Abstract
The tumor suppressor in lung cancer-1 (TSLC1) gene is frequently silenced in human lung carcinomas, and its expression suppresses tumorigenesis in nude mice. TSLC1 encodes a cell-surface protein called Necl-2 that belongs to the Nectin and Nectin-like (Necl) family of molecules. Necl-2 mediates epithelial cell junctions by homotypic contacts and/or heterotypic interactions with other Nectins and Necls. Thus, it inhibits tumorigenesis by ensuring that epithelial cells grow in organized layers. Here, we demonstrate that natural killer (NK) cells and CD8+ T cells recognize Necl-2 through a receptor known as class I-restricted T-cell–associated molecule (CRTAM), which is expressed only on activated cells. CRTAM–Necl-2 interactions promote cytotoxicity of NK cells and interferon γ (IFN-γ) secretion of CD8+ T cells in vitro as well as NK cell–mediated rejection of tumors expressing Necl-2 in vivo. These results provide evidence for an additional mechanism of tumor suppression mediated by TSLC1 that involves cytotoxic lymphocytes. Furthermore, they reveal Necl-2 as one of the molecular targets that allows the immunosurveillance network to distinguish tumor cells from normal cells.
Introduction
Studies over the past decade have established the notion that both the innate and adaptive arms of the immune system regulate cancer development via multiple mechanisms cumulatively referred to as immunosurveillance.1,2 Lymphocyte recognition of various molecular targets on tumor cells triggers the release of cytotoxic granules and interferon γ (IFN-γ), which suppress tumor growth.3 T lymphocytes recognize tumor-associated antigens in the context of major histocompatibility complex (MHC) class I and class II proteins.4-6 Alternatively, natural killer (NK) cells, CD8+ T lymphocytes, and γδ T cells recognize “alert” molecules expressed on cells undergoing neoplastic transformation, including MHC class I chain related (MIC) and UL16 binding protein (ULBP) in humans and Rae in mice, through the activating receptor natural killer group 2D (NKG2D).7-18 Moreover, tumor-cell apoptosis and damage of tumor-infiltrated tissues cause the release of heat shock proteins, extracellular matrix derivatives, and uric acid, which may initiate immune responses through toll-like receptor-dependent and independent pathways.19,20
A tumor suppressor gene, called tumor suppressor in lung cancer-1 (TSLC1), induces rejection of tumors generated by the cell line A549 in nude mice.21 TSLC1 is located on the long arm of human chromosome 11 (11q) and is functionally inactivated in many non-small-cell lung cancers (NSCLC), such as A549, and other types of epithelial cancers because of loss of heterozygosity for 11q. This results in deletion of one TSLC1 allele and methylation of the promoter of the other allele.21-27 TSLC1 encodes an immunoglobulin (Ig)–like cell-surface protein that belongs to the family of Nectin and Nectin-like (Necl) molecules and is called Necl-2.28 Nectins and Necls are Ca2+-independent cell-cell adhesion molecules that contribute to a variety of cell-cell junctions through homotypic contacts or heterotypic interactions with other Nectins and Necls in cis and in trans.29-31 Accordingly, Necl-2 interacts with Necl-2, Necl-1, and Nectin-328 and has been shown to mediate formation of synapses between neurons,32 adhesion of spermatogenic cells to Sertoli cells,33,34 adhesion of mast cells to fibroblasts, and junctions between epithelial cells.35 Given this, lack of Necl-2 function in lung epithelial cells may result in disruption of cell polarity and cell-cell adhesion, leading to epithelial neoplastic growth and metastasis. Consistent with this model, the Necl-2 cytoplasmic domain recruits intracellular proteins, including DAL-1/protein 4.1B,36 MAGUK (membrane-associated guanylate kinase) p55 subfamily member 3 (MPP3),37 and protein associated with Lin-seven 2 (Pals2),28 which are also involved in organizing epithelial- and neuronal-cell junctions and either mediate tumor suppression or interact with tumor suppressor proteins.
Recently, it was shown that CD226 (also called DNAM-1 [DNAX accessory molecule-1])38-41 and CD96 (also called tactile)42 are Ig-like receptors that mediate NK-cell recognition of Necl-5 (also called poliovirus receptor, PVR) and Nectin-2, promoting NK-cell lysis of tumor cells.43-45 However, since Necl-5 and Nectin-2 are highly expressed on tumor cells46-49 and Necl-5 is secreted in human serum,50 it is unclear whether they elicit immune responses in vivo, or, rather, they inhibit immunosurveillance by inducing a continuous down-regulation of their cognate receptors, as it was previously shown for MIC.9 CD226, CD96, Nectins, and Necls have structurally related extracellular regions, which consist of 3 Ig-like domains. Cognate interactions involve mainly the membrane distal Ig-like domains.44 Here, we investigate whether Necl-2 is recognized by a receptor on leukocytes and, consequently, suppresses tumors by inducing antitumor immune responses. We demonstrate that NK cells and CD8+ T cells recognize Necl-2 through a receptor known as class I–restricted T-cell–associated molecule (CRTAM).51 In contrast to CD226 and CD96, CRTAM is expressed only on activated NK cells and T cells. CRTAM–Necl-2 interactions promote cytotoxicity of NK cells and CD8+ T-cell secretion of IFN-γ in vitro as well as NK cell–mediated rejection of tumors expressing Necl-2 in vivo. These observations demonstrate that Necl-2 is a molecular target that allows the immunosurveillance network to distinguish tumor cells from normal cells and suggest that Necl-2 silencing may be an important mechanism for tumor immunoevasion.
Materials and methods
Cloning of human and mouse CRTAM and Necl-2
Sequence manipulations were made in the Bioedit program (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Multiple sequence alignments were made with the T-Coffee program.52 Sequence similarity searches were performed with NCBI BLAST53 and the HMMER package.54 CRTAM (class-I MHC-restricted T-cell–associated molecule; NCBI accession NP_062550) and human Necl-2 (NCBI accession NP_055148) were amplified by nested polymerase chain reaction (PCR) from human CD56+ NK-cell cDNA and 293T cDNA, respectively. CRTAM primers for nested PCR were 5′-CAGTATGTGGTGGAGAGTTCTCAGC-3′ to 5′-AGTGCTAGGATTACAGGCATGAGC-3′ and 5′-CAGTATGTGGTGGAGAGTTCTCAGC-3′ to 5′-GGCAACCCTGAAATCACATGTTCC-3′. Necl-2 primers for nested PCR were 5′-GACATGGCGAGTGTAGTGCTGC-3′ to 5′-AGTCTCACACCTTTCCACCCATTC-3′ and 5′-GACATGGCGAGTGTAGTGCTGC-3′ to 5′-GCCAGTTGGACACCTCATTGAAAC-3′. Similarly, mouse CRTAM (NCBI accession NM_019465) and mouse Necl-2 (NCBI accession NP_061240) were amplified by nested PCR from mouse NK-cell cDNA isolated from spleen and C57BL/6 cDNA from whole brain, respectively. CRTAM primers for nested PCR were 5′-AGAGACTCCCTCCGTTCAGCACAG-3′ to 5′-ACAGTACTTACGATCCTTTCCAGGGC-3′ and 5′-AGACTCCCTCCGTTCAGCACAG-3′ to 5′-GAACACTACACAATACTCTCCGGG-3′. Necl-2 primers for nested PCR were 5′-CACCATGGCGAGTGCTGTGCTGCC-3′ to 5′-CCTAGATGAAGTACTCTTTCTTTTC-3′ and 5′-CACCATGGCGAGTGCTGTGCTGCCGAG-3′ to 5′-CCTAGATGAAGTACTCTTTCTTTTCTTCGGAGTTGTTCTGTCC-3′. An additional primer was used to introduce a stop codon after the transmembrane domain of mouse Necl-2 5′-GCTATCTGGCAAAATAGCGGCCCAGAATG-3′. PCR products were cloned into the pEF6 expression plasmid (Invitrogen, Carlsbad, CA). Flag-tagged receptors were obtained by subcloning the inserts into pCMV3-Flag (Sigma, St Louis, MO). Sequencing services were provided by the Protein and Nucleic Acid Chemistry Laboratories (PNACL, Washington University Medical School, St Louis, MO). P815, EL4, and Daudi cells were transfected by electroporation with a BTX (San Diego, CA) electroporator. Positive clones were selected by flow cytometry with the M2 anti-flag antibody (KODAK) and sorted for high expressing on a MOFLO cell sorter (Siteman Cancer Center, Washington University School of Medicine, St Louis, MO). Human crystallizable fragment (Fc; IgG1) or mouse Fc (IgG1) fusion proteins were made as reported previously.44
Antibodies
Monoclonal antibody (mAb) Cr24.1 was generated by immunizing BALB/c mice with CRTAM-P815 cells. Cells were injected bilaterally 4 times at weekly intervals into the hind leg. Cytosine-phosphate-guanosine (CpG) oligonucleotide was used as adjuvant (CpG 1826, 25 μg/injection). One day after the fourth injection, popliteal and inguinal lymph nodes were removed and processed to a single-cell suspension. Cells were fused to the SP2/0 myeloma. Positive hybridomas were selected by flow cytometry based on their supernatant's ability to stain CRTAM-P815 cells as opposed to mock-transfected P815 cells. Two clones Cr24.1 and Cr16.1 were generated, both IgG2a. NKp46, NKp44, and NKp30 antibodies have been described.44
Conjugates
Conjugate assays were performed essentially as described.44 Briefly, P815–Necl-2 and P815–Necl-5 (PVR) as well as mock-transfected P815 were labeled for 15 minutes at 37°C with 0.5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) in RPMI-5% fetal cal serum (FCS). Labeling was blocked with 1:1 vol/vol FCS, and cells were washed 3 times with RPMI/10% FCS. The various P815 cells were mixed with CRTAM-Daudi cells or mock-transfected Daudi cells in complete medium in wells of a 96-well U-bottom plate, spun down, and incubated at 37°C for 20 to 30 minutes. Cells were then stained on ice for 15 minutes with allophycocyanin-conjugated anti–human CD45, washed once, and gently resuspended before flow cytometric analysis. For blocking experiments, CRTAM-Daudi or mock-Daudi were preincubated for 20 minutes at room temperature (RT) with mAb Cr24.1 or control IgG2a (PK136).
Binding assays
Binding was performed in complete RPMI medium containing 10% FCS. Soluble molecules were used at a final concentration of 50 to 100 μg/mL. Cells and soluble molecules were incubated 10 minutes at 37°C and then on ice for 30 to 40 minutes. After 1 wash with phosphate-buffered saline (PBS)/2% FCS, second-step antibodies were added. These included R-phycoerythrin (RPE)–labeled anti–human or anti–mouse IgG (10 μg/mL final concentration; Southern Biotechnologies Associates [SBA], Birmingham, AL), or biotin-conjugated anti–human Fc (10 μg/mL, SBA) followed by streptavidin-allophycocyanin (Molecular Probes, Eugene, OR).
Laminar flow assays
Glass cover slips were coated overnight with PBS, pH 7.4, containing 100 μg/mL protein A (Sigma) at 4°C, then washed, and finally incubated with Necl-2–hFc chimeric protein or control human IgG (Sigma) diluted to a concentration of 100 μg/mL (PBS, 0.1% bovine serum albumin [BSA], pH 7.4) for 2 hours at 37°C. Nonspecific interactions were blocked with PBS containing rabbit Ig (50 μg/mL) for 30 minutes at 37°C. Cover slips were then assembled in a parallel-plate laminar flow chamber (Glycotech, Gaithersburg, MD). Daudi cells transfected with CRTAM cDNA or control vector (2 × 106/mL; HBSS [Hanks balanced salt solution], 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 1 mM CaCl2, 0.5% BSA, pH 7.4) were infused over the protein substrates at a shear rate of 50 s–1 for 30 seconds and then allowed to remain in close proximity with the substrates for 10 minutes under static conditions. Flow was then reinstituted at a wall shear stress of 0.5 dyn/cm2 and doubled every 15 seconds to a maximum of 16 dyn/cm2. The number of cells remaining adherent to the surface-immobilized proteins was determined at each wall shear stress and expressed as a percentage of the initial population of cells prior to the application of flow. Cell-protein interactions were recorded on Hi-8 videotape using an inverted Nikon microscope (Nikon, Tokyo, Japan) with a plan 10 × objective. For CRTAM–Necl-2 blockade, transfected cells were incubated with mAb Cr24.1 prior to adhesion assays.
Generation of T-cell clones
To generate mixed lymphocyte reaction (MLR)–TF74, peripheral blood mononuclear cells (PBMCs) from a healthy donor (2 × 105) were cocultured with irradiated (50 Gy [5000 rad]) BM16 cells (5 × 104) in wells of a 96-well U-bottom plate. BM16 is an HLA-A2, -B18, -Cw7 homozygous B-cell line.66 At day 7 of culture interleukin 2 (IL-2) was added at a final concentration of 20 U/mL. Proliferating CD8+ T cells were sorted at day 13 and cloned by limiting dilution. MLR-TF74 clone was selected for the ability to proliferate in response to BM16 but not to the unrelated HLA-A3, -B7, -Cw7 B-cell line BM14. The clone was tested against a panel of HLA homozygous B-cell lines and shown to recognize HLA-B18 and -B35 alleles. Proliferation was specifically blocked by anti–class I antibody W6/32 (American Type Culture Collection [ATCC], Manassas, VA) (M.C., unpublished observations, 2003). Melanoma antigen-recognized by T cell 1 (MART-1)–specific CD8+ T-cell lines were generated by sorting MART-1 tetramer+ CD8+ T cells from an HLA-A2+ donor. Tetramers were purchased from Beckman Coulter Immunomics (San Diego, CA). T-cell and NK-cell cultures were performed as previously described.55
T-cell assays
MLR-TF74 T cells (2.5 × 104) were coincubated with graded numbers of BM16.1 and BM16.2 B cells in the presence of CRTAM-hFc (10 μg/mL) or human control (ctr) IgG1 in 200 μL final volume. After 48 hours, 50 μL supernatants were collected, and IFN-γ secretion was determined by microbead array (human Th1-Th2 kit; BD Pharmingen, San Diego, CA). 3H-Thymidine was added to the cultures for an additional 18 hours. MART-1–specific T cells (2 × 104) were coincubated with BM16.1 and BM16.2 cells (2.5 × 104/well) pulsed for 3 hours with graded dilutions of the MART-1–derived peptide 26-35. Free peptide was removed by 3 sequential washes of BM16 cells. Supernatants were collected for IFN-γ determination after 20 hours.
Tumor experiments
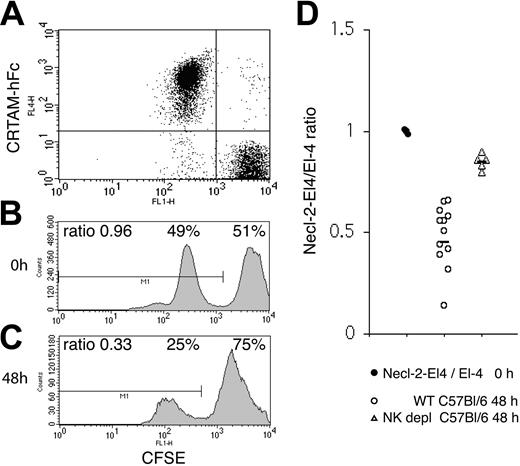
Tumor experiments were performed essentially as previously described.56 Necl-2– and mock-transfected EL-4 cells (107) were labeled with 1 μM (low peak) and 10 μM (high peak) CFSE for 15 minutes at 37°C in 5% FCS containing RPMI medium. Labeling was blocked with 1:1 vol/vol FCS, and cells were washed 3 more times with RPMI/10% FCS. Tumor cells were precisely counted, mixed at a 1:1 ratio, and injected intraperitoneally (8-10 × 106 cells/mouse in 200 μL volume). A sample was acquired at 0 time point to record the ratio between EL-4–Necl-2 and EL-4 cells. Additionally, Necl-2 expression was monitored by binding with CRTAM-hFc. CRTAM-hFc preferentially detects Necl-2, as no significant binding to cells expressing DNAM, CD96, Nectin1, Nectin2, Necl-1, Necl-4, and Necl-5 was observed (data not shown). Since human, mouse, and rat Necl-2 are virtually identical, it has been impossible to effectively immunize rodents to generate mAbs specific for human or mouse Necl-2. Mice were injected with 200 μg poly-I:C (polyinosinic:polycytidylic acid; InvivoGen, San Diego, CA) 24 hours before tumor-cell challenge. Cells were recovered by peritoneal lavage 48 hours after injection. The ratio between CFSE-low– and CFSE-high–labeled cells was determined by flow cytometry. In some experiments EL-4–Necl-2 and EL-4 cells were interchangeably labeled with high or low levels of CSFE. In both cases identical results were obtained. In depletion experiments, mice were injected intraperitoneally with 200 μg purified NK1.1 (PK136, mouse IgG2a) at day –2, followed by intraperitoneal injection of 100 μg PK136 and 200 μg poly-I:C at day –1 before tumor-cell challenge. NK cells were purified from the peritoneal lavage by positive selection with DX5 microbeads (Miltenyi Biotec, Auburn, CA). After blocking Fc receptors (mAb 4G2, rat IgG2b; ATCC), NK cells were stained with Necl-2–hFc or a control hFc, followed by biotin-conjugated anti–human Fc and streptavidin-APC as described for the binding assays. NK cells were counter-stained with NK1.1-RPE during the last 25 minutes of incubation with streptavidin-APC and analyzed by flow cytometry. Poly-I:C injection did not induce up-regulation of CRTAM on mouse NK cells, as assessed by Necl-2–hFc binding to NK cells obtained by peritoneal lavage (data not shown). However, it significantly accelerated tumor rejection, possibly by inducing recruitment of NK cells to the peritoneal cavity (data not shown). Indeed, rejection of cells injected intraperitoneally occurred as late as 72 of 96 hours after injection in the absence of poly-I:C treatment.
Results
The tumor suppressor Necl-2 binds CRTAM
The extracellular domains of Nectins and Necls consist of one membrane distal Ig-V-type domain followed by 2 membrane-proximal Ig-C-type domains.29,31,57 Previous studies indicated that both homotypic and heterotypic interactions of Nectins and Necls involve a binding interface in the C-C′-C″-D β-strands of the Ig-V-type domains.58 Amino acid alignment of this region from Nectins, Necls, and their homologues revealed 2 types of putative binding interfaces (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). While Necl-5, Nectin-2, and their counter receptors CD226 and CD96 had similar binding interfaces, Necl-2 did not, indicating a potential difference in binding specificity. To find a lymphocyte receptor with a binding surface similar to that of Necl-2, we performed similarity searches against the potential binding region of Necl-2 within the NCBI cDNA database and found a candidate receptor, CRTAM51 (Figure S1). CRTAM is an Ig-like cell-surface protein originally cloned from activated NK-T cells with no known function.51 Despite the similarity of the putative binding interface of CRTAM with that of Necl-2, the overall percentage of identity between CRTAM and Necl-2, other nectins, CD226, and CD96 is below 20%, and, indeed, CRTAM was not recognized as a member of the nectin family.
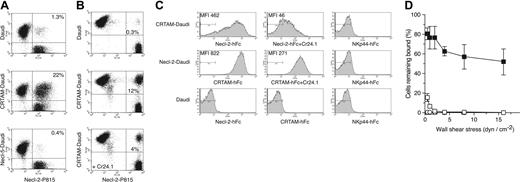
CRTAM and Necl-2 cDNAs were cloned and expressed in the human cell line Daudi (CRTAM-Daudi and Necl-2–Daudi) and the mouse cell line P815 (CRTAM-P815 and Necl-2–P815) (Figure S2). We measured formation of conjugates between CRTAM-Daudi and Necl-2–P815 cells by 2-color flow cytometry after coincubating cells at 37°C. We observed abundant conjugates between Necl-2–P815 and CRTAM-Daudi but not between Necl-2–P815 and Necl-5–Daudi (Figure 1A and 1B). Although Necl-2 has been reported to mediate homophilic cell-cell adhesion,28 Necl-2–P815 and Necl-2–Daudi did not form conjugates (data not shown), suggesting that CRTAM–Necl-2 interactions are stronger than Necl-2 homophilic interactions. To confirm the specificity of CRTAM–Necl-2 binding, we generated the CRTAM-specific mAb Cr24.1 (Figure S2) and demonstrated that this antibody significantly reduces conjugate formation, whereas an isotype-matched control antibody does not (Figure 1B and data not shown).
To further investigate Necl-2–CRTAM interactions, we synthesized a soluble Necl-2–hFc protein composed of the membrane distal Ig domain of Necl-2 fused to the Fc portion of human IgG1. We also produced soluble CRTAM consisting of the entire extracellular domain of CRTAM fused to the Fc portion of hIgG1. These fusion proteins specifically bound CRTAM-Daudi and Necl-2–Daudi, respectively (Figure 1C and data not shown). To determine the strength of Necl-2–CRTAM interactions, we measured the adhesion of CRTAM-Daudi cells to substrates coated with Necl-2–Fc or control IgG using a laminar flow assay. CRTAM-Daudi cells were attached to Necl-2–Fc and resisted detachment by shear forces up to 16 dyn/cm–2 (Figure 1D). Attachment of CRTAM-Daudi to Necl-2–Fc was virtually abrogated by the CRTAM mAb (Figure 1D). We conclude that Necl-2 binds specifically and robustly to CRTAM both under static and flow conditions and that the first domain of Necl-2 is sufficient for interaction with CRTAM.
Expression of CRTAM on NK cells and CD8+ T cells is tightly regulated by cell activation
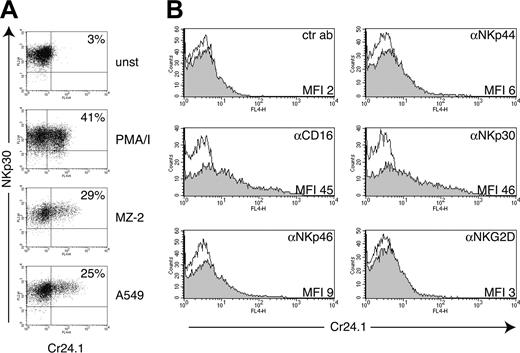
CRTAM mRNA has been detected in NK-T cells and CD8+ T cells.51 However, we were not able to detect CRTAM on the surface of peripheral blood mononuclear cells from healthy donors using our CRTAM mAb Cr24.1 (data not shown). Therefore, we asked whether expression of CRTAM requires cell activation. Human NK cells were treated with phorbol myristate acetate (PMA)/ionomycin for 4 hours or coincubated overnight with the tumor cell lines MZ-2 and A549, which are susceptible to NK cell–mediated lysis. Both treatments induced up-regulation of CRTAM on significant numbers of NK cells (20%-50% in different experiments) (Figure 2A). Furthermore, we triggered NK cell activation using plate-bound mAbs specific for NK cell–activating receptors.59 CD16 and NKp30 mAbs induced considerable CRTAM surface expression (Figure 2B). NKp44 and NKp46 mAbs induced only a slight increase in CRTAM expression, and an NKG2D mAb had no effect (Figure 2B). Among several cytokines, only IL-12 and tumor necrosis factor α (TNF-α) induced slight but reproducible up-regulation of CRTAM on IL-2–cultured NK cells (data not shown).
CRTAM binds Necl-2. (A) Conjugate analysis. Daudi, Daudi-CRTAM, or Daudi–Necl-5 and P815–Necl-2 were tested. Daudi cells were stained with anti–human CD45, while P815 cells were CFSE labeled. A high amount of conjugation (10%-25% of total cells) was reproducibly detected only when CRTAM-Daudi cells were coincubated with Necl-2–P815. The experiment represents 1 of 3 separate assays with comparable results. (B) Incubation of CRTAM-Daudi with mAb Cr24.1 prior to conjugation reduces conjugates between CRTAM-Daudi and Necl-2–P815. An isotype-matched control antibody had no effect (data not shown). One of 2 representative experiments is shown. (C) Soluble Necl-2–hFc binds to CRTAM-Daudi and soluble CRTAM-hFc binds to Necl-2–Daudi. Binding of soluble molecules to transfected cell lines was dramatically reduced by the addition of CRTAM mAb but not by an isotype-matched control antibody (data not shown). Soluble molecules did not bind to Daudi cells, and a control soluble molecule (NKp44-hFc) did not bind CRTAM- or Necl-2–transfected cells. One of 3 experiments with comparable results is shown. MFI indicates mean fluorescence intensity. (D) Detachment profile of CRTAM-Daudi cells interacting with surface-immobilized Necl-2–hFc. The percentage of CRTAM-Daudi or mock-Daudi cells capable of binding to Necl-2–hFc as a function of increasing wall shear stress. Cells were allowed to settle onto the indicated surface-immobilized protein substrates for 10 minutes prior to the application of flow (0.5-16 dyn cm–2). ▪ indicates CRTAM-Daudi/Nec1-2–hFc; □, CRTAM-Daudi/Nec1-2–hFc + Cr24.1; ○, Daudi/Nec1-2–hFc; and ▵, CRTAM-Daudi/hIgG1. Values represent the mean ± SD for 2 experiments performed in duplicate.
CRTAM binds Necl-2. (A) Conjugate analysis. Daudi, Daudi-CRTAM, or Daudi–Necl-5 and P815–Necl-2 were tested. Daudi cells were stained with anti–human CD45, while P815 cells were CFSE labeled. A high amount of conjugation (10%-25% of total cells) was reproducibly detected only when CRTAM-Daudi cells were coincubated with Necl-2–P815. The experiment represents 1 of 3 separate assays with comparable results. (B) Incubation of CRTAM-Daudi with mAb Cr24.1 prior to conjugation reduces conjugates between CRTAM-Daudi and Necl-2–P815. An isotype-matched control antibody had no effect (data not shown). One of 2 representative experiments is shown. (C) Soluble Necl-2–hFc binds to CRTAM-Daudi and soluble CRTAM-hFc binds to Necl-2–Daudi. Binding of soluble molecules to transfected cell lines was dramatically reduced by the addition of CRTAM mAb but not by an isotype-matched control antibody (data not shown). Soluble molecules did not bind to Daudi cells, and a control soluble molecule (NKp44-hFc) did not bind CRTAM- or Necl-2–transfected cells. One of 3 experiments with comparable results is shown. MFI indicates mean fluorescence intensity. (D) Detachment profile of CRTAM-Daudi cells interacting with surface-immobilized Necl-2–hFc. The percentage of CRTAM-Daudi or mock-Daudi cells capable of binding to Necl-2–hFc as a function of increasing wall shear stress. Cells were allowed to settle onto the indicated surface-immobilized protein substrates for 10 minutes prior to the application of flow (0.5-16 dyn cm–2). ▪ indicates CRTAM-Daudi/Nec1-2–hFc; □, CRTAM-Daudi/Nec1-2–hFc + Cr24.1; ○, Daudi/Nec1-2–hFc; and ▵, CRTAM-Daudi/hIgG1. Values represent the mean ± SD for 2 experiments performed in duplicate.
CRTAM expression on NK cells is induced by NK cell conjugation with tumor targets or engagement of NK cell–activating receptors. (A) CRTAM expression was analyzed with the CRTAM mAb Cr24.1 on resting NK cell bulk cultures (unst), NK cells stimulated with PMA/ionomycin for 4 hours, or NK cells conjugated for 20 hours with the NK-susceptible targets A549 (lung carcinoma cell line) and MZ-2 (melanoma cell line). Percentages of CRTAM-positive cells (indicated) varied in different experiments from 25% to 50% of total cells. One of 6 concordant experiments is shown. NKp30 was used as a NK cell–specific marker for counterstaining. (B) CRTAM expression is induced upon cross-linking of some NK cell–activating receptors. CRTAM expression was determined 20 hours after stimulation with plastic coated CD16, NKp44, NKp46, NKp30, and NKG2D mAbs (all mouse IgG1). NKp30 and CD16 mAbs always induced the strongest up-regulation of CRTAM. NKG2D cross-linking reproducibly did not up-regulate CRTAM expression in at least 3 separate experiments. CRTAM was detected with Cr24.1 followed by biotinylated anti–mouse IgG2a and streptavidin allophycocyanin. Shaded histograms represent staining with Cr24.1; open histograms, second- and third-step reagents alone. Comparable results were obtained at 20 and 42 hours of incubation.
CRTAM expression on NK cells is induced by NK cell conjugation with tumor targets or engagement of NK cell–activating receptors. (A) CRTAM expression was analyzed with the CRTAM mAb Cr24.1 on resting NK cell bulk cultures (unst), NK cells stimulated with PMA/ionomycin for 4 hours, or NK cells conjugated for 20 hours with the NK-susceptible targets A549 (lung carcinoma cell line) and MZ-2 (melanoma cell line). Percentages of CRTAM-positive cells (indicated) varied in different experiments from 25% to 50% of total cells. One of 6 concordant experiments is shown. NKp30 was used as a NK cell–specific marker for counterstaining. (B) CRTAM expression is induced upon cross-linking of some NK cell–activating receptors. CRTAM expression was determined 20 hours after stimulation with plastic coated CD16, NKp44, NKp46, NKp30, and NKG2D mAbs (all mouse IgG1). NKp30 and CD16 mAbs always induced the strongest up-regulation of CRTAM. NKG2D cross-linking reproducibly did not up-regulate CRTAM expression in at least 3 separate experiments. CRTAM was detected with Cr24.1 followed by biotinylated anti–mouse IgG2a and streptavidin allophycocyanin. Shaded histograms represent staining with Cr24.1; open histograms, second- and third-step reagents alone. Comparable results were obtained at 20 and 42 hours of incubation.
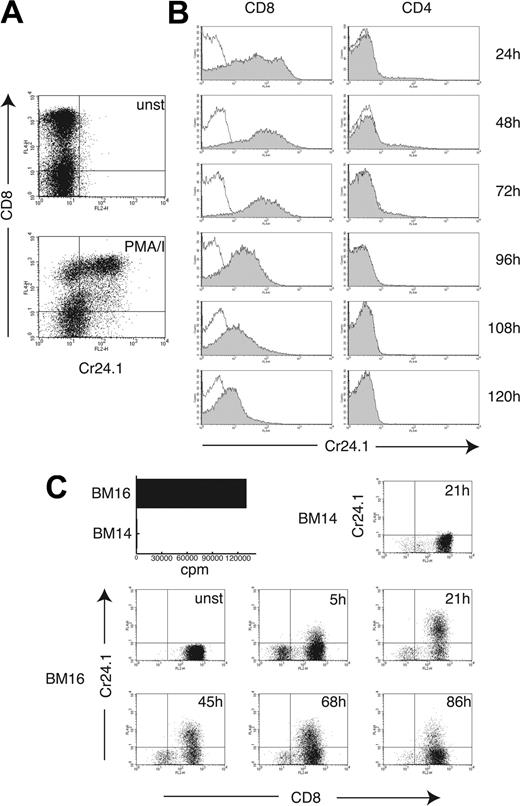
We also investigated the inducibility of CRTAM on T cells. Stimulation of a human polyclonal T-cell line with PMA/ionomycin induced expression of CRTAM on CD8+ T cells after 4 hours of stimulation (Figure 3A). To corroborate this result, CD4+ and CD8+ T cells were purified from peripheral blood and stimulated with plate-bound CD3 mAb. Significant CRTAM expression was observed on the majority of CD8+ T cells 24 hours after stimulation and was sustained for at least 72 to 96 hours (Figure 3B). CD4+ T cells did not express CRTAM 24 to 48 hours after stimulation, except for a small subset of cells (2%-5% in different experiments). Strong up-regulation of CRTAM on CD8+ T cells was also observed when the alloreactive CD8+ T-cell clone MLR-TF74 was stimulated with a B-cell line expressing the alloantigen HLA-B18 (BM16), whereas stimulation with a B-cell line expressing HLA-B7 (BM14) had no effect (Figure 3C). We conclude that CRTAM expression on NK cells and CD8+ T cells is tightly regulated by NK cell–activating receptors and T-cell receptor (TCR) triggering, respectively.
CRTAM promotes NK cell cytotoxicity and IFN-γ secretion by CD8+ T cells
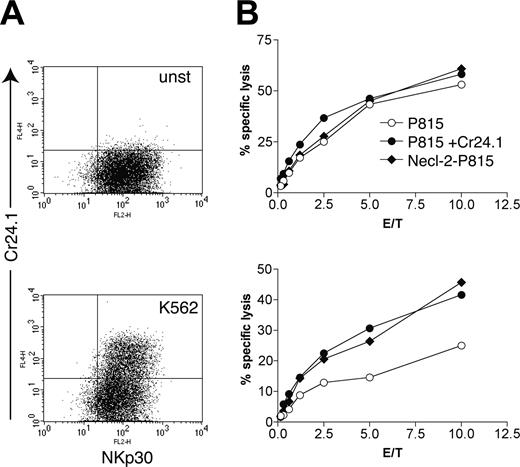
Does expression of CRTAM on activated human NK and CD8+ T cells have functional consequences? To address this question, we used the tumor cell line K562 as a stimulus to induce CRTAM expression on NK cells. After incubation with or without K562 cells for 24 hours, we tested NK cell–mediated cytotoxicity against Necl-2–P815. In addition, we tested redirected lysis against P815 cells coated with the CRTAM antibody, which binds the Fc receptor on P815 cells and CRTAM on NK cells. P815 cells coated with control mouse IgG were used as controls. NK cells incubated with K562 expressed an appreciable amount of CRTAM (Figure 4A) and preferentially lysed Necl-2–P815 cells and P815 cells coated with the CRTAM antibody (Figure 4B). NK cells that had not been incubated with K562 did not express CRTAM and lysed all targets equally well. Thus, triggering of CRTAM augments the cytotoxic function of activated NK cells. NK cells incubated overnight with K562 killed P815 targets slightly less efficiently than unstimulated NK cells (Figure 4). In addition, NK cells down-regulated expression of NKp30 after coculture with K562 (data not shown). Therefore, K562-induced reduction of NK cell–mediated cytotoxicity may be due to down-regulation of activating receptors participating in recognition of P815.
CRTAM expression on CD8+ T cells is tightly associated with TCR triggering. (A) A T-cell bulk culture was stimulated for 4 hours with PMA/ionomycin, and CRTAM expression was determined on unstimulated (unst, open histograms) and stimulated cells (shaded histograms) with Cr24.1. CD8 mAb was used as a counterstain to discriminate between CD4+ and CD8+ T cells. (B) CD8+ and CD4+ T cells were purified from PBMCs by positive selection (> 97% purity) and stimulated with plastic-bound CD3 mAb for the indicated time periods. CRTAM expression was detected as in Figure 2B. CRTAM was strongly up-regulated on CD8+ T cells 24 hours after stimulation and was expressed at high levels up to 72 hours after stimulation. At 96 hours, expression gradually decreased and completely disappeared 180 hours after stimulation (data not shown). CRTAM expression was detected only on a small percentage of CD4+ T cells 48 to 72 hours after stimulation. One of 2 similar experiments is shown. (C) Antigen recognition induces CRTAM expression in CD8+ T cells. CRTAM expression was determined over time on an alloreactive CD8+ T-cell clone, MLR-TF74, which recognizes HLA-B18 expressed by BM16, but not HLA-B7 expressed on BM14. The bar graph indicates proliferation of the T-cell clone stimulated with BM16 and BM14 as detected by standard 3H-Thymidine incorporation (cpm indicates counts per minute). CRTAM expression started to be detected as early as 5 hours after stimulation and peaked at 20 to 24 hours. Lack of expression of CRTAM in T cells incubated with the BM14 cell line, which does not induce MLR-TF74 activation, is shown as a control. CD8 was used as marker to distinguish between the responder T cells and the stimulator B cells.
CRTAM expression on CD8+ T cells is tightly associated with TCR triggering. (A) A T-cell bulk culture was stimulated for 4 hours with PMA/ionomycin, and CRTAM expression was determined on unstimulated (unst, open histograms) and stimulated cells (shaded histograms) with Cr24.1. CD8 mAb was used as a counterstain to discriminate between CD4+ and CD8+ T cells. (B) CD8+ and CD4+ T cells were purified from PBMCs by positive selection (> 97% purity) and stimulated with plastic-bound CD3 mAb for the indicated time periods. CRTAM expression was detected as in Figure 2B. CRTAM was strongly up-regulated on CD8+ T cells 24 hours after stimulation and was expressed at high levels up to 72 hours after stimulation. At 96 hours, expression gradually decreased and completely disappeared 180 hours after stimulation (data not shown). CRTAM expression was detected only on a small percentage of CD4+ T cells 48 to 72 hours after stimulation. One of 2 similar experiments is shown. (C) Antigen recognition induces CRTAM expression in CD8+ T cells. CRTAM expression was determined over time on an alloreactive CD8+ T-cell clone, MLR-TF74, which recognizes HLA-B18 expressed by BM16, but not HLA-B7 expressed on BM14. The bar graph indicates proliferation of the T-cell clone stimulated with BM16 and BM14 as detected by standard 3H-Thymidine incorporation (cpm indicates counts per minute). CRTAM expression started to be detected as early as 5 hours after stimulation and peaked at 20 to 24 hours. Lack of expression of CRTAM in T cells incubated with the BM14 cell line, which does not induce MLR-TF74 activation, is shown as a control. CD8 was used as marker to distinguish between the responder T cells and the stimulator B cells.
To evaluate the function of CRTAM on CD8+ T cells, we assessed proliferation and IFN-γ secretion of the alloreactive T-cell clone MLR-TF74, which recognizes HLA-B18 on the B-cell line BM16. MLR-TF74 up-regulates CRTAM after incubation with BM16 as a consequence of TCR-induced activation (Figure 3C). However it was unclear whether BM16 expresses Necl-2, engaging CRTAM. To address this, we evaluated the expression of CRTAM ligands on BM16 using soluble CRTAM fused to the Fc portion of mouse IgG1 (CRTAM-mFc). This reagent is preferable to CRTAM-hFc because it can be detected with anti–mouse IgG, which will not cross-react with human IgG on BM16. CRTAM-mFc stained several epithelial cell lines, but not A549, consistent with CRTAM's specificity for Necl-2 (Figure S3). It also stained a subset of tonsil B cells (Figure S3) and approximately 20% of BM16 cells (data not shown). We sorted BM16 into 2 fractions: BM16.1, which is highly positive for CRTAM ligand, and BM16.2, which expresses low or no CRTAM ligand (Figure 5A). When BM16.1 and BM16.2 were used to stimulate MLR-TF74, BM16.1 was markedly more effective than BM16.2 in inducing IFN-γ release, and secretion was specifically blocked by the presence of CRTAM-Fc (Figure 5B). On the contrary, no significant differences were noticed in T-cell proliferation when either of the 2 BM16 cell lines were used (Figure 5C). Incubation of activated MLRT-F74 clone with P815 cells coated with anti-CRTAM did not induce redirected lysis, despite substantial expression of CRTAM on CD8+ T cells. We conclude that CRTAM–Necl-2 interaction alone is not sufficient to induce CD8 T-cell cytotoxicity.
Since BM16.1 and BM16.2 also express HLA-A2, we evaluated the effects of CRTAM-CRTAM ligand interaction on IFN-γ production by HLA-A2–restricted CD8+ T cells specific for the tumor-associated antigen MART-1. In general, MART-1–specific CD8+ T cells produced higher levels of IFN-γ than did the alloreactive MLR-TF74 T-cell clone. Yet, IFN-γ production was much higher when the MART-1 26-35 peptide was presented by BM16.1 in comparison to BM16.2. Moreover, IFN-γ release was specifically reduced by addition of CRTAM-Fc. Therefore, we conclude that coengagement of TCR and CRTAM on CD8+ T cells is crucial for inducing IFN-γ release.
Necl-2–bearing tumors are lysed by NK cells in vivo
Restoration of Necl-2 expression by DNA complementation induces the rejection of A549 tumors in nude mice in vivo.21 Moreover, the Necl-2 intracellular domain recruits multiple proteins that suppress tumorigenesis by maintaining polarized, organized growth of epithelial cells.28,36,37 In the context of our data, we postulated that tumor suppression in nude mice might also be mediated by NK cells through CRTAM–Necl-2 interactions. That this interaction also occurs in mice is supported by the high homology between mouse and human Necl-2 proteins (98% identity), as well as mouse and human CRTAM (72%). Moreover, we demonstrated that Daudi cells transiently transfected with mouse CRTAM bound Necl-2–hFc (Figure S4). To assess CRTAM expression on mouse NK cells we used Necl-2–hFc. NK cells freshly isolated from mouse spleen or cultured for 5 to 10 days in IL-2 did not bind Necl-2–hFc. In contrast, some NK cells did bind Necl-2–hFc after incubation with the YAC-1 tumor cell line for 20 hours, indicating that CRTAM expression is associated with cell activation in mice as it is in humans (Figure S4). Although the percentage of CRTAM+ NK cells and the level of expression appeared to be lower in mice than in humans, this discrepancy may be due to a lower affinity of Necl-2–hFc for CRTAM in comparison to the CRTAM mAb, which allows detection of minimal levels of expression.
CRTAM expression on NK cells results in increased lysis of Necl-2–expressing target cells. (A) CRTAM expression is induced by incubation of a resting NK cell bulk culture with the NK-sensitive target cell line K562 (bottom graph). unst indicates unstimulated. (B) Cytotoxicity of unstimulated (top graph) and K562-activated NK cells (bottom graph) against Necl-2–transfected P815 cells, P815 cells coated with the CRTAM mAb Cr24, or control IgG2a Ab (PK136). Significant lysis of Necl-2–P815 and redirected activation by Cr24.1 were observed in at least 3 separate assays. E/T indicates effector-target ratio.
CRTAM expression on NK cells results in increased lysis of Necl-2–expressing target cells. (A) CRTAM expression is induced by incubation of a resting NK cell bulk culture with the NK-sensitive target cell line K562 (bottom graph). unst indicates unstimulated. (B) Cytotoxicity of unstimulated (top graph) and K562-activated NK cells (bottom graph) against Necl-2–transfected P815 cells, P815 cells coated with the CRTAM mAb Cr24, or control IgG2a Ab (PK136). Significant lysis of Necl-2–P815 and redirected activation by Cr24.1 were observed in at least 3 separate assays. E/T indicates effector-target ratio.
Necl-2–CRTAM interaction leads to strong IFN-γ secretion by CD8+ T cells. (A) BM16 cells were sorted into the BM16.1 and BM16.2 cell lines expressing high levels and very low levels of CRTAM ligand, respectively (open histograms are control; shaded histograms are CRTAM-hFc labeling). Necl-2 expression in BM16.1 was confirmed by immunoblot with a rabbit anti–Necl-2 antiserum (data not shown). (B-D) BM16.1 and BM16.2 were separately used to stimulate the MLR-TF74 alloreactive T-cell clone (B-C), and Mart1-specific HLA-A0201–restricted CD8+ T cells (D). INF-γ secretion was measured after 48 hours (B) or 20 hours (D) of stimulation in cell culture supernatants. Proliferation (C) was measured by a standard 3H-thymidine incorporation assay after 72 hours of stimulation. Graded numbers of BM16 were used (B-C) to generate a dose-response curve of antigen (APC, antigen-presenting cell). Serial dilutions of the MART-1 26-35 peptide were used (D) to pulse BM16 cells. Experiments were performed in the presence of CRTAM-hFc to specifically block CRTAM–Necl-2 interaction or hIgG1 as control. One of 3 representative experiments is shown.
Necl-2–CRTAM interaction leads to strong IFN-γ secretion by CD8+ T cells. (A) BM16 cells were sorted into the BM16.1 and BM16.2 cell lines expressing high levels and very low levels of CRTAM ligand, respectively (open histograms are control; shaded histograms are CRTAM-hFc labeling). Necl-2 expression in BM16.1 was confirmed by immunoblot with a rabbit anti–Necl-2 antiserum (data not shown). (B-D) BM16.1 and BM16.2 were separately used to stimulate the MLR-TF74 alloreactive T-cell clone (B-C), and Mart1-specific HLA-A0201–restricted CD8+ T cells (D). INF-γ secretion was measured after 48 hours (B) or 20 hours (D) of stimulation in cell culture supernatants. Proliferation (C) was measured by a standard 3H-thymidine incorporation assay after 72 hours of stimulation. Graded numbers of BM16 were used (B-C) to generate a dose-response curve of antigen (APC, antigen-presenting cell). Serial dilutions of the MART-1 26-35 peptide were used (D) to pulse BM16 cells. Experiments were performed in the presence of CRTAM-hFc to specifically block CRTAM–Necl-2 interaction or hIgG1 as control. One of 3 representative experiments is shown.
We next sought to determine whether tumor cells expressing Necl-2 are efficiently rejected by NK cells in vivo. To test this, we challenged C57BL/6 with the syngeneic lymphoma cell line EL-4 transfected with a plasmid encoding mouse Necl-2 (EL-4–Necl-2) or a control plasmid (EL-4–mock). Both transfectants exhibited similar growth rate, as assessed by 3[H]-thymidine incorporation (data not shown). Mice were stimulated with poly-I:C 24 hours prior to intraperitoneal injection of tumor cells to accelerate NK cell activation and recruitment into the peritoneal cavity. EL-4–ecl-2 and EL-4–mock cells were labeled with 2 different concentrations of CFSE and mixed at a 1:1 ratio. Additionally, Necl-2 expression was monitored using CRTAM-hFc (Figure 6A-B). Forty-eight hours after intraperitoneal injection of the tumor cell mixture, surviving cells were recovered by peritoneal lavage. Flow cytometric analysis revealed a ratio of approximately 0.5 EL-4–Necl-2 to 1 EL-4–mock cells, indicating preferential rejection of EL-4–Necl-2 (Figure 6C). Furthermore, no proliferation of injected cells occurred during the 48 hours. Identical results were obtained when Necl-2–transfected/Baf-3 and mock-transfected/Baf-3 cells were injected intraperitoneally into BALB/c mice (data not shown). Importantly, when C57BL/6 mice were depleted of NK cells with an anti-NK1.1 antibody (> 99% depletion; data not shown), the rejection of Necl-2–expressing cells was dramatically reduced (Figure 6D). In addition, NK cells recovered and purified from the peritoneal cavities of mice that had efficiently rejected Necl-2–EL-4 cells specifically bound Necl-2–hFc (Figure S4), indicating expression of CRTAM. Altogether, these data demonstrate that NK cells play a crucial role in clearing Necl-2–expressing tumors in vivo. Although definitive demonstration of a specific function for CRTAM–Necl-2 interactions in this process will require the use of a CRTAM-blocking antibody, our results are consistent with CRTAM–Necl-2-interactions playing a decisive role in antitumor immunosurveillance.
Discussion
This study demonstrates that activated NK cells and CD8+ T cells recognize the tumor suppressor Necl-2 through the cell surface receptor CRTAM. NK cells express CRTAM upon encountering tumor cells susceptible to lysis and upon engagement of their activating receptors. De novo expression of CRTAM promotes NK cell killing of tumor cells expressing Necl-2. In light of the considerable strength of CRTAM–Necl-2 interaction both under static and flow conditions, the function of CRTAM in this process is most likely to consolidate adhesion of NK cells to target cells, facilitating sustained engagement of activating receptors, such as NKp46, NKp44, and NKp30, with their cognate ligands. This function may be essential when the activating receptors have low affinity for their ligands. Moreover, CRTAM–Necl-2 interactions may directly deliver activating signals that trigger exocytosis of cytolytic granules. Consistent with this, it has been shown that engagement of adhesion molecules is sufficient to trigger NK cell–mediated cytotoxicity.60,61 Incubation of NK cells expressing CRTAM with P815 cells coated with the CRTAM antibody or Necl-2 transfectants did not elicit significant IFN-γ release (data not shown). This result suggests that CRTAM–Necl-2 interaction alone is insufficient to trigger IFN-γ in NK cells and may require cooperation with other activating interactions for cytokine induction.
In vivo tumor challenge with Necl-2+ EL-4 cells. (A-B) EL-4 cells transfected with mouse Necl-2 or EL-4 cells were labeled with 2 different concentrations of CFSE and mixed at a 1:1 ratio. Necl-2 expression was assessed by labeling with CRTAM-hFc, which binds Necl-2 but not CD96, DNAM, Nectin-1, Nectin-2, Necl-1, Necl-4, or Necl-5. (C) The cell mixture was injected intraperitoneally into C57BL/6 mice that had been previously treated with poly-I:C 24 hours earlier. Forty-eight hours after tumor challenge, cells were recovered by peritoneal lavage and examined by flow cytometry. Approximately half of the CFSE low population (EL-4–Necl-2) was rejected. One representative profile of 12 mice is shown. (D) Summary of EL-4–Necl-2/EL-4 ratios in 12 non–NK cell–depleted and 5 NK cell–depleted mice. The slight decrease in fluorescence observed in both CFSE peaks after intraperitoneal injection is due to a fluorescence quenching that occurs within the first 4 hours after labeling all cells tested. Poly-I:C injection did not induce up-regulation of CRTAM on mouse NK cells, as assessed by Necl-2–hFc binding to NK cells obtained by peritoneal lavage (data not shown). However, it significantly accelerated tumor rejection, possibly by inducing recruitment of NK cells to the peritoneal cavity (data not shown). Indeed, rejection of cells injected intraperitoneally occurred as late as 72 of 96 hours after injection in the absence of poly-I:C treatment. Horizontal bars indicate the mean ratio.
In vivo tumor challenge with Necl-2+ EL-4 cells. (A-B) EL-4 cells transfected with mouse Necl-2 or EL-4 cells were labeled with 2 different concentrations of CFSE and mixed at a 1:1 ratio. Necl-2 expression was assessed by labeling with CRTAM-hFc, which binds Necl-2 but not CD96, DNAM, Nectin-1, Nectin-2, Necl-1, Necl-4, or Necl-5. (C) The cell mixture was injected intraperitoneally into C57BL/6 mice that had been previously treated with poly-I:C 24 hours earlier. Forty-eight hours after tumor challenge, cells were recovered by peritoneal lavage and examined by flow cytometry. Approximately half of the CFSE low population (EL-4–Necl-2) was rejected. One representative profile of 12 mice is shown. (D) Summary of EL-4–Necl-2/EL-4 ratios in 12 non–NK cell–depleted and 5 NK cell–depleted mice. The slight decrease in fluorescence observed in both CFSE peaks after intraperitoneal injection is due to a fluorescence quenching that occurs within the first 4 hours after labeling all cells tested. Poly-I:C injection did not induce up-regulation of CRTAM on mouse NK cells, as assessed by Necl-2–hFc binding to NK cells obtained by peritoneal lavage (data not shown). However, it significantly accelerated tumor rejection, possibly by inducing recruitment of NK cells to the peritoneal cavity (data not shown). Indeed, rejection of cells injected intraperitoneally occurred as late as 72 of 96 hours after injection in the absence of poly-I:C treatment. Horizontal bars indicate the mean ratio.
We show that CD8+ T cells up-regulate CRTAM upon TCR triggering, which allows them to effectively adhere to Necl-2+ antigen-presenting cells and target cells, generating a sustained interaction that enhances CD8+ T-cell secretion of IFN-γ. Interestingly, it has been shown that CD8+ T-cell cytotoxicity occurs at a relatively low threshold of activation and does not require establishment of a mature immunologic synapse (IS) with target cells. In contrast, T-cell secretion of IFN-γ requires a high-activation threshold with sustained calcium mobilization and a mature IS.62-64 Thus, CRTAM–Necl-2 interactions may be crucial for the establishment of a mature IS. Moreover, since CRTAM ligand is expressed on B cells and epithelial cells but not on dendritic cells (Figure S3), it may also serve to enhance T-cell stimulation by nonprofessional antigen-presenting cells.
The CRTAM–Necl-2 interaction reported in this study appears to be essential for immune responses against tumors in vivo. Tumor cells expressing Necl-2 were rejected more rapidly than tumor cells lacking Necl-2 by a host response mediated primarily by NK cells. This result parallels previous studies demonstrating that Necl-2 expression is necessary for rejection of the NSCLC A549 cell line in athymic (nu/nu) nude mice.21 It has been proposed that Necl-2 suppresses tumorigenesis by recruiting intracellular proteins that regulate epithelial cell polarization, adhesion, and formation of organized epithelial tissues. This nonimmunologic mechanism certainly contributes to tumor suppression by Necl-2. However, because nude mice do have functional NK cells, A549 cells transfected with Necl-2 may be less tumorigenic because they can also be lysed by NK cells expressing CRTAM. Our data show that Necl-2 is one of the molecular targets that allows the immunosurveillance network to distinguish nascent transformed or established tumor cells from normal cells.
Immunogenicity of tumors depends on multiple mechanisms. Tumor-associated antigens are generated by mutational events that occur during tumorigenesis as well as dysregulated expression of genes that are either inactive in the normal tissue of origin or expressed at relatively low levels.2,4-6 MIC, ULBP, and Rae molecules are up-regulated on target cells upon neoplastic transformation, alerting lymphocytes through NKG2D.7-18 MHC class I molecules that normally inhibit NK cell activation can be down-regulated, signaling lack of “self” to NK cells and releasing them from inhibition.65 By engaging CRTAM, Necl-2 may tip the balance between activating signals (tumor-associated antigens and NKG2D ligands) and inhibitory signals (MHC class I) toward activation, promoting immune responses. Necl-2 may elicit immunogenicity in yet another manner. Since Necl-2 is normally expressed in epithelial cell junctions and mediates homotypic contact or heterotypic interactions with Necl-1 and Nectin 3,28 it may not be readily accessible to the immune system. However, when epithelial cells begin to transform, lose their polarization, invade surrounding tissues, and metastasize, Necl-2 may be unmasked and recognized by activated NK cells and CD8+ T cells expressing CRTAM, thereby promoting immune responses. Whatever the mechanisms, the importance of Necl-2 in immunosurveillance is underscored by the finding that NSCLC and many other epithelial tumors silence Necl-2 by deleting the gene or inhibiting transcription, most likely to escape immune responses.
Our study, together with previous reports,43-45 demonstrates that the multiplicity of Nectins and Necls in epithelial cells is matched by a comparably diverse assortment of receptors and adhesion molecules in the immune system. While CD226 (DNAM-1) and CD96 (tactile) bind Nectin 2 and Necl-5, CRTAM recognizes Necl-2. However, these receptor-ligand pairs may have different functions in tumor immunosurveillance. CRTAM is tightly regulated by lymphocyte activation, and its ligand Necl-2 is silenced in tumor progression. Thus, Necl-2 appears to mediate immunogenicity. In contrast, CD226 and CD96 are constitutively expressed on almost all lymphocytes,38,42 and their ligand, Necl-5, is overexpressed in tumors.46-48 Given this, Necl-5 may mediate immunoevasion. It is possible that NK cell and CD8+ T cells express additional receptors for other Nectins and Necls and that Necl-encoding genes other than TSLC1 are silenced in epithelial tumors. Unlocking these silenced Necl genes may represent an important strategy for tumor immunotherapy.
Prepublished online as Blood First Edition Paper, April 5, 2005; DOI 10.1182/blood-2005-02-0817.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank William Eades and Jackie Hughes (Siteman Cancer Center Core Sorting facility) for excellent cell sorting and Susan Gilfillan for helpful discussion and critical review of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal