Immunoglobulin genotyping of Epstein-Barr virus (EBV)-positive posttransplantation lymphoproliferative disease has suggested that such lesions often arise from atypical post-germinal center B cells, in some cases carrying functionally inactivated immunoglobulin genes. To investigate whether EBV can rescue cells that are failed products of the somatic hypermutation process occurring in germinal centers (GCs), we isolated GC cells from tonsillar cell suspensions and exposed them to EBV in vitro. Screening more than 100 EBV-transformed cell lines of GC origin identified 6 lines lacking surface immunoglobulin, a phenotype never seen among lines derived from circulating naive or memory B cells. Furthermore, 3 of the 6 surface immunoglobulin-negative GC lines carried inactivating mutations in the immunoglobulin H (IgH) variable gene sequence. The ability of EBV to rescue aberrant products of the germinal center reaction in vitro strengthens the probability that a parallel activity contributes to EBV's lymphomagenic potential in vivo.
Introduction
Epstein-Barr virus (EBV) has B-cell growth-transforming ability and is linked to a range of B-cell malignancies.1,2 Of these, posttransplantation lymphoproliferative disease (PTLD) is pathogenetically the least complex. Thus, PTLD tumors arising early after transplantation, when immunosuppression is greatest, resemble EBV-positive in vitro-transformed lymphoblastoid cell lines (LCLs) in cellular phenotype and in expressing the full range of EBV-latent proteins.3,4 Here, viral transformation appears to be sufficient for tumor growth, whereas in certain late-onset PTLD lesions, viral gene expression is more restricted and tumor evolution has involved additional cellular genetic changes.5-7 Recent studies have found that most PTLD tumors carry hypermutated immunoglobulin sequences, many of which are atypical of conventional antigen-selected memory cells.8-10 Indeed, some tumors lacked surface immunoglobulin and had functionally inactivated immunoglobulin sequences,8 highlighting parallels with another EBV-associated B-cell malignancy, Hodgkin lymphoma (HL), in which the tumor cells are again surface immunoglobulin negative and often carry similarly “crippled” immunoglobulin genes.11
These findings suggest that EBV infection can promote the survival of atypical post-germinal center (GC) B cells carrying unfavorable or even inactivating immunoglobulin gene mutations. Such cells, arising as failed products of the somatic hypermutation process that occurs during GC transit, normally die by apoptosis12 ; however, if rescued by EBV, they might form a pool of cells particularly prone to tumor development. Here we ask whether EBV infection of GC B cells in vitro leads to the outgrowth of LCLs with crippled immunoglobulin genes.
Study design
Target cell transformation
Fresh tonsils were obtained with informed consent from 3 adult patients after tonsillectomy. Mononuclear cells were isolated by Ficoll-Isopaque centrifugation and then T-depleted using CD3 Dynabeads (Dynal Biotech, Bromborough, United Kingdom). The resultant B-cell population was stained for the CD10 marker of GC origin13 using phycoerythrin (PE)-Cy5-labeled anti-CD10 (BD Pharmingen, Oxford, United Kingdom) monoclonal antibody (mAb) and CD10+ cells collected by fluorescence-activated cell sorter (FACS) on a Mo-Flo sorter (Dako Cytomation, Ely, United Kingdom). In parallel experiments involving different adult donors, naive, and memory B cells were isolated from peripheral blood B cells by FACS sorting of immunoglobulin D+ (IgD+)CD27- and IgD-CD27+ subsets, respectively, after staining with FITC-labeled anti-IgD (Caltag, Buckingham, United Kingdom) and RPE (R-phycoerythrin)-labeled anti-CD27 (BD Pharmingen) mAbs. Target B cells were exposed to B95.8 EBV for 1 hour at 37°C and then were seeded at limiting dilutions onto wells containing a human fibroblast feeder layer. Cells were maintained in RPMI 1640 medium supplemented with 2 mM glutamine and 10% vol/vol fetal calf serum (FCS) for 3 to 4 weeks, at which time isolated wells showing LCL outgrowth were harvested and expanded. These studies were approved by the University of Birmingham and the South Birmingham Research Ethics Committee, United Kingdom. Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Immunostaining, immunoblotting, and IgH PCR analysis
Surface immunoglobulin, CD10, and CD23 expression was measured by immunofluorescence FACS analysis using a Coulter Epics XL-MCL flow cytometer (Beckman Coulter, Fullerton, CA). EBV latent antigen expression was analyzed by immunoblotting with mAbs to the nuclear antigens (EBNAs) and latent membrane proteins (LMPs), as described.14 EBV genome load was determined as described.15 IgH sequences spanning FR1, CDR1, FR2, CDR2, FR3, and CDR3 were PCR amplified from genomic DNA using primers specific for the FR1 and JH regions and analyzed, as described.8
Results and discussion
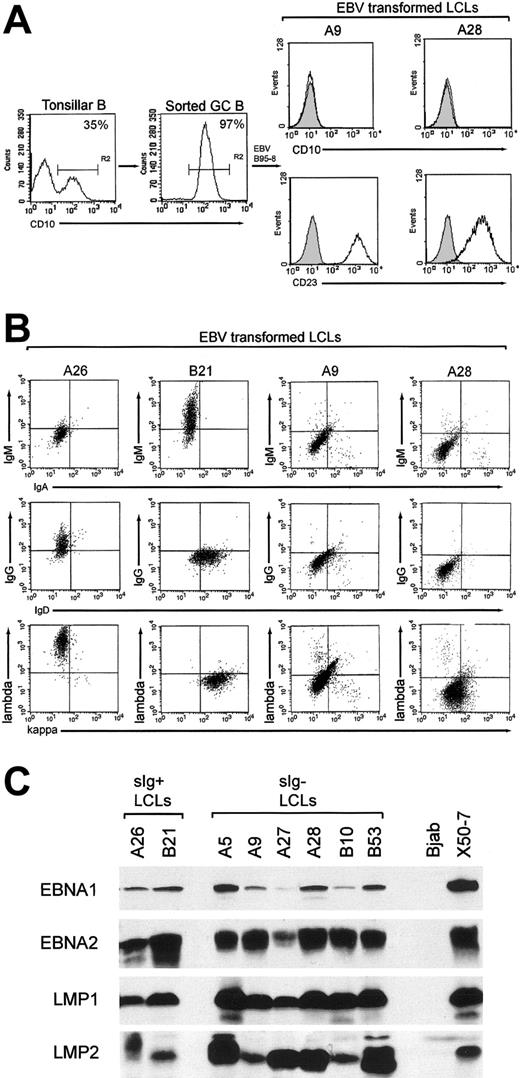
CD10+ GC cells, constituting 10% to 35% tonsillar B-cell preparations, were enriched to purities of 95% to 99% by FACS sorting (Figure 1A, left panels). Using EBV doses infecting approximately 10% target cells15 and seedings of 250 cells/well, we isolated emergent cell lines from 106 GC cell cultures. All GC-derived lines (GC-LCLs) examined had down-regulated the CD10 marker of GC origin and had up-regulated the LCL-associated CD23 activation marker (Figure 1A, right panels). These lines were then screened for surface immunoglobulin expression by staining with heavy- and light-chain immunoglobulin isotype-specific antibodies. We found that 100 of the lines were clearly surface immunoglobulin positive, of which 24 coexpressed IgM and IgD, 57 expressed IgG, and 19 expressed IgA. However, 6 GC-LCLs had little or no surface immunoglobulin staining with any of the heavy or light chain-specific reagents. Figure 1B illustrates the results from 2 such lines, A9 and A28, compared with 2 surface immunoglobulin-positive GC-LCLs, A26 and B21. The appearance of surface immunoglobulin-negative lines in these experiments contrasted with the results of parallel studies in which we had established an almost equal number of LCLs by EBV infection of the naive and memory B-cell subsets from adult peripheral blood samples. All these peripheral blood-derived lines expressed detectable surface immunoglobulin, predominantly IgM/IgD coexpression for naive cell transformants and IgG or IgA expression for memory cell transformants (N.B.P., Gouri Baldwin, A.B., Claire Shannon-Lowe, Regina Feederle, S.C., A.B.R., and Henri-Jacques Deleclose, manuscript submitted August 2005). This strongly suggests that the surface immunoglobulin-negative LCLs are indeed derived from GC cells and not from other B cells contaminating the GC preparations. These lines were then examined for EBV genome load and latent cycle antigen expression. All 6 surface immunoglobulin-negative GC-LCLs carried episomal genome loads in the normal range (data not shown) and expressed the same full spectrum of 6 EBNAs and 2 LMPs seen in conventional surface immunoglobulin-positive transformants; representative immunoblots for 4 key latent proteins—EBNA1, EBNA2, LMP1, and LMP2—are shown in Figure 1C.
We next analyzed the IgH sequences of these 6 surface immunoglobulin-negative GC-LCLs. Three of these lines clearly carried hypermutated nonfunctional IgH genes (Figure 2). Thus, the A5 line, monoclonal by immunoglobulin genotyping, carried a 7-nucleotide insertion resulting in a frameshift of the downstream IgH sequence, whereas the A28 and B53 lines carried mutated immunoglobulin genes containing stop codons in the CDR1 or the CDR2 regions of the IgH sequence. In each case these inactivating mutations appeared to have been introduced into the GC cell's originally functional IgH allele since the downstream VDJ junction sequences carried in-frame productive rearrangements. In the 3 other surface immunoglobulin-negative GC-LCLs, the IgH reading frame was mutated but still intact, and we infer that their IgH genes might have experienced deleterious mutations outside the sequenced region or might have been silenced by other means.16 The frequent identification of inactivating mutations among surface immunoglobulin-negative lines is clearly significant because we did not observe such sequence inactivation in a number of surface immunoglobulin-positive GC lines analyzed in parallel (data not shown) or, as reported in detail elsewhere (N.B.P., Gouri Baldwin, A.B., Claire Shannon-Lowe, Regina Feederle, S.C., A.B.R., and Henri-Jacques Deleclose, manuscript submitted August 2005), in any naive or memory B cell-derived LCLs of peripheral blood origin.
Purification of GC cells and phenotype of GC-derived transformants. (A) Tonsillar B-cell suspensions were stained with PE-Cy5-labeled anti-CD10 mAb, and CD10+ GC cells were positively selected by FACS sorting. Percentages of CD10+ cells in the unsorted and enriched populations are indicated; note that such CD10+ cells lack CD23 expression13 (left panels). The resultant GC-derived LCLs were stained with PE-Cy5-labeled anti-CD10 and FITC-labeled anti-CD23 mAbs; CD10 and CD23 expression (open profiles) are shown relative to isotype control values (shaded profiles) for 2 surface immunoglobulin-negative GC-LCLs, A9 and A28 (right panels); all GC-LCLs examined, irrespective of surface immunoglobulin status, were CD10-CD23+. (B) Immunoglobulin isotype expression in GC-LCLs was analyzed by 2-color flow cytometry after staining with the following combinations of isotype-specific antibodies: RPE-labeled anti-IgM and FITC-labeled anti-IgA, RPE-labeled anti-IgG and FITC-labeled anti-IgD, and RPE-labeled anti-λ and FITC-labeled anti-κ (Dako). The A26 (IgG+, λ+) and B21 (IgM+, IgD+, κ+) GC-LCLs served as positive controls, and A9 and A28 were representative surface immunoglobulin-negative GC-LCLs. Note that A9 and A28 were also negative for surface IgE and for the CD79b component of the surface immunoglobulin complex (data not shown). (C) EBV latent antigen expression in the 6 surface immunoglobulin-negative LCLs (A5, A9, A27, A28, B10, B53) was analyzed by immunoblotting protein extracts of these lines with mAbs to EBNA1, EBNA2, LMP1, and LMP2. Included as controls are 2 surface immunoglobulin-positive GC-LCLs from the same experiment (A26, B21), the EBV-negative B-lymphoma line BJAB and the EBV-transformed cord blood LCL X50-7.
Purification of GC cells and phenotype of GC-derived transformants. (A) Tonsillar B-cell suspensions were stained with PE-Cy5-labeled anti-CD10 mAb, and CD10+ GC cells were positively selected by FACS sorting. Percentages of CD10+ cells in the unsorted and enriched populations are indicated; note that such CD10+ cells lack CD23 expression13 (left panels). The resultant GC-derived LCLs were stained with PE-Cy5-labeled anti-CD10 and FITC-labeled anti-CD23 mAbs; CD10 and CD23 expression (open profiles) are shown relative to isotype control values (shaded profiles) for 2 surface immunoglobulin-negative GC-LCLs, A9 and A28 (right panels); all GC-LCLs examined, irrespective of surface immunoglobulin status, were CD10-CD23+. (B) Immunoglobulin isotype expression in GC-LCLs was analyzed by 2-color flow cytometry after staining with the following combinations of isotype-specific antibodies: RPE-labeled anti-IgM and FITC-labeled anti-IgA, RPE-labeled anti-IgG and FITC-labeled anti-IgD, and RPE-labeled anti-λ and FITC-labeled anti-κ (Dako). The A26 (IgG+, λ+) and B21 (IgM+, IgD+, κ+) GC-LCLs served as positive controls, and A9 and A28 were representative surface immunoglobulin-negative GC-LCLs. Note that A9 and A28 were also negative for surface IgE and for the CD79b component of the surface immunoglobulin complex (data not shown). (C) EBV latent antigen expression in the 6 surface immunoglobulin-negative LCLs (A5, A9, A27, A28, B10, B53) was analyzed by immunoblotting protein extracts of these lines with mAbs to EBNA1, EBNA2, LMP1, and LMP2. Included as controls are 2 surface immunoglobulin-positive GC-LCLs from the same experiment (A26, B21), the EBV-negative B-lymphoma line BJAB and the EBV-transformed cord blood LCL X50-7.
Studies of EBV's interaction with GC cells were first prompted by the realization that Burkitt lymphoma (BL), an EBV-associated tumor with highly restricted latent antigen expression, displays a GC phenotype.17 However, as confirmed here, viral transformation of GC cell preparations in vitro produced lines with a typical LCL phenotype expressing all the latent antigens18 ; indeed expression of the full spectrum of latent genes appears to be incompatible with retention of the GC phenotype.19 Interest in viral infection of GC cells was reawakened by the observation that EBV-positive PTLD tumors, many of which had full latent antigen expression, often arise from B cells with atypical, even nonfunctional, IgH alleles.8-10 The present work provides strong evidence that EBV infection in vitro can indeed rescue the failed products of GC reactions, leading to the outgrowth of surface immunoglobulin-negative LCLs with functionally inactivated IgH alleles; we recently learned of similar findings in 2 other laboratories.20,21 Such activity likely reflects not just the well-documented growth-transforming function of EBV but also the ability of certain EBV latent proteins to protect B cells from apoptosis. Thus, prosurvival functions have now been demonstrated for EBNA3 proteins,22 for LMP1 through mimicry of CD40 signaling,23 and for LMP2 through mimicry of surface immunoglobulin engagement.24 The present work, and the recent detection of EBV in a population of CD19+ surface immunoglobulin-negative cells in the blood of patients after transplantation,25 strengthen the view that virus-mediated rescue of aberrant GC products contributes directly to the pathogenesis of PTLD. The relevance of these findings to the pathogenesis of HL, an EBV-associated tumor similarly derived from atypical post-GC B cells often carrying crippled immunoglobulin genes,11 is less clear. Both the cellular phenotype of HL, with suppression of many B-lineage genes,26 and the viral phenotype with antigen expression limited to EBNA1, LMP1, and LMP2 in the absence of the other EBNAs,1 are different from that of the GC-derived surface immunoglobulin-negative LCLs. It is nevertheless possible that EBV-positive HL and often PTLD arise by different pathways from the same pool of atypical post-GC survivors.
IgH sequence analysis of 3 GC-LCLs lacking surface immunoglobulin expression.IgH sequences (corresponding to codons 9-94) of the surface immunoglobulin-negative GC-LCLs A5, A28, and B53 aligned to the corresponding most homologous germline sequence. The IgH sequence of the A5 cell line contains a 7-nt insertion (shaded) downstream of CDR2; A28 sequence has stop codons (boxed) introduced within CDR1 and CDR2, and B53 sequence has stop codon (boxed) introduced in CDR1.
IgH sequence analysis of 3 GC-LCLs lacking surface immunoglobulin expression.IgH sequences (corresponding to codons 9-94) of the surface immunoglobulin-negative GC-LCLs A5, A28, and B53 aligned to the corresponding most homologous germline sequence. The IgH sequence of the A5 cell line contains a 7-nt insertion (shaded) downstream of CDR2; A28 sequence has stop codons (boxed) introduced within CDR1 and CDR2, and B53 sequence has stop codon (boxed) introduced in CDR1.
Prepublished online as Blood First Edition Paper, August 25, 2005; DOI 10.1182/blood-2005-06-2327.
Supported by a Leukaemia Research Fund Clinical Research Fellowship (S.C.) and by Cancer Research UK.
S.C., A.I.B., N.B.P., A.E.M., and M.D. performed the experiments. J.G. and A.B.R. contributed to the experimental design. S.C., A.I.B., M.D., and A.B.R. analyzed the data. S.C., A.I.B., and A.B.R. wrote the manuscript.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Debbie Croom-Carter for help with immunoblotting and David Lloyd for help with FACS analysis.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal