MALT1, BCL10 (B-cell lymphoma 10), and API2 (apoptosis inhibitor 2)-MALT1 are key molecules in mucosa-associated lymphoid tissue (MALT) lymphomagenesis. We previously reported that MALT1 and API2-MALT1 were localized only in cytoplasm, where we suggested that both molecules were likely to be active. In the study presented here, we further examined the localization-determining region by generating various mutants and were able to demonstrate that there were nuclear export signal (NES)-containing domains in the MALT1 C-terminal region. The use of leptomycin B, an NES-specific inhibitor, demonstrated that both MALT1 and API2-MALT1 were predominantly retained in the nuclei, indicating that these molecules were shuttling between nucleus and cytoplasm in an NES-dependent manner. It was also found that MALT1 was involved in the nuclear export of BCL10, which is originally localized in both nucleus and cytoplasm. These results correlate well with the nuclear BCL10 expression pattern in both t(1;14) and t(11;18) MALT lymphomas. The nucleocytoplasmic shuttling of MALT1 and BCL10 complex may indicate that these molecules are involved not only in the nuclear factor κB (NF-κB) pathway but also in other biologic functions in lymphocytes.
Introduction
Two characteristic chromosome aberrations, t(11;18)(q21;q21) and t(1;14)(p22;q32), have been implicated in the pathogenesis of malignant lymphoma of mucosa-associated lymphoid tissue (MALT). t(11;18)(q21;q21) results in the fusion of 2 genes, the API2 gene at 11q21 and the MALT1 gene at 18q21.1-6 In t(1;14)(p22; q32), the entire BCL10 gene at 1p22 is juxtaposed to the Ig-heavy-chain-gene enhancer at 14q32, resulting in its overexpression.7,8
Given that these seemingly unrelated translocations lead to the same clinicopathologic entity—that is, MALT lymphoma—it has been suspected that they share a common oncogenic activity. Recently, 2 common features among these chromosomal translocations have been identified.9-15 The first one is that BCL10 (B-cell lymphoma 10) and MALT1 both activate the antigen-receptor-mediated nuclear factor κB (NF-κB) signaling pathway in cytoplasm. MALT1 possesses one caspaselike domain (CLD) in its C-terminal region, and one death domain (DD) and 2 immunoglobulin-like domains (Ig-like) in its N-terminal region.16 The Ig-like domain of MALT1 reportedly binds to BCL10,10 and such complexes liberate the cytoplasmic sequestration of NF-κB, resulting in rapid translocation of NF-κB into the nucleus. In t(1;14) MALT lymphoma, overexpressed BCL10 is thought to accelerate the level of NF-κB activation.10 In t(11;18) MALT lymphoma, API2 (apoptosis inhibitor 2)-MALT1 is very stable and expressed in cytoplasm,17 the function of which is to activate NF-κB without the aid of BCL10 and to provide resistance to apoptosis.10,18,19
The second common feature of t(11;18) and t(1;14) MALT lymphomas is nuclear localization of BCL10. In marginal zone B cells, thought to be the normal counterpart of MALT lymphoma, BCL10 is immunostained primarily in the cytoplasm.20 However, BCL10 is strongly immunostained in the nuclei in t(1;14) MALT lymphoma.20 Although the BCL10 gene is not involved in t(11;18), moderate nuclear immunostaining of BCL10 has been detected with high frequency in t(11;18) MALT lymphomas.21-23 It has also been reported that nuclear immunostaining of BCL10 occurs in 20% to 50% of MALT lymphomas without t(11;18) and t(1;14).21-23 These observations suggested that BCL10 displayed its oncogenic activity in the nuclei.20,24 It was also shown that nuclear localization of BCL10 was associated with advanced MALT lymphoma.21 The immunohistochemistry of BCL10 for MALT lymphoma patients is therefore clinically very important, but the mechanism of the subcellular localization of BCL10 has remained unknown.
We previously demonstrated that MALT1 and API2-MALT1 were localized in cytoplasm and that the C-terminal region of MALT1 was responsible for their cytoplasmic localization.17 In the study reported here, we investigated this C-terminal region of MALT1 in detail to understand the mechanism of subcellular localization. We found that MALT1 and API2-MALT1 were shuttling between nucleus and cytoplasm. Furthermore, our investigation of the mechanism of BCL10 subcellular localization disclosed that MALT1 played a role in its localization.
Materials and methods
Plasmid constructions and site-directed mutagenesis
The expression vectors pcDNA3-MALT1-flag, pcDNA3-flag-MALT1-CLD(328-813), and pcDNA3-flag-API2-MALT1 were described previously.17 To generate pcDNA3-myc-BCL10, the fragments were amplified by reverse-transcription-polymerase chain reaction (RT-PCR) from the RNA of human peripheral mononuclear cells using sense primer 5′-CTAGCTAGCGAGCCCACCGCACCGTCCCTC-3′ and antisense primer 5′-TACGGGCCCTTATTGTCGTGAAACAGTACG-3′. The amplified fragments were digested with NheI/ApaI and cloned into the XbaI/ApaI sites of pcDNA3-Myc vectors. To generate all the MALT1 deletion mutants, the fragments were amplified by PCR from the pcDNA3-MALT1-flag vector using KOD-Plus DNA polymerase (Toyobo, Osaka, Japan) according to the manufacturer's instructions. The following were the sense and antisense primers of each of the constructs: pcDNA3-flag-MALT1(1-562): sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-GGGGGGCCCTCAAGATTCAGCAGAATATTCTGT-3′; pcDNA3-flag-MALT1(1-460): sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-AAAGGGCCCTCAATCGTAGTCATTTCTTTTCCTACACAT-3′; pcDNA3-flag-MALT1(1-420): sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-AAAGGGCCCTCATGGAGCATCAACGGGGACCATGAAGCT-3′; pcDNA3-flag-MALT1(1-380): sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-AAAGGGCCCTCACATCTCATATTCAGTAAGATCCAACAG-3′; pcDNA3-flag-MAL-T1(1-328): sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-AAAGGGCCCTCACAAAGGCTGGTCAGTTGTTTGCTCTTT-3′. The amplified fragments for the MALT1 deletion mutants were digested with NheI/ApaI and cloned into the XbaI/ApaI sites of pcDNA3-flag vectors. To generate the green fluorescent protein (GFP) fusion MALT1 deletion mutants, the fragments were amplified by PCR from the pcDNA3-MALT1-flag vector using KOD-Plus DNA polymerase. The following were the sense and antisense primers of each of the constructs: pEGFP-C1-NES1: sense primer 5′-CCGCTCGAGCTGCGAAGGACAAGGTTGCCC-3′, antisense primer 5′-CTCGGATCCTCACATCTCATATTCAGTAAGATCC-3′; pEGFP-C1-NES2: sense primer 5′-CCGCTCGAGCTGATACCATTCCAATCTTGG-3′, antisense primer 5′-CTCGGATCCTCAAGATTCAGCAGAATATTCTGTTCCC-3′; pEGFP-C1-NES1 + NES1R: sense primer 5′-CCGCTCGAGCTGAGAAGGACAAGGTTGCCC-3′, antisense primer 5′-CGCGGATCCTCATGGAGCATCAACGGG-3′; pEGFP-C1-NES1R: sense primer 5′-CCGCTCGAGCTCGTAATGCTGTGGATGAG-3′, antisense primer 5′-CGCGGATCCTCATGGAGCATCAACGGG-3′. The amplified fragments for the GFP fusion MALT1 deletion mutants were digested with XhoI/BamHI and cloned into the XhoI/BamHI sites of pEGFP-C1 vectors. For site-directed mutagenesis, we first amplified 2 fragments by PCR with mutated primers from the pcDNA3-MALT1-flag vector using KOD-Plus DNA polymerase. The following were the sense and antisense primers of each of the constructs: pcDNA3-flag-MALT1(1-380)-NES1 mutant: one fragment, sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-AAGTCCGCCTGTCTCGCTAAGTTAG-3′, and the other fragment, sense primer 5′-CTAACTTAGCGAGACAGGCGGACTT-3′, antisense primer 5′-AAAGGGCCCTCACATCTCATATTCAGTAAGATCCAACAG-3′; pcDNA3-flag-MALT1(1-562)-NES2 mutant: one fragment, sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-ACTTTTGCTGCATCCGCGATTGGAA-3′, and the other fragment, sense primer 5′-TTCCAATCGCGGATGCAGCAAAAGT-3′, antisense primer 5′-GGGGGGCCCTCAAGATTCAGCAGAATATTCTGT-3′. The 2 amplified fragments were mixed and diluted to 1:1000 and then amplified again by PCR using KOD-Plus DNA polymerase. The following were the sense and antisense primers of each of the constructs: pcDNA3-flag-MALT1(1-380)-NES1 mutant: sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-AAAGGGCCCTCACATCTCATATTCAGTAAGATCCAACAG-3′; pcDNA3-flag-MAL-T1(1-562)-NES2 mutant: sense primer 5′-CTAGCTAGCTCGCTGTTGGGGGACCCGCTACAG-3′, antisense primer 5′-GGGGGGCCCTCAAGATTCAGCAGAATATTCTGT-3′. The amplified fragments for the MALT1 site-directed mutants were digested with NheI/ApaI and cloned into the XbaI/ApaI sites of pcDNA3-flag vectors. To generate the pMALT1-flag-SV40NLS vector, the fragments were amplified by PCR from the pcDNA3-MALT1-flag vector using KOD-Plus DNA polymerase. The sense and antisense primers used were 5′-CTAGCTAGCCCGAGGCCCGTGACGGGG-3′ and 5′-GGAAGATCTAGACTTGTCATCGTCGTC-3′, respectively. The amplified fragments were digested with NheI/BglII and cloned into the NheI/BglII sites of pEYFP-NUC vectors (BD Bioscience Clontech, San Jose, CA). The competent cells transformed with all the constructs were cultured at 30°C. The sequences of all the constructs were confirmed with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA). pBIIx-Luc vectors were kindly provided by Dr T. Koseki of Tohoku University and Dr N. Inohara of the University of Michigan Medical School.25 phRL-SV40 vectors were purchased from Promega (Madison, WI).
Cell culture and transfection
COS7 cells were grown in Iscoves medium supplemented with 5% fatal calf serum (FCS) and maintained in a 5% CO2 incubator at 37°C. For immunofluorescence staining studies and NF-κB reporter assays, COS7 cells were transiently transfected with Effectene (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
Immunofluorescence and intrinsic fluorescence microscopy
COS7 cells were grown on circular slide glasses (12 mm in diameter; IWAKI, Tokyo, Japan) in a 24-well dish (Corning, Corning, NY), and transfection was carried out with the Effectene method (QIAGEN). At 2.5 hours before fixation, 25 μM MG132 was added only to cells transfected with MALT1 and MALT1 deletion mutants. For nuclear export signal (NES) experiments, 4 ng/mL leptomycin B (LMB; Biomol, Plymouth Meeting, PA) was added to cells transfected with MALT1, API2-MALT1, BCL10, GFP, and MALT1 deletion mutants for 6 hours before fixation. The cells transfected with GFP fusion MALT1 deletion mutants were treated with 4 ng/mL LMB for 18 hours before fixation. At 24 hours after transfection, the cells were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 minutes at room temperature, and after washing with PBS, they were permealized in 100% methyl alcohol for 8 minutes at -20°C. For studying subcellular localization of endogenous MALT1, COS7 cells were grown, treated with 4 ng/mL LMB for 6 hours, fixed, and permealized as described above. After PBS washing, the first antibodies were applied for 1.5 hours at room temperature. Anti-flag M2 monoclonal antibody (Sigma, St Louis, MO) at 4.9 μg/mL in PBS, anti-BCL10 polyclonal antibody (Sigma) at 0.5 μg/mL in PBS, and anti-MALT1 polyclonal antibody (kindly provided by Dr Zhijian J. Chen of University of Texas Southwestern Medical Center)15 at 1.0 μg/mL in PBS were used to visualize flag-tag constructs, BCL10, and MALT1. The cells were then washed 3 times with PBS and incubated with the second antibodies for 1.5 hours at room temperature. Fluorescein goat anti-mouse IgG (Molecular Probes, Leiden, the Netherlands), diluted to 1:100 in PBS, and Alexa Fluor 546 goat anti-rabbit IgG (Molecular Probes), diluted to 1:600 in PBS, were used. The specimens, except for those incubated with Alexa Fluor 546 goat anti-rabbit IgG, were then incubated in 0.1 μg/mL propidium iodide in PBS for 5 minutes, washed 3 times with PBS, and then mounted in PermaFluor Aqueous Mounting Medium (Immunon, Pittsburgh, PA). Finally, the specimens were examined by confocal fluorescence microscopy (GB200X-SP; Olympus, Tokyo, Japan) using a 60× oil/1.10 NA objective (Olympus) and a laser scanning system (Radiance 2100; BIO-RAD Japan, Tokyo, Japan). Images were processed using Photoshop (Adobe Systems, San Jose, CA). The percentage of positive cells was calculated by counting 1000 cells. The relative proportion of cells with specific subcellular localization patterns of positive cells was calculated by counting 50 to 200 cells.
NF-κB reporter assay
COS7 cells (1.0 × 105) were grown in a 6-well dish (Corning) and transfected with the Effectene method. pBIIx-Luc (0.05 μg), phRL-SV40 (0.02 μg), and the following expression vectors were cotransfected: control, 0.4 μg pcDNA3-myc; MALT1, 0.4 μg pcDNA3-MALT1-flag; MALT1-SV40NLS, 0.4 μg pMALT1-flag-SV40NLS; BCL10, a mixture of 0.04 μg pcDNA3-myc-BCL10 and 0.36 μg pcDNA3-myc; MALT1 and BCL10, a mixture of 0.36 μg pcDNA3-MALT1-flag and 0.04 μg pcDNA3-myc-BCL10; MALT1-SV40NLS and BCL10, a mixture of 0.36 μg pMALT1-flag-SV40NLS and 0.04 μg pcDNA3-myc-BCL10. At 24 hours after transfection, the cells were harvested with the method of the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured with the Luminescence Reader (Berthold, Bad Wildbad, Germany).
Western blot analysis
Western blot analysis was carried out essentially as previously described.17 The transferred polyvinylidenefluoride (PVDF) membranes were incubated for 1 hour with 4.9 μg/mL anti-flag M2 monoclonal antibody (Sigma) or 3.3 μg/mL anti-myc monoclonal antibody (9E10; Invitrogen, Tokyo, Japan) in blocking buffer at room temperature. They were then washed extensively in PBS containing 0.05% Tween 20 T-PBS and incubated for 1 hour with horseradish peroxidase-conjugated anti-mouse IgG (Amersham, Arlington Heights, IL) diluted at 1:1000 in blocking buffer without 0.05% NaN3. Antibody binding was visualized with an enhanced chemiluminescence (ECL) detection kit (Amersham).
Results
Subcellular localization of MALT1, API2-MALT1, and BCL10
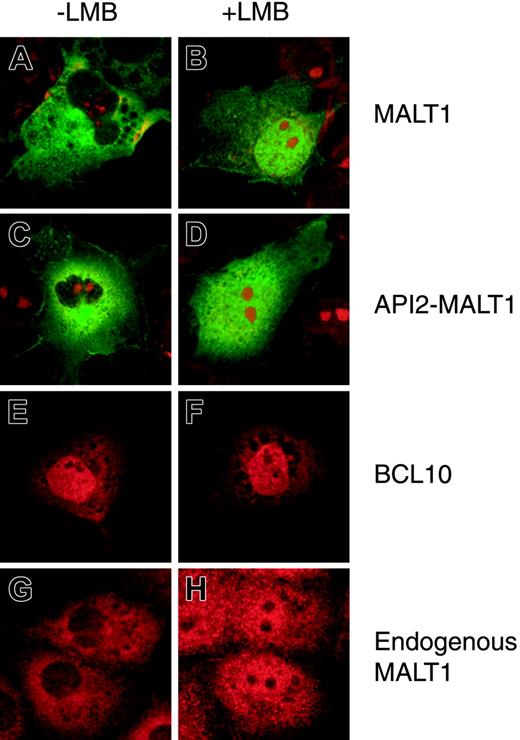
Investigation of the subcellular localization of MALT1 and API2-MALT1 in COS7 cells by means of immunofluorescence analysis disclosed that these 2 molecules were localized only in the cytoplasm of transfected cells (Figure 1A,C). To examine the mechanism of subcellular localization, the NES-specific inhibitor LMB was used.26,27 MALT1 and API2-MALT1 accumulated in the nuclei after treatment with 4 ng/mL LMB for 6 hours, indicating that MALT1 and API2-MALT1 contained a functional NES (Figure 1B,D). BCL10 was localized in both nuclei and cytoplasm (Figure 1E). Treatment with LMB did not change the subcellular localization, indicating that BCL10 did not contain any functional NES (Figure 1F). With another cell line, 293T cells, we were able to obtain similar results (data not shown).
Since COS7 cells were found to express endogenous MALT1 when examined with affinity-purified polyclonal anti-MALT1 antibody, endogenous MALT1 localization was examined by means of immunofluorescence analysis. As expected from findings in the transient expression system with anti-flag antibody (Figure 1A-B), endogenous MALT1 accumulated in the nuclei when treated with LMB (Figure 1G-H).
Identification of functional NES in MALT1
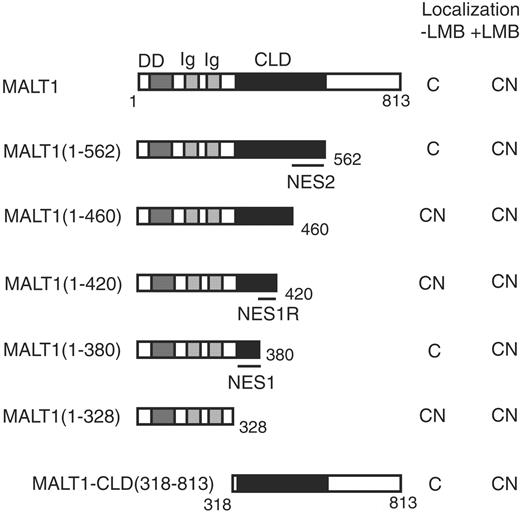
Several MALT1 deletion mutants were constructed to identify a region in MALT1 that might contain an NES (Figure 2). MALT1-CLD(318-813) was localized in cytoplasm as previously described17 and treatment with 4 ng/mL LMB for 6 hours led to nuclear retention of MALT1-CLD(318-813), indicating that this region contained an NES responsible for the cytoplasmic localization of MALT1 (Figure 2, MALT1-CLD(318-813)). MALT1(1-328) was localized in nuclei but MALT1(1-380) was not, while treatment with LMB led to nuclear retention of MALT1(1-380), indicating that the NES1 region (329-380AA) contained an NES (Figure 2, MALT1(1-328) and MALT1(1-380)). MALT1(1-380) was localized only in cytoplasm, but MALT1(1-420) was localized in nuclei and cytoplasm (Figure 2, MALT1(1-380) and MALT1(1-420)). Further investigation disclosed that the region of MALT1(381-420AA) contained a regulatory region for the NES1 region (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). MALT1(1-460) was localized in nuclei, but MALT1(1-562) was not, indicating that the NES2 region (461-562AA) contained another NES (Figure 2, MALT1(1-460) and MALT1(1-562)).
Subcellular localization of MALT1, API2-MALT1, and BCL10 with or without LMB treatment. Subcellular localization of MALT1-flag, flag-API2-MALT1, and myc-BCL10 transiently expressed in COS7 cells (A-F): 2 × 104 COS7 cells were transfected with the following expression vectors: MALT1-flag, 0.2 μg pcDNA3-MALT1-flag without (A) or with 4 ng/mL LMB (B); flag-API2-MALT1, 0.2 μg pcDNA3-flag-API2-MALT1 without (C) or with 4 ng/mL LMB (D); myc-BCL10, a mixture of 0.02 μg pcDNA3-myc-BCL10 and 0.18 μg pcDNA3 without (E) or with 4 ng/mL LMB (F). The LMB treatment was conducted at 18 hours after transfection. At 21.5 hours after transfection, 25 μM MG132 was added only to the wells for MALT1-flag (A-B), and 24 hours after transfection, the cells were examined with an immunofluorescence microscope. Mouse anti-flag antibody was used for flag-tagged constructs; fluorescein goat anti-mouse IgG, as the second antibody for the mouse anti-flag antibody; rabbit anti-human BCL10 antibody, for myc-BCL10; and Alexa Fluor 546 goat anti-rabbit IgG, as the second antibody for the rabbit anti-BCL10 antibody. Nuclei were stained with propidium iodide except for the cells expressing myc-BCL10. Subcellular localization of endogenous MALT1 in COS7 cells (G-H): COS7 cells treated without (G) or with 4 ng/mL LMB (H) for 6 hours were examined. Rabbit anti-MALT1 polyclonal antibody was used as the first antibody and Alexa Fluor 546 goat anti-rabbit IgG as the second antibody for the rabbit anti-MALT1 polyclonal antibody.
Subcellular localization of MALT1, API2-MALT1, and BCL10 with or without LMB treatment. Subcellular localization of MALT1-flag, flag-API2-MALT1, and myc-BCL10 transiently expressed in COS7 cells (A-F): 2 × 104 COS7 cells were transfected with the following expression vectors: MALT1-flag, 0.2 μg pcDNA3-MALT1-flag without (A) or with 4 ng/mL LMB (B); flag-API2-MALT1, 0.2 μg pcDNA3-flag-API2-MALT1 without (C) or with 4 ng/mL LMB (D); myc-BCL10, a mixture of 0.02 μg pcDNA3-myc-BCL10 and 0.18 μg pcDNA3 without (E) or with 4 ng/mL LMB (F). The LMB treatment was conducted at 18 hours after transfection. At 21.5 hours after transfection, 25 μM MG132 was added only to the wells for MALT1-flag (A-B), and 24 hours after transfection, the cells were examined with an immunofluorescence microscope. Mouse anti-flag antibody was used for flag-tagged constructs; fluorescein goat anti-mouse IgG, as the second antibody for the mouse anti-flag antibody; rabbit anti-human BCL10 antibody, for myc-BCL10; and Alexa Fluor 546 goat anti-rabbit IgG, as the second antibody for the rabbit anti-BCL10 antibody. Nuclei were stained with propidium iodide except for the cells expressing myc-BCL10. Subcellular localization of endogenous MALT1 in COS7 cells (G-H): COS7 cells treated without (G) or with 4 ng/mL LMB (H) for 6 hours were examined. Rabbit anti-MALT1 polyclonal antibody was used as the first antibody and Alexa Fluor 546 goat anti-rabbit IgG as the second antibody for the rabbit anti-MALT1 polyclonal antibody.
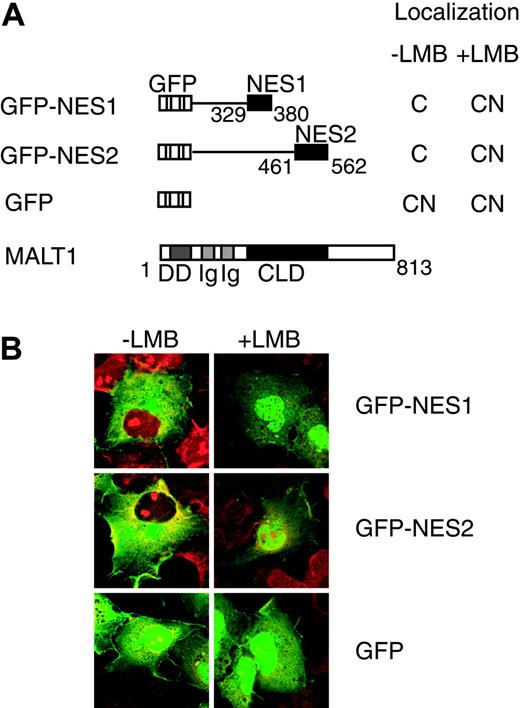
To determine whether the NES1 and NES2 regions contained functional NESs, each region was fused to GFP (Figure 3A). GFP-NES1 and GFP-NES2 were localized exclusively in cytoplasm, whereas a control protein, GFP, was localized in both nuclei and cytoplasm (Figure 3B). Treatment with 4 ng/mL LMB for 18 hours led to nuclear retention of GFP-NES1 and GFP-NES2, but did not change subcellular localization of GFP alone. These results showed that both NES1 and NES2 regions contained functional NESs. We noticed a short, hydrophobic, leucine-rich motif resembling an NES in the NES1 region (LTNLLRQL (358-365AA)).28-32 Sequence alignment showed that the leucine-rich motif was well conserved among species (Figure 4A). To determine whether this motif is critical for MALT1 nuclear export, we constructed the MALT1(1-380)-NES1 mutant, which contains mutations of the 2 leucine residues (Leu362 and Leu365) into alanines. The MALT1(1-380)-NES1 mutant was localized in both nuclei and cytoplasm, while the wild-type protein MALT1(1-380) was localized exclusively in cytoplasm, indicating that these leucines were critical for the function of NES1 (Figure 4B). We also noticed an NES-like leucine-rich motif in the NES2 region (IPILDAL (463-469AA)). We then constructed the MALT1(1-562)-NES2 mutant, which contains mutations of the 2 leucine residues (Leu466 and Leu469) into alanines. However this mutant did not change its cytoplasmic localization and showed a similar pattern to that of the wild type (data not shown). Furthermore, 3 other NES-like motifs were recognized in the NES2 region. Two hydrophobic residues in each motif were changed into alanines (F500A and L504A; I539A and L543A; L549A and I553A), but none of these mutants changed its cytoplasmic localization, suggesting that these leucines within the motifs were not involved in subcellular localization (data not shown).
MALT1 deletion mutant structures and their subcellular localization with or without LMB treatment. Protein structures of various deletion mutants are shown. Numbers in the parentheses and below the structures indicate the amino acid number of MALT1. NES1 and NES2 represent the NES1 and NES2 regions (329-380AA and 461-562AA), respectively, while NES1R indicates an NES1 regulatory region (381-420AA). These regions are shown by bars below the constructs. For the nuclear localization study, 2 × 104 COS7 cells were transfected with 0.2 μg of each of the expression vectors. Treatment with 4 ng/mL LMB was conducted 18 hours after transfection, and 21.5 hours after transfection, 25 μM MG132 was added to all of the wells for the visualization of unstable proteins. At 24 hours after transfection, the cells were examined with an immunofluorescence microscope (see Figure 1). Subcellular localization is shown on the right of each structure. C indicates cytoplasm; CN, both cytoplasm and nuclei.
MALT1 deletion mutant structures and their subcellular localization with or without LMB treatment. Protein structures of various deletion mutants are shown. Numbers in the parentheses and below the structures indicate the amino acid number of MALT1. NES1 and NES2 represent the NES1 and NES2 regions (329-380AA and 461-562AA), respectively, while NES1R indicates an NES1 regulatory region (381-420AA). These regions are shown by bars below the constructs. For the nuclear localization study, 2 × 104 COS7 cells were transfected with 0.2 μg of each of the expression vectors. Treatment with 4 ng/mL LMB was conducted 18 hours after transfection, and 21.5 hours after transfection, 25 μM MG132 was added to all of the wells for the visualization of unstable proteins. At 24 hours after transfection, the cells were examined with an immunofluorescence microscope (see Figure 1). Subcellular localization is shown on the right of each structure. C indicates cytoplasm; CN, both cytoplasm and nuclei.
Subcellular localization of GFP fusion proteins containing NES regions with or without LMB treatment. (A) Protein structures of various GFP fusion MALT1 deletion mutants; GFP and full-length MALT1 expression vector are shown. NES1 and NES2 indicate the NES1 and NES2 regions, respectively. Numbers below the structures indicate the amino acid number of MALT1, and the subcellular localization pattern is shown on the right of each structure. C indicates cytoplasm; CN, both cytoplasm and nuclei. (B) COS7 cells (2 × 104) were transfected with 0.2 μg of each of the expression vectors. The cells were treated with 4 ng/mL LMB for 18 hours before examination with an immunofluorescence microscope. Nuclei were stained with propidium iodide.
Subcellular localization of GFP fusion proteins containing NES regions with or without LMB treatment. (A) Protein structures of various GFP fusion MALT1 deletion mutants; GFP and full-length MALT1 expression vector are shown. NES1 and NES2 indicate the NES1 and NES2 regions, respectively. Numbers below the structures indicate the amino acid number of MALT1, and the subcellular localization pattern is shown on the right of each structure. C indicates cytoplasm; CN, both cytoplasm and nuclei. (B) COS7 cells (2 × 104) were transfected with 0.2 μg of each of the expression vectors. The cells were treated with 4 ng/mL LMB for 18 hours before examination with an immunofluorescence microscope. Nuclei were stained with propidium iodide.
NES1 region of MALT1 and its role in subcellular localization. (A) Amino acid sequence of the NES1 region in alignment with MALT1 homologs of various species. The amino acid numbers are based on amino acid sequences from accession numbers BAA83099 (Homo sapiens), AAG38590 (Danio rerio), AAG38591 (Caenorhabditis elegans), and AAG38592 (Dictyostelium discoideum) in GenBank. Residues that resemble the consensus sequence of leucine-rich NES are boxed. Alanine substitutions were made at Leu362 and Leu365 for studying its role in subcellular localization (MALT1[1-380]-NES1 mutant). NES1 indicates the NES1 region. (B) Cells transfected with either MALT1(1-380)-NES1 mutant or wild type are shown. The cells were processed in the same manner as those in Figure 2. Relative proportions of cells with specific subcellular localization patterns are shown below each panel. NES1 mutant represents MALT1(1-380)-NES1 mutant and wild-type MALT1(1-380). C indicates cytoplasm; CN, both cytoplasm and nuclei.
NES1 region of MALT1 and its role in subcellular localization. (A) Amino acid sequence of the NES1 region in alignment with MALT1 homologs of various species. The amino acid numbers are based on amino acid sequences from accession numbers BAA83099 (Homo sapiens), AAG38590 (Danio rerio), AAG38591 (Caenorhabditis elegans), and AAG38592 (Dictyostelium discoideum) in GenBank. Residues that resemble the consensus sequence of leucine-rich NES are boxed. Alanine substitutions were made at Leu362 and Leu365 for studying its role in subcellular localization (MALT1[1-380]-NES1 mutant). NES1 indicates the NES1 region. (B) Cells transfected with either MALT1(1-380)-NES1 mutant or wild type are shown. The cells were processed in the same manner as those in Figure 2. Relative proportions of cells with specific subcellular localization patterns are shown below each panel. NES1 mutant represents MALT1(1-380)-NES1 mutant and wild-type MALT1(1-380). C indicates cytoplasm; CN, both cytoplasm and nuclei.
Nuclear export of BCL10
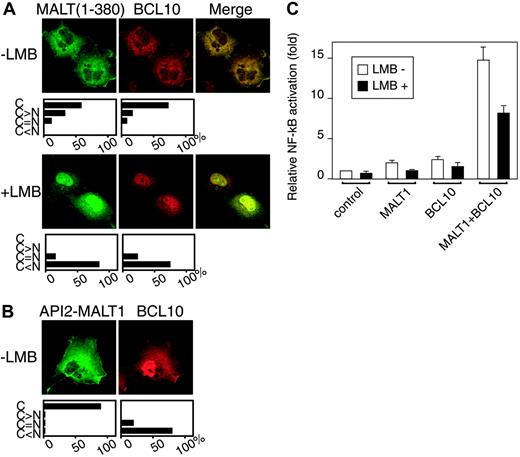
Since BCL10 is known to bind to the Ig-like domain of MALT1,10 we investigated BCL10 subcellular localization in the presence of MALT1. MALT1(1-380) was used instead of full-length MALT1 because the former is more stable (data not shown). In the presence of MALT1(1-380), BCL10 changed its nucleocytoplasmic localization (Figure 1E) to predominantly cytoplasmic, while MALT1(1-380) did not change its cytoplasmic localization in the presence of BCL10 (Figure 5A, upper panel). Treatment with 4 ng/mL LMB for 6 hours led to nuclear retention of not only MALT1(1-380) but also of BCL10, suggesting that MALT1(1-380) had exported BCL10 from nucleus to cytoplasm (Figure 5A, lower panel). A similar result was obtained with BCL10 and full-length MALT1, although the percentage of BCL10 and full-length MALT1 double-positive cells was low (< 0.1%) because of the unstable nature of MALT117 (data not shown). In COS7 cells cotransfected with BCL10 and API2-MALT1, the nucleocytoplasmic localization of BCL10 did not change in the presence of API2-MALT1, indicating that the API2-MALT1 did not export BCL10 from nucleus to cytoplasm (Figure 5B). As expected, the cytoplasmic localization of API2-MALT1 did not change (Figure 5B).
Subcellular localization of BCL10 cotransfected with MALT1(1-380) and API2-MALT1. COS7 cells (2 × 104) were transfected with the following expression vectors: (A) myc-BCL10 and flag-MALT1(1-380), a mixture of 0.02 μg pcDNA3-myc-BCL10 and 0.18 μg pcDNA3-flag-MALT1(1-380) with or without 4 ng/mL LMB; (B) myc-BCL10 and flag-API2-MALT1, a mixture of 0.02 μg pcDNA3-myc-BCL10 and 0.18 μg pcDNA3-flag-API2-MALT1 without 4 ng/mL LMB. For each experiment, the cells were processed as described in Figure 2 before fixation. For the first antibodies, mouse anti-flag antibody and rabbit anti-human BCL10 antibody were used, and for the second antibodies, fluorescein goat anti-mouse IgG and Alexa Fluor 546 goat anti-rabbit IgG. Below the immunofluorescence images, the relative proportions of cells with specific subcellular localization patterns are shown. C indicates cytoplasm only; C > N, predominantly cytoplasm; C = N, evenly distributed between cytoplasm and nuclei; C < N, predominantly nuclei. (C) COS7 cells (1 × 105) were transfected with NF-κB-dependent luciferase reporter vector, an internal control vector (phRL SV40), and the following expression vectors: for control, pcDNA3-myc; for MALT1, pcDNA3-MALT1-flag; for BCL10, pcDNA3-myc-BCL10; for MALT1 + BCL10, pcDNA3-MALT1-flag and pcDNA3-myc-BCL10. Treatment with 4 ng/mL LMB was performed 18 hours after transfection, and luciferase activity was measured using the method of the Dual-Luciferase Reporter Assay System (Promega) 24 hours after transfection. The figure shows the results of representative experiments performed in triplicate. Error bars indicate standard deviation
Subcellular localization of BCL10 cotransfected with MALT1(1-380) and API2-MALT1. COS7 cells (2 × 104) were transfected with the following expression vectors: (A) myc-BCL10 and flag-MALT1(1-380), a mixture of 0.02 μg pcDNA3-myc-BCL10 and 0.18 μg pcDNA3-flag-MALT1(1-380) with or without 4 ng/mL LMB; (B) myc-BCL10 and flag-API2-MALT1, a mixture of 0.02 μg pcDNA3-myc-BCL10 and 0.18 μg pcDNA3-flag-API2-MALT1 without 4 ng/mL LMB. For each experiment, the cells were processed as described in Figure 2 before fixation. For the first antibodies, mouse anti-flag antibody and rabbit anti-human BCL10 antibody were used, and for the second antibodies, fluorescein goat anti-mouse IgG and Alexa Fluor 546 goat anti-rabbit IgG. Below the immunofluorescence images, the relative proportions of cells with specific subcellular localization patterns are shown. C indicates cytoplasm only; C > N, predominantly cytoplasm; C = N, evenly distributed between cytoplasm and nuclei; C < N, predominantly nuclei. (C) COS7 cells (1 × 105) were transfected with NF-κB-dependent luciferase reporter vector, an internal control vector (phRL SV40), and the following expression vectors: for control, pcDNA3-myc; for MALT1, pcDNA3-MALT1-flag; for BCL10, pcDNA3-myc-BCL10; for MALT1 + BCL10, pcDNA3-MALT1-flag and pcDNA3-myc-BCL10. Treatment with 4 ng/mL LMB was performed 18 hours after transfection, and luciferase activity was measured using the method of the Dual-Luciferase Reporter Assay System (Promega) 24 hours after transfection. The figure shows the results of representative experiments performed in triplicate. Error bars indicate standard deviation
Cancellation of NF-κB activity by nuclear localization of MALT1 in the presence of BCL10
It is known that coexpression of MALT1 and BCL10 in transiently transfected cells leads to a higher level of NF-κB activation than does expression of either MALT1 or BCL10 alone.10 We therefore investigated the NF-κB activation in the COS7 cells cotransfected with MALT1 and BCL10 with or without LMB. The level of NF-κB activation in the cells treated with LMB was 45% lower than in those not treated with LMB (Figure 5C), indicating that nuclear localization of MALT1 and BCL10 by LMB treatment affected the NF-κB activation.
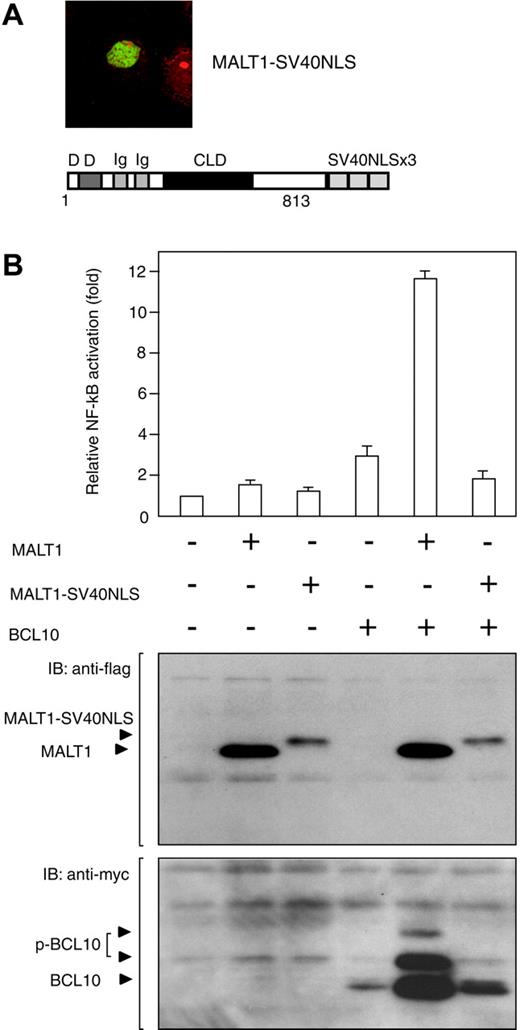
To confirm this observation, we attempted to relocalize MALT1 into the nucleus. A fusion construct of MALT1 with 3 tandem repeats of simian virus 40 large T-antigen nuclear localization signal (SV40NLS) was generated in order to force MALT1 to locate in the nucleus. Since, as expected, immunofluorescence analysis showed that the MALT1-SV40NLS was localized in the nucleus only in COS7 cells (Figure 6A), we conducted an NF-κB reporter assay using the MALT1-SV40NLS. The COS7 cells cotransfected with the MALT1-SV40NLS and BCL10 showed significantly lower levels of NF-κB activation than did those cotransfected with MALT1 and BCL10 (Figure 6B), indicating that cytoplasmic localization of MALT1 is involved in NF-κB activation.
Discussion
We previously reported that MALT1 and API2-MALT1 were localized in cytoplasm and that the MALT1 C-terminal region was responsible for their subcellular localization.17 In the study reported here, we showed that MALT1 and API2-MALT1 contained NES and shuttled between nuclei and cytoplasm. NES1 and NES2 regions in MALT1 were proved to be functional when fused to GFP. The active transport mechanism of macromolecules between nuclei and cytoplasm has recently been characterized. In this mechanism, NES-containing molecules are recognized by the export receptor chromosomal region maintenance 1 (CRM1) and then exported to cytoplasm in a CRM1-dependent manner.33,34 Leucine-rich NESs are composed of 4 hydrophobic amino acids, usually leucines, with variable spacing. The leucine-rich NES consensus sequence is defined as Lx(1-3)Lx(2-3)LxL (x indicates any residues), with allowance for certain variations.28-32 We demonstrated in this study that the NES1 region contained a motif that closely resembled the leucine-rich NES consensus sequence. It is interesting to note that the leucine-rich motif of the NES1 region was preserved among the species. It was shown that GFP-NES2 was exported to cytoplasm in a CRM1-dependent manner, although we could not identify the leucine-rich consensus sequence in the NES2 region, which suggests that the other sequences recognized by CRM1 were localized in the NES2 region.
The shuttling of MALT1 between nuclei and cytoplasm may have a biologic significance for lymphocytes. In support of this notion, Fas-associated death domain (FADD) and tumor necrosis factor receptor-associated death domain (TRADD) were recently found to shuttle between nuclei and cytoplasm during receptor signaling.35-38 In this context, it is noteworthy that FADD and TRADD as well as MALT1 contain a DD.
MALT1 and API2-MALT1 may possess a nuclear localization signal (NLS) because the molecular weights of MALT1 (90 kDa) and API2-MALT1 (116 kDa) were over the diffusion limit of the nuclear pores (40-60 kDa).34 However, we could not detect the NLS-like motif described in previous reports, indicating the need for further studies into the import mechanism of MALT1 and API2-MALT1.
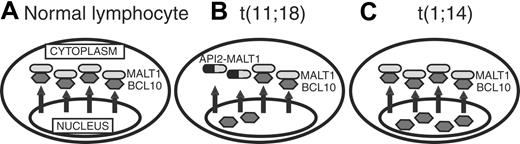
In normal B cells, BCL10 is expressed primarily in the cytoplasm, but in t(11;18) and t(1;14) MALT lymphoma, it is expressed mostly in the nuclei.20-23 However, the mechanism of subcellular localization of BCL10 is not well understood, and this is the first report to show that BCL10 is exported from the nucleus to the cytoplasm with the aid of MALT1. Based on our new findings, we propose a model for subcellular localization mechanisms of BCL10 (Figure 7), which correlates well with the histopathologic findings for BCL10.
Cancellation of NF-κB activity by nuclear localization of MALT1 in the presence of BCL10. (A) The cells transfected with MALT1-SV40NLS expression vectors were examined by means of immunofluorescence analysis. The protein structure of the MALT1-SV40NLS expression vector is shown below the immunofluorescence image. Numbers below the structures represent the amino acid number of MALT1. (B) COS7 cells (1 × 105) were transfected with NF-κB-dependent luciferase reporter vectors, an internal control vector (phRL SV40), and the following expression vectors: for control, pcDNA3-myc; for MALT1, pcDNA3-MALT1-flag; for MALT1-SV40NLS, pMALT1-flag-SV40NLS; for BCL10, pcDNA3-myc-BCL10; for MALT1 + BCL10, pcDNA3-MALT1-flag and pcDNA3-myc-BCL10; for MALT1-SV40NLS + BCL10, pMALT1-flag-SV40NLS and pcDNA3-myc-BCL10. At 24 hours after transfection, total extracts were used to measure luciferase activity, using the method of the Dual-Luciferase Reporter Assay System (Promega) (top panel) and to detect each of the proteins by means of Western blotting (bottompanel). NF-κB activity was examined 3 times, and one set of representative data is shown. IB indicates immunoblot. Error bars indicate standard deviation.
Cancellation of NF-κB activity by nuclear localization of MALT1 in the presence of BCL10. (A) The cells transfected with MALT1-SV40NLS expression vectors were examined by means of immunofluorescence analysis. The protein structure of the MALT1-SV40NLS expression vector is shown below the immunofluorescence image. Numbers below the structures represent the amino acid number of MALT1. (B) COS7 cells (1 × 105) were transfected with NF-κB-dependent luciferase reporter vectors, an internal control vector (phRL SV40), and the following expression vectors: for control, pcDNA3-myc; for MALT1, pcDNA3-MALT1-flag; for MALT1-SV40NLS, pMALT1-flag-SV40NLS; for BCL10, pcDNA3-myc-BCL10; for MALT1 + BCL10, pcDNA3-MALT1-flag and pcDNA3-myc-BCL10; for MALT1-SV40NLS + BCL10, pMALT1-flag-SV40NLS and pcDNA3-myc-BCL10. At 24 hours after transfection, total extracts were used to measure luciferase activity, using the method of the Dual-Luciferase Reporter Assay System (Promega) (top panel) and to detect each of the proteins by means of Western blotting (bottompanel). NF-κB activity was examined 3 times, and one set of representative data is shown. IB indicates immunoblot. Error bars indicate standard deviation.
A model for subcellular localization of BCL10 under various conditions. (A) Normal lymphocyte: BCL10 is localized in cytoplasm because nuclear BCL10 is exported by MALT1. (B) t(11;18) MALT lymphoma: Normal MALT1 products are reduced by half because the translocated allele generates API2-MALT1. Since the API2-MALT1 cannot export nuclear BCL10, the reduction of normal MALT1 proteins will be less efficient in BCL10 exportation, resulting in nuclear retention of BCL10. (C) t(1;14) MALT lymphoma: Overexpressed BCL10 results in nuclear retention because of a relative shortage of MALT1.
A model for subcellular localization of BCL10 under various conditions. (A) Normal lymphocyte: BCL10 is localized in cytoplasm because nuclear BCL10 is exported by MALT1. (B) t(11;18) MALT lymphoma: Normal MALT1 products are reduced by half because the translocated allele generates API2-MALT1. Since the API2-MALT1 cannot export nuclear BCL10, the reduction of normal MALT1 proteins will be less efficient in BCL10 exportation, resulting in nuclear retention of BCL10. (C) t(1;14) MALT lymphoma: Overexpressed BCL10 results in nuclear retention because of a relative shortage of MALT1.
As shown in Figure 5B, API2-MALT1 could not export BCL10 from nucleus to cytoplasm, indicating that API2-MALT1 does not bind to BCL10. The API2-MALT1 construct used in this study contains one Ig-like domain, while MALT1 contains 2, a difference that may affect the interaction between API2-MALT1 and BCL10. This is supported by our previous data, obtained by means of immunoprecipitation, that showed loss of interaction between the API2-MALT1 construct and endogenous BCL10 in 293T cells.18
Recently, t(14;18)(q32;q21) was identified as the third translocation in MALT lymphoma.39,40 This juxtaposes the Ig-heavy-chain-gene promoter at 14q32 to the MALT1 gene at 18q21, resulting in overexpression of the latter.40 In most MALT lymphomas with t(14;18), both MALT1 and BCL10 were immunostained in the cytoplasm.40-42 This observation also correlates well with our finding that BCL10 is exported from nucleus to cytoplasm by MALT1. In the report from Remstein et al,41 only one case with t(14;18) showed nuclear BCL10. The subcellular localization mechanism of BCL10 in this case still needs to be investigated in detail. It is also important to study further a biologic significance of the subcellular localization of endogenous MALT1 and BCL10.
It has recently been shown that the MALT1-BCL10 complex activates the NF-κB signaling pathway in cytoplasm,10 while we demonstrated in this study that the nuclear localization of MALT1 cancelled NF-κB activation. This suggests that the nucleocytoplasmic shuttling of MALT1 has an important function in MALT lymphomagenesis.
It has been suggested that nuclear BCL10 expression may be involved in MALT lymphomagenesis,20,24 and more recently, Liu et al43 reported that BCL10 interacts with transcription factor IIB (TFIIB) and functions as a potential transcriptional activator. However, the oncogenic role of BCL10 in the nucleus remains to be investigated. Further characterization of the subcellular localization mechanism of BCL10, MALT1, and API2-MALT1 can be expected to provide new insights into MALT lymphomagenesis.
Prepublished online as Blood First Edition Paper, August 25, 2005; DOI 10.1182/blood-2004-12-4785.
Supported in part by a Grant-in-Aid for the Second-Term Comprehensive 10-Year Strategy for Cancer Control from the Japan Ministry of Health, Labor and Welfare; a Grant-in-Aid for Science on Primary Areas (Cancer Research) from the Japan Ministry of Education, Culture, Sports, Science and Technology; a Grant-in Aid from the New Energy and Industrial Technology Development Organization; and a Grant-in-Aid for Cancer Research from the Princess Takamatsu Cancer Research Fund (03-23503).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Ms H. Suzuki and Ms Y. Kasugai for their outstanding technical assistance. We also would like thank Dr Zhijian J. Chen of University of Texas Southwestern Medical Center for providing anti-MALT1 polyclonal antibodies, Dr T. Koseki of Tohoku University and Dr N. Inohara of the University of Michigan Medical School for providing pBIIx-Luc vector, and Dr R. Ohno of the Aichi Cancer Center Hospital for his encouragement and support.

![Figure 4. NES1 region of MALT1 and its role in subcellular localization. (A) Amino acid sequence of the NES1 region in alignment with MALT1 homologs of various species. The amino acid numbers are based on amino acid sequences from accession numbers BAA83099 (Homo sapiens), AAG38590 (Danio rerio), AAG38591 (Caenorhabditis elegans), and AAG38592 (Dictyostelium discoideum) in GenBank. Residues that resemble the consensus sequence of leucine-rich NES are boxed. Alanine substitutions were made at Leu362 and Leu365 for studying its role in subcellular localization (MALT1[1-380]-NES1 mutant). NES1 indicates the NES1 region. (B) Cells transfected with either MALT1(1-380)-NES1 mutant or wild type are shown. The cells were processed in the same manner as those in Figure 2. Relative proportions of cells with specific subcellular localization patterns are shown below each panel. NES1 mutant represents MALT1(1-380)-NES1 mutant and wild-type MALT1(1-380). C indicates cytoplasm; CN, both cytoplasm and nuclei.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/13/10.1182_blood-2004-12-4785/2/m_zh80240587890004.jpeg?Expires=1767717918&Signature=zoF7F7R4M0j~t4jD0McB7OT0t0V4A2QZk~NMuIFLgTij-MId50BENss-sJ2~otxf8PpTuox8EhjRYq-ZL8wgo9JXDncDPdfrhX5Gi9f2kYUZoKW5TARhUsPs0L964Irf164wno85J48UXGd0XIyM39gBK9n5pCq6TJJDdjLZVx1-JLrLPQM1vdrQTuJxYol3PQ6u5mc7uSj2xwykZQ1h2CXxJqQmGT1tuczySH0MzQwNa~XV7qoCh7T7HROMeCUfYBvloMnYDywzVgPCS~H0yKa5KPLvHM3JXB9lVg~M9-h1PJ6fx~fwFNza7kVjRohoz4P58UMVpkbagf6HOzcPzQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal