining with streptavidinCychrome (BD Pharmingen) was required.
Cell suspensions were stained with the antibodies for 30 minutes on ice in 50 μL Dulbecco modified medium (Life Technologies, Gaithersburg, MD), supplemented with 5% fetal calf serum (FCS) and 0.01% sodium azide. Cells were washed in this medium between incubations and prior to analysis on the FACScan (fluorescence-activated cell scan; Becton Dickinson, Franklin Lakes, NJ). Events were collected in list mode using CellQuest software (Becton Dickinson), and data were analyzed using CellQuest Pro software. Live cells were gated according to their FSC and SSC profiles. Data are representative of at least 3 experiments.
Intracellular staining for TCRβ and CD3 was performed on cells stained for surface markers as described at the beginning of this section following fixation and permeabilization with the Fix/Perm solutions (BD Biosciences, Mountain View, CA) according to the manufacturer's instructions.
Propidium iodide (Sigma, St Louis, MO) staining was carried out on cells treated with 100 μg/mL RNAse (Sigma) and permeabilized in 0.1% Triton X-100, as described previously.36 DRAQ5 (Alexis Biochemicals, Paris, France) staining was carried out according to the manufacturer's instructions at 10 μM staining concentration. Cell-cycle analysis was carried out using a doublet-discrimination module on the FACScan.
Annexin V staining was carried out using an annexin V–FITC apoptosis detection kit (BD Pharmingen), according to the manufacturer's instructions. Prior to annexin V staining, cells were stained as described at the beginning of this section.
Fetal thymic organ culture (FTOC)
Fetal thymi were cultured on 8-μm pore size Millipore filters (Millipore, Bedford, MA) in AIM-V serum-free medium (Life Technologies) at 37°C and 5% CO2, with 1 μg/mL azide-free anti-CD3 (BD Pharmingen).
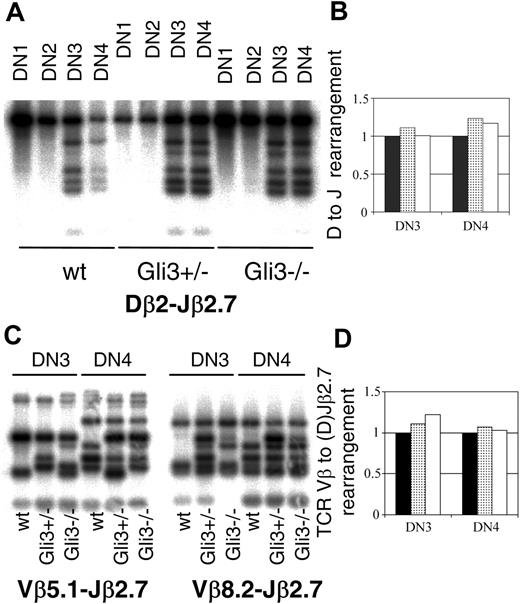
Quantification of TCRβ locus rearrangements
DN1 to DN4 thymocyte subsets were sorted from E15.5 fetal thymi as described in “Real-time reverse transcription–polymerase chain reaction (RT-PCR) analysis,” and DNA was extracted using the DNeasy tissue kit (Qiagen, Valencia, CA). PCR for TCRβ rearrangements was performed using the following primers: Dβ2 (5′) GTA GGC ACC TGT GGG GAA GAA ACT; Jβ2.7 (3′) TGA GAG CTG TCT CCT ACT ATC GAT T; Vβ5.1 (5′) GTC CAA CAG TTT GAT GAC TAT CAC; Vβ8.2 (5′) CCT CAT TCT GGA GTT GGC TAC CC.37,38 We performed 24 cycles of amplification for the D to J rearrangement and 28 for the V(D)J rearrangement. To determine the number of amplification cycles needed to maintain all reactions in the exponential phase the templates were titrated. Sampling was performed at 2-cycle intervals from 20 to 30 cycles, and the amplified products were quantified by Southern. Reactions were run on a Stratagene Robocycler (La Jolla, CA) as follows: 3 minutes at 94°C; 24 of 28 cycles of 45 seconds at 94°C, 90 seconds at 65°C, and 150 seconds at 72°C; and finally 10 minutes at 72°C. The PCR products were separated on a 1% agarose gel and blotted onto a Hybond N′ membrane. The blots were hybridized with TCRβ probe labeled using the Prime-a-gene labeling system (Promega, Madison, WI) according to manufacturer's instructions and α-32P-dATP (deoxyadenosine triphosphate). Bands were visualized using a phosphoimager system (Fuji Film, Tokyo, Japan) and the image was analyzed with the 1D Image Analysis software (Kodak, Rochester, NY) to calculate the intensity of the bands.
The TCRβ probe was a PCR product amplified from germ line (non-rearranged) TCRβ locus with Dβ2 and Jβ2.7 primers, corresponding to the genomic region between the TCRDβ2 and TCRJβ2.7. The genomic DNA concentration of the templates for the V(D)J PCR were quantified by real-time PCR on an iCycler using a single copy genomic locus-specific primers 5′G2F AGA ACC TGA AGA CAC ACC TGC G and 3′G2R GAG GCA TTG GAG AAG GCT TTG.
Results
Gli3 is differentially expressed in fetal DN thymocytes and absent from adult thymocytes
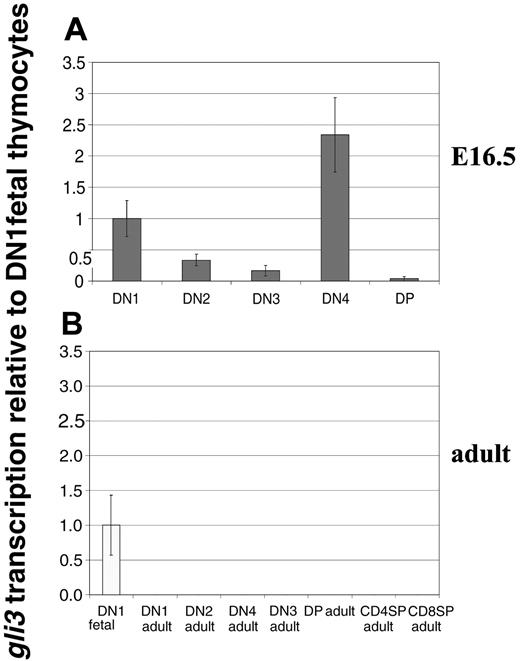
To study the expression pattern of Gli3 in developing thymocytes, we carried out quantitative RT-PCR on RNAs prepared from FACS-sorted thymocyte populations from E16.5 thymus and adult thymus. Transcription of Gli3 was assessed in sorted DN1 to DN4 and DP cells from E16.5 fetal thymi (Figure 1A). Differential transcription of Gli3 was observed in the fetal DN subsets. Gli3 transcripts were present in DN1 cells, their level was then reduced in DN2 and further reduced in the DN3 subset. Transcription reached its maximum in fetal DN4 thymocytes but was very low in the subsequent DP population. Surprisingly, we were unable to detect Gli3 transcripts in RNAs prepared from adult thymocytes. Gli3 was not detected in the DN or DP populations of adult thymi (Figure 1B), revealing a distinct difference in expression of gli3 between fetal and adult thymocytes. These data suggest a possible role for Gli3 in regulation of early fetal thymocyte development.
Transcription of Gli3 in fetal and adult thymic subsets. (A) Transcription of Gli3 was assessed by quantitative real-time RT-PCR on RNAs prepared from DN1, DN2, DN3, DN4, and DP thymocyte populations sorted from E16.5 wild-type C57BL/6 fetal thymi. Levels of Gli3 transcription shown are relative to that observed in the DN1 population and normalized for HPRT mRNA content. Maximal expression was seen in the DN4 subset. (B) Transcription of Gli3 was assessed as in panel A on RNAs prepared from adult thymocyte populations sorted from wild-type C57BL/6 mice. Gli3 transcription is absent or below levels of detection in all adult thymocyte populations.
Transcription of Gli3 in fetal and adult thymic subsets. (A) Transcription of Gli3 was assessed by quantitative real-time RT-PCR on RNAs prepared from DN1, DN2, DN3, DN4, and DP thymocyte populations sorted from E16.5 wild-type C57BL/6 fetal thymi. Levels of Gli3 transcription shown are relative to that observed in the DN1 population and normalized for HPRT mRNA content. Maximal expression was seen in the DN4 subset. (B) Transcription of Gli3 was assessed as in panel A on RNAs prepared from adult thymocyte populations sorted from wild-type C57BL/6 mice. Gli3 transcription is absent or below levels of detection in all adult thymocyte populations.
Interestingly, the pattern of Gli3 transcription in fetal DN thymocytes was inverse to the pattern of cell-surface expression of the Hh signal receptor molecule Smoothened (Smo), which was most highly expressed in DN2 and DN3 cells.34 Hh signaling promotes accumulation of Smo on the cell surface in Drosophila,39 whereas Gli3 transcription has been shown to be down-regulated by Hh signaling.40
Gli3 is involved in the transition from fetal DN1 to DN2 thymocyte
To assess the role of the Gli3 transcription factor in early thymocyte development in vivo, we analyzed the thymi of gli3Xt-J homozygote (Gli3–/–) and heterozygote (Gli3+/–) mutant embryos on E13.5, E14.5, E15.5, and E16.5.
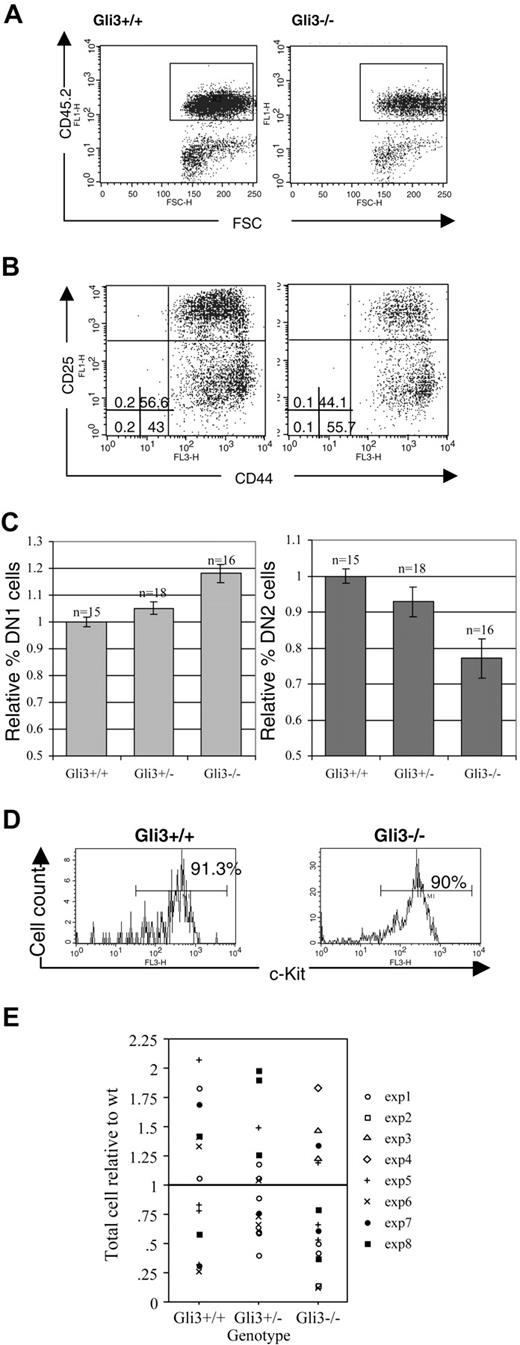
Analysis of E13.5 thymocytes revealed an increased percentage of DN1 cells in Gli3–/– compared with wild-type and Gli3+/– thymi and a correspondingly decreased percentage of DN2 cells (Figure 2A-C). The thymocytes analyzed were gated for positive expression of the leukocyte lineage-specific marker CD45.2, thereby excluding cells of nonhematopoietic origin from the analysis (Figure 2A). In a typical experiment, 55.7% of thymocytes were CD44+CD25– (DN1) in a Gli3–/– embryo, compared with 43% in a Gli3+/+ littermate. The DN2 population was correspondingly decreased from 56.6% in the littermate thymus to 44% in the Gli3–/– thymus (Figure 2B). The differences between knock-out and wild-type mice in the percentages of both DN1 and DN2 were highly significant (Student t test, P < .001 for DN1 and P = .001 for DN2).
The DN1 population contains cells that are not fully committed to the T-cell lineage.25,41,42 In mice in which the Notch1 gene was inactivated in lymphocyte progenitors in the bone marrow, there was an increase in CD44+CD25– thymocytes because of a failure of T-cell lineage commitment.43 In these mice the CD44+CD25– cells that accumulated in the thymus were not equivalent to the DN1 population in wild-type mice because the cells expressed the B-lineage marker B220 and did not express CD117 (c-kit).43 It seemed possible, therefore, that the increase in the proportion of CD44+CD25– cells in the Gli3–/– thymus was the result of a failure of T-cell lineage specification, and that the cells were not true DN1 cells. To test this we stained with anti-CD45.2, anti-CD117, and anti-B220. There was no difference in the proportion of CD45.2+ cells that stained positive with anti-CD117 (Figure 2D), and likewise there was no increase in the proportion of cells that stained positive with anti-B220 (data not shown), indicating that the increase in CD44+CD25– cells probably did not reflect a failure of T-cell lineage commitment, but rather a reduction in differentiation from DN1 to DN2.
No difference was observed in the number of thymocytes isolated from Gli3–/– and Gli3+/– thymi compared with wild-type littermates (Figure 2E), suggesting that the reduction in the proportion of DN2 cells in the Gli3-deficient thymus was unlikely to be due to a reduction in survival or proliferation of the DN2 population.
When we analyzed thymocyte development on E14.5 and E15.5, we found that the Gli3–/– thymi had recovered normal homeostasis, as there were no differences in the composition of the DN subsets or in the number of thymocytes recovered among Gli3–/–, Gli3+/–, and Gli3+/+ thymi (data not shown). Thus, although the reduction in differentiation from DN1 to DN2 cell indicates that Gli3 is involved at this transition, it does not seem to be essential at this stage, and redundancy may exist with other Gli family members.
Expression of Gli3 is required for efficient transition from DN1 to DN2 in thymocyte development. (A) E13.5 thymocytes isolated from Gli3+/+ and Gli3–/– mice were gated on positive expression of the hematopoietic lineage-specific marker, CD45.2, to exclude cells of a nonhematopoietic origin from the analysis. (B) DN1 is overrepresented and DN2 underrepresented in E13.5 Gli3–/– mice. CD45.2+ cells were analyzed for expression of CD44 and CD25. The percentages of DN1 to DN4 subsets are shown. Quadrants were defined according to DN subsets in E16.5 wild-type thymus. (C) Data from 8 litters showing the mean relative percentages of DN1 and DN2 cells in Gli3–/–, Gli3+/–, and Gli3+/+ E13.5 embryos, expressed relative to the mean of the Gli3+/+ littermates. Error bars represent standard error of the mean. (D) The c-kit expression was analyzed on E13.5 CD45.2+ thymocytes from Gli3+/+ (left) and Gli3–/– embryos (right). Percentages of CD45.2+c-Kit+ thymocytes are shown. (E) Thymocyte number isolated from E13.5 Gli3+/+, Gli3+/–, and Gli3–/– embryos. There were no significant differences between genotypes.
Expression of Gli3 is required for efficient transition from DN1 to DN2 in thymocyte development. (A) E13.5 thymocytes isolated from Gli3+/+ and Gli3–/– mice were gated on positive expression of the hematopoietic lineage-specific marker, CD45.2, to exclude cells of a nonhematopoietic origin from the analysis. (B) DN1 is overrepresented and DN2 underrepresented in E13.5 Gli3–/– mice. CD45.2+ cells were analyzed for expression of CD44 and CD25. The percentages of DN1 to DN4 subsets are shown. Quadrants were defined according to DN subsets in E16.5 wild-type thymus. (C) Data from 8 litters showing the mean relative percentages of DN1 and DN2 cells in Gli3–/–, Gli3+/–, and Gli3+/+ E13.5 embryos, expressed relative to the mean of the Gli3+/+ littermates. Error bars represent standard error of the mean. (D) The c-kit expression was analyzed on E13.5 CD45.2+ thymocytes from Gli3+/+ (left) and Gli3–/– embryos (right). Percentages of CD45.2+c-Kit+ thymocytes are shown. (E) Thymocyte number isolated from E13.5 Gli3+/+, Gli3+/–, and Gli3–/– embryos. There were no significant differences between genotypes.
Gli3 is necessary for efficient differentiation of fetal DN thymocytes to DP
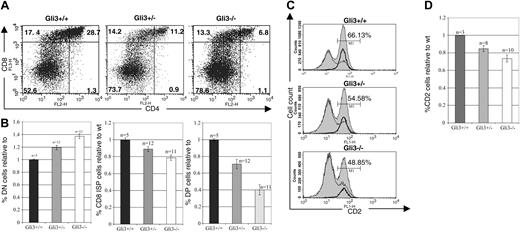
As Gli3 transcription was highest in the DN4 thymocyte subset (Figure 1A), we investigated the effect of defective Gli3 expression at the transition from DN to DP cells. Analysis of the Gli3–/– thymus on E16.5, the earliest embryonic day on which DP thymocytes appear, revealed a partial arrest in thymocyte development at the DN to DP transition. In a typical experiment 6.8% of Gli3–/– thymocytes were DP, compared with 28.7% in the wild-type littermate thymus and 11.2% in Gli3+/– littermate thymus. The proportion of CD8 ISP thymocytes was also reduced in the Gli3–/– thymus, to 13.3% compared with 17.4% in the wild-type littermate thymus, whereas the proportion of DN cells was increased to 78.8%, compared with 52.6% in the wild-type thymus (Figure 3A). Thus, the arrest in thymocyte development was occurring at the transition from DN to DP cells. Overall, the differences in the mean relative percentages of DN, CD8 ISP, and DP thymocytes between all genotypes were statistically significant based on Student t test P values (Figure 3B). Interestingly, the reduction in the maturation to the DP and CD8 ISP stage in heterozygote thymi was intermediate to that in Gli3–/– littermates, suggesting a dose-dependent effect of the expression of Gli3 on the transition from DN to DP thymocytes.
We also analyzed the expression of the CD2 maturation marker on thymocytes isolated from wild-type, Gli3+/–, and Gli3–/– mice. Expression of CD2 is up-regulated on the cell surface following pre-TCR signaling,44,45 and its expression is then sustained throughout thymocyte differentiation to the single-positive stage. In a typical experiment 66.13% of wild-type thymocytes expressed CD2, compared with only 54.58% and 48.85% of heterozygote and knock-out thymocytes, respectively (Figure 3C-D).
Gli3 does not affect survival or proliferation of immature fetal thymocytes
The reduced production of DP thymocytes observed in the Gli3–/– and Gli3+/– thymus could be the result of one or more of the following processes: increased cell death of DN, ISP, or DP thymocytes; reduced proliferation of ISP and/or DP thymocytes; overcommitment to non–αβT-cell lineages; or reduced differentiation from DN to DP cells.
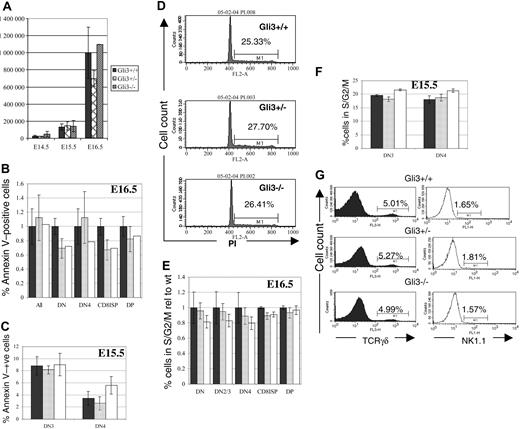
Increased cell death or reduced proliferation in any thymocyte subset would be likely to result in reduction in the number of thymocytes recovered from Gli3–/– and Gli3+/– thymi. We did not, however, observe any significant differences in total thymocyte number among Gli3–/–, Gli3+/–, and wild-type littermates on any embryonic day tested (Figure 4A). To confirm this, we measured survival and cell-cycle status in the thymocyte subsets. We used annexin V staining to measure directly apoptosis in thymocyte populations on E15.5 and E16.5. No significant differences in the percentages of apoptotic cells were observed in any of the thymocyte populations tested (Figure 4B). On E15.5 no DP cells had appeared, so we measured cell death in the DN3 and DN4 populations and found no statistically significant differences between genotypes (Figure 4C).
We assessed the cell-cycle status of thymocytes from E16.5 and E15.5 Gli3–/–, Gli3+/–, and wild-type littermates using DNA binding dyes. We detected no differences between mutant and littermate thymocytes overall (Figure 4D) or in any subset (Figure 4E-F). Thus, we could detect no influence of Gli3 on the survival or proliferation of fetal thymocytes, and changes in these processes could not account for the reduced proportion of DP cells in the Gli3–/– and Gli3+/– thymi.
Reduced transition from DN to DP thymocytes in E16.5 Gli3+/– and Gli3–/– mice. (A) Thymocytes from Gli3+/+, Gli3+/–, and Gli3–/– mice were isolated and stained for expression of the cell surface markers, CD4 and CD8. Percentage of thymocytes in each quadrant is shown. (B) Data from multiple litters represented in panel A are shown graphically. The mean percentage of DN, CD8+ ISP, and DP thymocytes relative to the corresponding mean percentage in wild-type littermates for each litter are shown in the left, middle, and right panels, respectively. P values for the comparisons between the mean values of Gli3–/– and Gli3+/–, between Gli3+/– and Gli3+/+, and between Gli3–/– and Gli3+/+ were all less than .05. (C) Expression of CD2 on thymocytes isolated from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– mice. A reduction in CD2 expression accompanies the reduction in CD8 ISP and DP thymocyte populations seen in Gli3+/– and Gli3–/– mice. Histogram overlays show the expression of CD2 on total E16.5 thymocytes (shaded curves), DN cells (thin lines), and CD8 ISP cells (bold lines). Percentages of total thymocytes positive for CD2 are shown. (D) Data from multiple experiments as shown in panel C. The results are expressed as relative mean percentage of CD2 expression in thymocyte DN, CD8 ISP, and DP populations relative to the wild-type control. Error bars in panels B and D represent the standard error of the mean.
Reduced transition from DN to DP thymocytes in E16.5 Gli3+/– and Gli3–/– mice. (A) Thymocytes from Gli3+/+, Gli3+/–, and Gli3–/– mice were isolated and stained for expression of the cell surface markers, CD4 and CD8. Percentage of thymocytes in each quadrant is shown. (B) Data from multiple litters represented in panel A are shown graphically. The mean percentage of DN, CD8+ ISP, and DP thymocytes relative to the corresponding mean percentage in wild-type littermates for each litter are shown in the left, middle, and right panels, respectively. P values for the comparisons between the mean values of Gli3–/– and Gli3+/–, between Gli3+/– and Gli3+/+, and between Gli3–/– and Gli3+/+ were all less than .05. (C) Expression of CD2 on thymocytes isolated from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– mice. A reduction in CD2 expression accompanies the reduction in CD8 ISP and DP thymocyte populations seen in Gli3+/– and Gli3–/– mice. Histogram overlays show the expression of CD2 on total E16.5 thymocytes (shaded curves), DN cells (thin lines), and CD8 ISP cells (bold lines). Percentages of total thymocytes positive for CD2 are shown. (D) Data from multiple experiments as shown in panel C. The results are expressed as relative mean percentage of CD2 expression in thymocyte DN, CD8 ISP, and DP populations relative to the wild-type control. Error bars in panels B and D represent the standard error of the mean.
To test whether the reduction in the percentage of DP thymocytes was due to commitment to other lymphoid lineages we analyzed the percentages of γδT and natural killer (NK) cells. No differences were observed in percentages of TCRγδ-positive cells and NK1.1-positive cells between genotypes (Figure 4G).
We, therefore, conclude that the dose-dependent influence of Gli3 on the production of DP thymocytes was due to direct involvement of Gli3 in the differentiation mechanism.
TCRβ locus rearrangement is not reduced in Gli3-deficient mice
Maturation of DN to DP thymocytes is dependent on pre-TCR signaling,2 so a reduction in DP thymocytes could be the result of defective signaling downstream of the pre-TCR or could be the result of earlier events such as reduction in TCRβ locus rearrangement.
To test whether Gli3 is involved in the control of TCRβ locus rearrangement, we analyzed TCRβ locus rearrangements in DN1 to DN4 subsets sorted from Gli3+/+, Gli3+/–, and Gli3–/– E15.5 thymi. We performed Southern hybridizations with PCR products amplified with TCRDβ2 and TCRJβ2.7 primers to allow relative quantification of genomic DNA target sequences38,46 (Figure 5A). For every sample, the intensity of the 6 bands corresponding to the rearranged TCRβ locus was calculated relative to the intensity of the band corresponding to the non-rearranged TCRβ locus allele, as a measure of the D to J rearrangement of the TCRβ locus. We did not detect rearrangement in the DN1 and DN2 subsets. D to J rearrangement was observed in the DN3 and DN4 subsets sorted from Gli3–/–, Gli3+/–, and Gli3+/+ mice (Figure 5A). We observed no reduction in the amount of D to J rearrangement in either DN3 or DN4 populations sorted from Gli3+/– or Gli3–/– embryos, relative to their wild-type littermates (Figure 5B). We also compared the amount of V to (D)J rearrangement of the TCRβ locus in DN3 and DN4 thymocytes sorted from Gli3–/–, Gli3+/–, and Gli3+/+ embryos. We used primers that recognized 2 different V segments (Vβ5.1 and Vβ8.2).37 This assay would only produce bands for the rearranged TCRβ locus, so the quantity of genomic DNA used as template was normalized, relative to another genomic sequence. We quantified the genomic DNA samples by real-time PCR and used the same amount of starting material for all samples. We did not detect a reduction in the amount of V(D)J rearrangement in Gli3+/– and Gli3–/– DN3 or DN4 thymocytes as assessed by the total intensity of the rearranged Vβ5.1/Vβ8.2 (D)Jβ2.7 bands, relative to the intensity of the rearranged bands in the wild type (Figure 5C-D).
Likewise, we did not detect a reduction in TCRβ chain protein expression in the Gli3–/– embryos. We analyzed the intracellular (ic) expression of 2 pre-TCR components, TCRβ and CD3ζ proteins in DN3 and DN4 cells on day E16.5. The percentages of icTCRβ+ and icCD3+ DN3 and DN4 cells were very similar in all genotypes (Table 1).
Relative percentage of icTCRβ+ cells in DN3 and DN4 thymocytes of Gli3+/+, Gli3+/–, and Gli3–/– thymi at day E16.5 and relative percentages of icCD3 high-expressing cells in DN3 and icCD3 intermediate-expressing DN4 subsets of Gli3+/+, Gli3+/–, and Gli3–/– thymi at day E16.5
Genotype . | icTCRβ+ DN3 cells relative to wt . | icTCRβ+ DN4 cells relative to wt . | No. embryos analyzed . | icCD3 high DN3 cells relative to wt . | icCD3 intermediate DN4 cells relative to wt . | No. embryos analyzed . |
|---|---|---|---|---|---|---|
| Gli3+/+ | 1.00 ± 0.06 | 1.00 ± 0.04 | 4 | 1.00 ± 0.1 | 1.00 ± 0.006 | 3 |
| Gli3+/- | 0.92 ± 0.03 | 0.96 ± 0.03 | 11 | 1.08 ± 0.072 | 1.01 ± 0.002 | 5 |
| Gli3-/- | 0.90 ± 0.06 | 0.97 ± 0.05 | 5 | 0.94/1.24* | 1.016/1.004* | 2 |
Genotype . | icTCRβ+ DN3 cells relative to wt . | icTCRβ+ DN4 cells relative to wt . | No. embryos analyzed . | icCD3 high DN3 cells relative to wt . | icCD3 intermediate DN4 cells relative to wt . | No. embryos analyzed . |
|---|---|---|---|---|---|---|
| Gli3+/+ | 1.00 ± 0.06 | 1.00 ± 0.04 | 4 | 1.00 ± 0.1 | 1.00 ± 0.006 | 3 |
| Gli3+/- | 0.92 ± 0.03 | 0.96 ± 0.03 | 11 | 1.08 ± 0.072 | 1.01 ± 0.002 | 5 |
| Gli3-/- | 0.90 ± 0.06 | 0.97 ± 0.05 | 5 | 0.94/1.24* | 1.016/1.004* | 2 |
Values shown are mean relative percentages ± SE.
Two values only for the Gli3-/- genotype in ic CD3 high DN3 and intermediate DN4 cells; therefore, SE is not applicable.
Gli3 is involved in the differentiation of DN to DP thymocytes after pre-TCR signaling
To ask whether Gli3 functions after pre-TCR signaling in the control of differentiation we crossed the Gli3+/– mice onto a Rag1–/– background. Rag1–/– mice have arrested T-cell development at the DN3 stage because they are unable to rearrange the TCRβ locus35 and express the pre-TCR complex. This arrest in thymocyte development can be overcome when pre-TCR signaling is mimicked by treatment with anti-CD3 antibody.47
Gli3 is not involved in survival, proliferation, or lineage commitment of fetal thymocytes. (A) Thymocyte number isolated from Gli3–/–, Gli3+/–, and Gli3+/+ embryos is not significantly different at embryonic days E14.5, E15.5, and E16.5. (B-C,E-F) ▪ indicates Gli3+/+; ▦, Gli3+/–; and □, Gli3–/–. Error bars represent the standard error of the mean. (B) Cell death (as measured by annexin V staining) was not significantly different in any thymocyte population isolated from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– mice. (C) Cell death measured as in panel B of DN3 and DN4 cells isolated from E15.5 Gli3+/+, Gli3+/–, and Gli3–/–. No significant differences were observed. (D) Propidium iodide staining was used to assess cell-cycle status of total thymocytes isolated from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– mice. Percentages of thymocytes at the G2/S phases of the cell cycle are shown. (E) Cell-cycle status analyzed by staining with DRAQ5 simultaneously with cell-surface markers for a combination of CD4, CD8, CD44, and CD25. No significant differences in cell-cycle status was observed in any thymocyte subset analyzed from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– embryos. (F) Cell-cycle status of E15.5 DN3 and DN4 was analyzed as in panel E. There were no significant differences observed between Gli3+/+, Gli3+/–, and Gli3–/– embryos. (G) E16.5 thymocytes were stained with anti-TCRγδ and anti-NK1.1 antibodies. No significant differences were observed. Percentages of TCRγδ or NK1.1 CD44–CD25– DN cells are shown.
Gli3 is not involved in survival, proliferation, or lineage commitment of fetal thymocytes. (A) Thymocyte number isolated from Gli3–/–, Gli3+/–, and Gli3+/+ embryos is not significantly different at embryonic days E14.5, E15.5, and E16.5. (B-C,E-F) ▪ indicates Gli3+/+; ▦, Gli3+/–; and □, Gli3–/–. Error bars represent the standard error of the mean. (B) Cell death (as measured by annexin V staining) was not significantly different in any thymocyte population isolated from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– mice. (C) Cell death measured as in panel B of DN3 and DN4 cells isolated from E15.5 Gli3+/+, Gli3+/–, and Gli3–/–. No significant differences were observed. (D) Propidium iodide staining was used to assess cell-cycle status of total thymocytes isolated from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– mice. Percentages of thymocytes at the G2/S phases of the cell cycle are shown. (E) Cell-cycle status analyzed by staining with DRAQ5 simultaneously with cell-surface markers for a combination of CD4, CD8, CD44, and CD25. No significant differences in cell-cycle status was observed in any thymocyte subset analyzed from E16.5 Gli3+/+, Gli3+/–, and Gli3–/– embryos. (F) Cell-cycle status of E15.5 DN3 and DN4 was analyzed as in panel E. There were no significant differences observed between Gli3+/+, Gli3+/–, and Gli3–/– embryos. (G) E16.5 thymocytes were stained with anti-TCRγδ and anti-NK1.1 antibodies. No significant differences were observed. Percentages of TCRγδ or NK1.1 CD44–CD25– DN cells are shown.
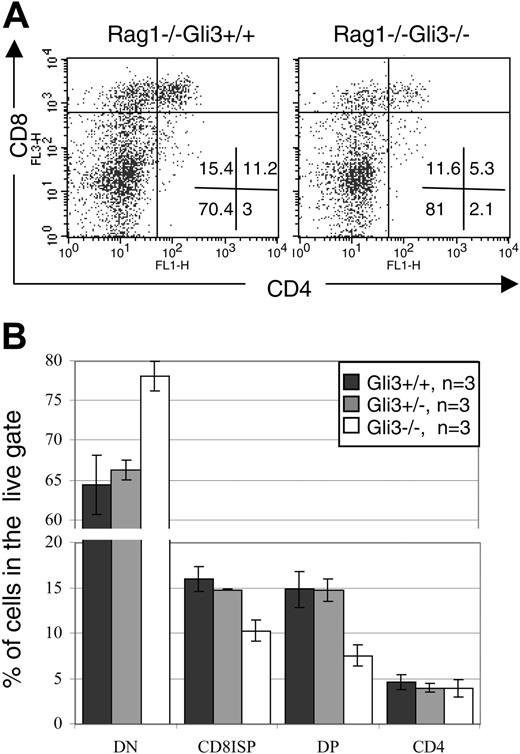
We cultured E15.5 Rag1–/–Gli3–/– and Rag1–/–Gli3+/– and Rag1–/–Gli3+/+ thymi for 2 days with anti-CD3 antibody. This culture period was long enough to allow the Rag1–/–Gli3+/+ FTOCs to produce ISP and DP cells at a similar rate to E16.5 wild-type thymi. The production of DP thymocytes was reduced in the Rag1–/–Gli3–/– FTOC relative to control. In a representative experiment, 5.3% of thymocytes were DP in the anti-CD3–treated Rag1–/–Gli3–/–, compared with 11.2% in the anti-CD3–treated Rag1–/–Gli3+/+ control cultures (Figure 6A). There was a relative accumulation of DN cells in the Gli3–/– FTOC, indicating that the partial arrest occurred at the DN to DP transition. The mean percentages of DN, CD8 ISP, and DP cells were statistically significantly different between cultures from Rag1–/–Gli3–/– and littermate embryos (Figure 6B). A small percentage of CD4 single-positive cells were present in all FTOCs, but no difference was observed between the different genotypes. In Rag1–/– FTOC mature CD4 SP cells are likely to be immature cells, and these cells could be either very immature pre-DN cells (reviewed by Ceredig and Rolink48 ) or cells that arise at the transition between DN and DP (equivalent to CD8 ISP).48
In conclusion, the induction of differentiation of Rag1–/– thymocytes upon anti-CD3 antibody stimulation was dependent on the expression of Gli3, demonstrating that Gli3 acts independent of rearrangement and after pre-TCR signaling in its regulation of differentiation of DN to DP thymocyte.
Discussion
Here, we show that the transcription factor Gli3 is differentially expressed during the development of fetal DN thymocytes and that it is involved in differentiation from DN1 to DN2 cells and from DN to DP cells, downstream of pre-TCR signaling. The expression pattern of Gli3 is consistent with the data from ex vivo analysis of Gli3–/– thymi, as the reduction in differentiation from DN1 to DN2 cells, and from DN to DP cells, coincided with increased levels of expression in the DN1 population relative to DN2, and in the DN4 population relative to the DP population. The stage-specific impairment of differentiation in Gli3-deficient mice was dose dependent, as heterozygote thymi showed intermediate phenotypes to wild-type and Gli3–/– thymi. This dose dependence indicates that the quantity of Gli3 protein present is rate limiting in the differentiation of fetal DN thymocytes and suggests that Gli3 is acting directly to modulate target genes involved in differentiation. Interestingly, we were unable to detect any changes in thymocyte number, survival, or cell-cycle status, indicating that the role of Gli3 in thymocyte development is to regulate the expression of stage-specific genes, allowing thymocyte differentiation. Thus, we show that Gli3 acts after pre-TCR signaling for differentiation only, but it is not involved in pre-TCR–induced proliferation or survival.
TCRβ locus rearrangement does not require the presence of Gli3. (A) Southern blot hybridizations were performed on PCR products amplified using primers specific for TCRDβ2 and TCRJβ2.7 and DNA templates from DN subsets sorted from E15.5 thymus. The probe used corresponded to a germ line (unrearranged) TCRβ fragment and was obtained by radio-labeling a PCR product amplified from genomic DNA using the TCRDβ2 and TCRJβ2.7 specific oligos. PCR was performed on DN1, DN2, DN3, and DN4 thymocyte populations sorted from wt, Gli3+/–, and Gli3–/– mice. The PCR reactions were stopped when all were still in the exponential phase as determined by the titration of templates. (B) The extent of TCRβ rearrangement observed in panel A was quantified by measuring the total intensity of the 6 rearranged bands and relating back to the intensity of the germ line band (uppermost band in each panel) by dividing the total intensity of the bands corresponding to the rearranged locus by the intensity of the band corresponding to nonrearranged locus. The levels of rearrangement observed in the wt mice (▪) was set to 1 and the levels of rearrangement observed in Gli3+/– (▦) and Gli3–/– mice (□) expressed relative to this level. No reduction in TCRDβ2-Jβ2.7 rearrangement was observed in DN3 and DN4 thymic subsets isolated from Gli3+/– or Gli3–/– mice. (C) TCRβ V-(D)J rearrangements were analyzed using the same technique as in panel A, except that the PCR products were generated using 5′ primers specific to TCR Vβ5.1 (left) and TCR Vβ8.2. (right) together with the same 3′ primer to Jβ2.7 as in panel A. In addition the genomic DNA content for each extraction was calculated by real-time PCR, and the same starting template for the rearrangement PCRs was used in all reactions. The probe used was the same as in panel A. TCRβ V(D)J rearrangement had occurred in DN3 and DN4 thymic subsets isolated from wt, Gli3+/–, and Gli3–/– mice. The PCR reactions were stopped when all were still in the exponential phase as determined by the titration of templates. (D) The levels of rearrangement observed in panel C was quantified by using equivalent starting amounts of genomic DNA as determined by real-time PCR and measuring the total intensity of all the bands and relating this amount back to that observed in DNA isolated from the wt mouse. No significant differences were observed in the level of TCRβ V(D)J rearrangement observed in wt (▪), Gli3+/– (▦), and Gli3–/– (□) mice.
TCRβ locus rearrangement does not require the presence of Gli3. (A) Southern blot hybridizations were performed on PCR products amplified using primers specific for TCRDβ2 and TCRJβ2.7 and DNA templates from DN subsets sorted from E15.5 thymus. The probe used corresponded to a germ line (unrearranged) TCRβ fragment and was obtained by radio-labeling a PCR product amplified from genomic DNA using the TCRDβ2 and TCRJβ2.7 specific oligos. PCR was performed on DN1, DN2, DN3, and DN4 thymocyte populations sorted from wt, Gli3+/–, and Gli3–/– mice. The PCR reactions were stopped when all were still in the exponential phase as determined by the titration of templates. (B) The extent of TCRβ rearrangement observed in panel A was quantified by measuring the total intensity of the 6 rearranged bands and relating back to the intensity of the germ line band (uppermost band in each panel) by dividing the total intensity of the bands corresponding to the rearranged locus by the intensity of the band corresponding to nonrearranged locus. The levels of rearrangement observed in the wt mice (▪) was set to 1 and the levels of rearrangement observed in Gli3+/– (▦) and Gli3–/– mice (□) expressed relative to this level. No reduction in TCRDβ2-Jβ2.7 rearrangement was observed in DN3 and DN4 thymic subsets isolated from Gli3+/– or Gli3–/– mice. (C) TCRβ V-(D)J rearrangements were analyzed using the same technique as in panel A, except that the PCR products were generated using 5′ primers specific to TCR Vβ5.1 (left) and TCR Vβ8.2. (right) together with the same 3′ primer to Jβ2.7 as in panel A. In addition the genomic DNA content for each extraction was calculated by real-time PCR, and the same starting template for the rearrangement PCRs was used in all reactions. The probe used was the same as in panel A. TCRβ V(D)J rearrangement had occurred in DN3 and DN4 thymic subsets isolated from wt, Gli3+/–, and Gli3–/– mice. The PCR reactions were stopped when all were still in the exponential phase as determined by the titration of templates. (D) The levels of rearrangement observed in panel C was quantified by using equivalent starting amounts of genomic DNA as determined by real-time PCR and measuring the total intensity of all the bands and relating this amount back to that observed in DNA isolated from the wt mouse. No significant differences were observed in the level of TCRβ V(D)J rearrangement observed in wt (▪), Gli3+/– (▦), and Gli3–/– (□) mice.
We demonstrate a difference in both the expression pattern of Gli3 and the requirement for Gli3 during fetal and adult development, as we were unable to detect Gli3 transcription in adult DN subsets, and differentiation from the DN to DP stage in Gli3+/– adult thymi was not affected (data not shown). Few knock-out mice have revealed differences between the molecular requirements for fetal and for adult thymocyte development.48 In the case of T-cell factor 1 (Tcf1) and the interleukin-7 receptor α chain (IL-7Rα) knock-outs, the thymus phenotype was more severe in the adult than in the fetus,49-51 whereas Ikaros is essential in fetal but not adult thymocyte development.52
Gli3 is downstream of pre-TCR signaling for thymocyte differentiation. (A) Two-day FTOCs of E15.5 Rag1–/–Gli3–/– and Rag1–/–Gli3+/+ thymi with anti-CD3 antibody were performed and stained for CD4 and CD8 expression. The percentage of DP thymocytes was reduced in Rag1–/–Gli3–/– relative to the Rag1–/–Gli3+/+ control. Percentages of thymocytes in each quadrant are shown. (B) Mean percentages of DN, CD8 ISP, DP, and SP CD4 cells induced in the Rag1–/–Gli3+/+, Rag1–/–Gli3+/–, and Rag1–/–Gli3–/– FTOCs treated with anti-CD3 are shown. The differences observed between Rag1–/–Gli3+/+ and Rag1–/–Gli3–/– are statistically significant according to Student t test P values (< .05). Error bars represent the standard error of the mean.
Gli3 is downstream of pre-TCR signaling for thymocyte differentiation. (A) Two-day FTOCs of E15.5 Rag1–/–Gli3–/– and Rag1–/–Gli3+/+ thymi with anti-CD3 antibody were performed and stained for CD4 and CD8 expression. The percentage of DP thymocytes was reduced in Rag1–/–Gli3–/– relative to the Rag1–/–Gli3+/+ control. Percentages of thymocytes in each quadrant are shown. (B) Mean percentages of DN, CD8 ISP, DP, and SP CD4 cells induced in the Rag1–/–Gli3+/+, Rag1–/–Gli3+/–, and Rag1–/–Gli3–/– FTOCs treated with anti-CD3 are shown. The differences observed between Rag1–/–Gli3+/+ and Rag1–/–Gli3–/– are statistically significant according to Student t test P values (< .05). Error bars represent the standard error of the mean.
The way in which the function of Gli3 during thymocyte development relates to the Hh signaling pathway is unclear. Evidence from many different tissues suggests that Gli3 does not require Hh signaling for initiation of its expression or for its repressor activity but that Hh signaling is essential for formation of the correct Gli3 repressor-Gli3 activator ratio, resulting in normal target gene regulation and pattern formation.18,53,54
In mammals there are 3 hedgehog proteins (Shh, Indian Hh [Ihh], and Desert Hh [Dhh]; reviewed by Ingham and McMahon5 ), and Shh–/– embryos have defective thymocyte development. Their thymus is about 10-fold smaller than that of littermates, and there is a reduction in proliferation and survival of DN cells.55 As there was no difference in number or cell-cycle status of Gli3–/– thymocytes relative to littermates, Gli3 is not acting downstream of Shh signaling for proliferation and control of cell number. The Shh–/– thymus showed reduced differentiation at the transition from DN1 to DN2. This phenotype was more pronounced than that observed in the Gli3–/– thymus; therefore, it seems that Gli3 is downstream of Shh at this transition, but that there may also be input from other Gli family members. The fact that both the Shh–/– and Gli3–/– thymus have the same phenotype at this developmental stage is most simply interpreted by suggesting that Gli3 is acting as a transcriptional activator (in response to Hh signaling) at this transition.
The way in which Gli3 and Shh interact at the transition from DN to DP is difficult to interpret because of conflicting data from different experimental systems. The Shh–/– thymus showed reduced differentiation to DP cells compared with wild type,55 but in vitro treatment of FTOC with recombinant Shh arrested development at the DN stage, whereas removal of Shh by treatment with a neutralizing antibody increased differentiation from DN to DP cell.34 Likewise, the development of human CD34+ thymocyte progenitors was arrested in vitro by recombinant Shh treatment and accelerated by neutralization of Hh signaling.56 When these in vitro studies are considered,34,56 the fact that Gli3 is involved after pre-TCR signaling for differentiation from DN to DP thymocyte would suggest that Gli3 is acting as a transcriptional repressor at this developmental stage. However, the fact that the Shh–/– and Gli3–/– thymus have the same phenotype at the DN to DP transition is most simply interpreted as suggesting that Gli3 is acting as a transcriptional activator. The data presented here do not show whether Gli3 has activator or repressor function at this transition.
All the experimental data from the 4 studies34,55,56 (and this study) are, however, compatible with a model in which Shh is necessary before transduction of the pre-TCR signal, but Shh signaling must cease after pre-TCR signaling, allowing formation of the repressor form of the Gli3. Two pieces of evidence suggest that Shh does indeed act before pre-TCR signaling: (1) The fact that the cell-surface expression of the Hh signal transduction molecule Smo is largely restricted to DN2 and DN3 populations34 suggests that Shh is required before pre-TCR signaling. (2) The Shh–/– thymus showed increased cell death at the DN4 stage,55 compatible with failure to express TCRβ/γδ proteins, leading to apoptotic cell death at the DN4 stage.57 Taken together, these studies would be consistent with a model in which Hh signaling must cease after pre-TCR signal transduction to allow formation of the repressor form of Gli3 and differentiation to DP cell. Clearly, the relationship between the requirement for Gli3 and the function of the Hh signaling pathway during thymocyte development will require further study.
Other signaling pathways and transcription factors first discovered in Drosophila, including the Notch pathway,43 the Winglesstype (Wnt) proteins and their downstream transcription factors Tcf1 and lymphoid enhancer binding factor 1 (Lef1)50,58-60 (reviewed by Staal and Clevers60 ), and the bone morphogenetic protein 2 and 4 (BMP2/4) signaling pathway,36,61 have been shown to be involved both at the transition from DN1 to DN2 thymocytes, and from DN to DP thymocytes. Gli3 activity interacts with BMP2/4 signaling and Wnt signaling in the development of other tissues,62-64 and the Gli3R corepressors, skinny hedgehog (Ski) and Ski-related novel protein (Sno), repress BMP2/4 signaling.65,66 It will be important to determine the relationship between Gli3 function and these other signaling pathways in the regulation of thymocyte development.
In summary, we have identified the transcription factor Gli3 as a regulator of fetal thymocyte differentiation. Gli3 acts in a dose-dependent manner, down-stream of pre-TCR signaling, to allow differentiation from DN to DP thymocytes. In addition, Gli3 is involved in fetal thymocyte differentiation at the transition from DN1 to DN2.
Prepublished online as Blood First Edition Paper, April 26, 2005; DOI 10.1182/blood-2005-03-0998.
Supported by the Wellcome Trust and the Medical Research Council (MRC).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Derek Davies and the Cancer Research UK flow cytometry facility for cell sorting.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal