Protein ubiquitination plays important roles in a variety of basic cellular processes. Proteins are ubiquitinated by E2-E3 ubiquitin ligase complexes. Depending on the type of ubiquitin chain conjugated, proteins are either targeted for degradation by the proteasome or their activity is specifically altered. We describe a novel conserved nuclear protein, Triad1 (2 RING [really interesting new gene] fingers and DRIL [double RING finger linked] 1), which is strongly induced during myeloid differentiation. Triad1 contains a TRIAD motif that harbors 2 RING finger structures. Triad1 binds the E2 ubiquitin-conjugating enzyme UbcH7 as well as ubiquitinated proteins and supports the formation of ubiquitin chains that are recognized by the proteasome. The biologic function of Triad1 in myelopoiesis was studied by performing granulocyte-macrophage colony-forming unit (CFU-GM) assays using retrovirally transduced primary murine bone marrow cells. Triad1 severely inhibited myeloid colony formation. In contrast, 2 Triad1 RING finger point mutants that failed to bind UbcH7 did not affect colony formation. Moreover, proteasome inhibition counteracted the inhibition of colony formation exerted by wild-type Triad1. In liquid cultures, Triad1 did not influence differentiation but strongly inhibited proliferation resulting in a G0/G1 accumulation. We conclude that proteasomal degradation of proteins that are ubiquitinated by Triad1 affects the clonogenic growth of primary myeloid progenitor cells.
Introduction
Granulocytes and monocytes develop from common myeloid progenitor cells through a complex network of cell growth, differentiation, and apoptosis-regulating factors. Alterations in these processes may cause acute myeloid leukemia, which is characterized by uncontrolled proliferation of immature myeloid cells that fail to differentiate toward mature functional cells.1,2 In leukemia, mutations occur in genes encoding tyrosine kinases such as Fms-like tyrosine kinase 3 (Flt3), c-kit, neuroblastoma ras (N-ras) and transcription factors such as AML1 (acute myeloid leukemia 1), CBFB (core binding factor β), GATA1 (GATA binding protein 1), PU.1, C/EBPα (CCAAT/enhancer binding protein α), and RARα (retinoic acid receptor α).1,2 Although the transcription factors predominantly play a role in lineage commitment, activation of tyrosine kinases is thought to result in proliferative and/or survival signals.1,2 Recent studies have shown that several proteins involved in myelopoiesis (including proteins mutated in leukemia) are inactivated through ubiquitin (Ub)-proteasomal degradation pathways.3-7 This form of targeted protein degradation is accomplished by the covalent conjugation of Ub to substrate proteins, usually in the form of a multi-Ub chain, which marks these proteins for progressive degradation by the 26S proteasome.8,9 Protein ubiquitination is catalyzed through a cascade of reactions. Ub is first activated by the adenosine triphosphate (ATP)-dependent Ub-activating E1 enzyme and subsequently transferred to one of a set of E2 Ub-conjugating enzymes. The E2 enzymes act in conjunction with accessory E3 Ub protein ligases. In the E2-E3 complex, the E3 component binds to protein substrates, allowing the E2 to form a multi-Ub chain linked to a lysine of the substrate protein.8,9 Thus, the E3 Ub ligases play a crucial role in this process because these proteins recognize the cellular proteins destined for ubiquitination.10 To date, several types of E3 Ub ligases have been described. These include HECT (homologous to E6-AP carboxyl terminus), RING (really interesting new gene) finger, U-box, SOCS (suppressor of cytokine signaling)-box, F-box, and cullin ligases.11-20 Although the first 3 types of proteins are actively involved in substrate ubiquitination, the last 3 do not ubiquitinate substrate proteins themselves but are part of larger protein complexes that exhibit Ub ligase activity.15-19 Despite several proteins that play an important role in myelopoiesis are known to be targeted by the Ub-proteasome pathway, only a few E3 Ub ligases involved in myelopoiesis have been described. These include the cullin Ub ligase Cul-4A implicated in myelopoiesis.4,21 Cul-4a heterozygous mice have diminished multipotent progenitors, and enforced Cul-4A expression inhibits granulocytic differentiation of myeloid PLB-985 cells.21 An important target of Cul-4A in myelopoiesis may be the transcription factor HoxA9 (homeo box A9) that is targeted by Cul-4A for ubiquitination resulting in its proteasomal degradation.4 Another group of Ub ligases involved in myelopoiesis are the SOCS proteins. All of these proteins contain a SOCS box that provides in the scaffold for interaction with Ub ligase complexes.15,16 One of the main functions of the SOCS box proteins is to recruit substrate proteins to ligase complexes resulting in their ubiquitination. SOCS3 is induced on granulocyte colony-stimulating factor (G-CSF) treatment, binds to the activated G-CSF receptor, and negatively regulates G-CSF receptor-mediated STAT (signal transducer and activator of transcription) activation.22 In addition, mice with loss of SOCS3 function in the hematopoietic compartment develop neutrophilia.23 Although it is not exactly clear how SOCS proteins inhibit cytokine signaling, it has been suggested that the ubiquitination and subsequent degradation of signaling molecules may be involved in this process.24 Finally, another important E3 Ub ligase involved in myelopoiesis is the RING finger protein Cbl (CAS-BRM murine ecotropic retroviral transforming sequence homolog). Cbl catalyzes the ubiquitination of several activated receptor tyrosine kinases, including the colony-stimulating factor 1 (CSF-1) receptor.25 The ligand binding stimulated ubiquitination of these receptors results in their internalization and targeting to lysosomes where they are degraded. As this inhibits these receptors to recycle through early endosomes to the cell surface, Cbl negatively regulates receptor signaling.25 Here, we identify a novel E3 Ub ligase, Triad1 (2 RING fingers and DRIL [double RING finger linked] 1), to be strongly induced during granulocytic and monocytic differentiation and show that the Ub ligase activity of Triad1 inhibits the proliferation of committed myeloid progenitor cells.
Materials and methods
Subtractive PCR, 5′- and 3′-RACE and classic and real-time reverse transcriptase (RT)-PCR
Subtractive polymerase chain reaction (PCR) was performed using polyA+ mRNA isolated from 0 and 18 hours all-trans retinoic acid (ATRA)-treated NB4 cells (PCR-select cDNA subtraction kit; Clontech, Heidelberg, Germany). Subtracted radiolabeled PCR fragments were isolated from polyacrylamide gel, reamplified, cloned in pCR2.1, and sequenced. Methods for cDNA synthesis and classic and real-time RT-PCR were as described.26 5′-Rapid amplification of cDNA ends (RACE) was performed using the Marathon cDNA amplification kit (Clontech) on 1.0 μg human spleen mRNA. Relevant primers are indicated in Table 1.
Sequences of Triad1 oligonucleotides
Name . | Sequence (5′-3′) . |
|---|---|
| AO | GTCGCGAATTCGTCGACGCG (T)15 |
| AP1 | GTCGCGAATTCGTCGACGCG |
| AP2 | ACTCACTATAGGGCTCGAGCGGC |
| T1B | TGCCTGGTTCCAGCTGTCAGA |
| T1D | TGCTCTGAGGCTGAGGTGTG |
| T1K | GCAGACAGCTATGACAGAGG |
| T1O | AAAGGTGAGCCACAGACATG |
| T1P | TGCCAGTGTCTCTCAGGAGT |
| T1V | TTTGATCCCGAGGAGTACCA |
| T1W | GTTCACTTGCTTGACCTACA |
| T1X | TGTAGGTCAAGCAAGTGAAC |
| T1Y | TGGTACTCCTCGGGATCAAA |
| T1Z | TGATCTGCAACTTGGCTGGA |
| T1AB | GATGGGATGTGGGAACATGT |
| T1exp1 | GATCGGATCCATGTCAGTGGACATGAATAGCCAG |
| T1exp2 | TCGCGAATTCTTAGGTGTCATGGAAATCTTTCAGC |
| T1G1 | GGATCCTTAGGATGGGATGTGGGAACATGT |
| T1G2 | GGATCCTACAAGTCCAATTCTGCTCA |
| T1M4 | TCGCGGATCCTTACAGGTCCTCCTCGCTGATCAGCTTCTGCTCGGTGTCATGGAAATCTTTCAGC |
| T1M5 | GATCGAATTCATGCTAAGATGTCAGTGG |
| T1qF | TGGTCCTTTCTTTTGTGCCAGTA |
| T1qR | GCAGCAAGACAGTGTCAGGTTT |
| T1qP | CCAAATGTGACCCTGCTCAGAGCTATACCAC |
| H158AF | TCTCTGGCCTGTCAGGCCCAGTTTTGCCGCAGC |
| H158AR | GCTGCGGCAAAACTGGGCCTGACAGGCCAGAGA |
| C161AF | TGTCAGCACCAGTTTGCCCGCAGCTGCTGGGAG |
| C161AR | CTCCCAGCAGCTGCGGGCAAACTGGTGCTGACA |
Name . | Sequence (5′-3′) . |
|---|---|
| AO | GTCGCGAATTCGTCGACGCG (T)15 |
| AP1 | GTCGCGAATTCGTCGACGCG |
| AP2 | ACTCACTATAGGGCTCGAGCGGC |
| T1B | TGCCTGGTTCCAGCTGTCAGA |
| T1D | TGCTCTGAGGCTGAGGTGTG |
| T1K | GCAGACAGCTATGACAGAGG |
| T1O | AAAGGTGAGCCACAGACATG |
| T1P | TGCCAGTGTCTCTCAGGAGT |
| T1V | TTTGATCCCGAGGAGTACCA |
| T1W | GTTCACTTGCTTGACCTACA |
| T1X | TGTAGGTCAAGCAAGTGAAC |
| T1Y | TGGTACTCCTCGGGATCAAA |
| T1Z | TGATCTGCAACTTGGCTGGA |
| T1AB | GATGGGATGTGGGAACATGT |
| T1exp1 | GATCGGATCCATGTCAGTGGACATGAATAGCCAG |
| T1exp2 | TCGCGAATTCTTAGGTGTCATGGAAATCTTTCAGC |
| T1G1 | GGATCCTTAGGATGGGATGTGGGAACATGT |
| T1G2 | GGATCCTACAAGTCCAATTCTGCTCA |
| T1M4 | TCGCGGATCCTTACAGGTCCTCCTCGCTGATCAGCTTCTGCTCGGTGTCATGGAAATCTTTCAGC |
| T1M5 | GATCGAATTCATGCTAAGATGTCAGTGG |
| T1qF | TGGTCCTTTCTTTTGTGCCAGTA |
| T1qR | GCAGCAAGACAGTGTCAGGTTT |
| T1qP | CCAAATGTGACCCTGCTCAGAGCTATACCAC |
| H158AF | TCTCTGGCCTGTCAGGCCCAGTTTTGCCGCAGC |
| H158AR | GCTGCGGCAAAACTGGGCCTGACAGGCCAGAGA |
| C161AF | TGTCAGCACCAGTTTGCCCGCAGCTGCTGGGAG |
| C161AR | CTCCCAGCAGCTGCGGGCAAACTGGTGCTGACA |
For amplification of overlapping Triad1 RT-PCR fragments 2, 3, 4 (Figure 1) primer combinations T1V-T1AB, T1Z-T1P, and T1K-T1O were used, respectively. For 3′-RACE primers AO was used for cDNA synthesis followed by PCR using primers AP1 and T1D, followed by heminested PCR using primers AP1 and T1B. For 5′-RACE primers AP2 and T1X were used in PCR followed by heminested PCR using primers AP2 and T1Y. T1M4 and T1M5 were used to for the construction of a C-terminally myc tagged Triad1 PCR construct. The 30-bp myc-tag encoding sequence is underlined. Primers H158AF, H158AR, C161AF, and C161AR were used to construct Triad1 H158A and Triad1 C161, respectively. For generation of GST-Triad1 and GFP-Triad1 primer pairs T1exp1 and T1exp2 were used. For cloning full-length Triad1 in pCDNA3.1 and retroviral vectors primers T1M4 and T1exp2 were used. For cloning Triad1-119 and Triad1109-493 in pGAD vector primer combinations T1exp1-T1G1 and T1G2-T1exp2 were used, respectively. For Triad1 qPCR primers T1qF and T1qR and probe (TET-label) T1qP were used. BamHI and EcoRI sites introduced by PCR for cloning purposes are indicated (bold italics).
Plasmid constructs
Triad1 coding sequences were PCR amplified using cDNA derived from NB4 cells and cloned in pCR2.1. A 10 amino acid (aa) human myc epitope was introduced at the 3′ end of the coding sequence of Triad1 by PCR and cloned into pSG5, and nontagged Triad1 was cloned into pcDNA3.1. Triad1 coding sequences were generated by PCR and cloned in plasmid vector red-shifted green fluorescence protein (pEGFP)-C1 to generate GFP-Triad1, pFastBacHT to generate His-Triad1 in Hi5 cells (Baculovirus expression system; Invitrogen, Karlsruhe, Germany) and in pGEX-2TK to generate glutathione S transferase (GST)-Triad1. For retroviral transduction Triad1 coding sequences were inserted into the retroviral pLRZS vector. For yeast 2-hybrid assays sequences coding for Triad1 aa 1 to 119 and 109 to 493 were cloned in pGAD-C. All primers used for cloning purposes and for generating the Triad1 point mutants H158A and C161A by site-directed mutagenesis are indicated in Table 1. The integrity of all Triad1 sequences was checked by sequence analysis.
Immunofluorescence, Northern blotting, GST- and His-captures, and immunoprecipitations
In these studies antibodies against CD34, CD3, CD14, CD15, CD45 (Beckman Coulter, Brea, CA), Mac1 (BD Biosciences, Heidelberg, Germany), GFP (7.1/13.1; Roche, Basel, Switzerland), myc (9E10; Santa Cruz Biotechnology, Heidelberg, Germany), UbcH7 and UbcH9 (Transduction Laboratories, Lexington, KY), Ub (6C1; Sigma, Saint Louis, MO), Flag (M2; Sigma), annexinV (AlexaFluor 647; Molecular Probes, Leiden, The Netherlands), and Triad1 were used. Triad1 antibodies were generated by immunizing rabbits with a keyhole limpet hemocyanin (KLH)-coupled peptide corresponding to Triad1 aa 473 to 486, followed by affinity purification using peptides immobilized on Affigel 10 columns (BioRad, Munich, Germany). Human hematopoietic subsets were isolated by fluorescence-activated cell sorting (FACS) with appropriate antibodies. Granulocytes were selected based on CD45 positivity and scatter plot. Cell purity was confirmed to be greater than 95% by flow cytometric analysis. For immunofluorescence experiments cells were stained with indicated primary antibodies followed by staining with a secondary antibody. Samples were mounted in Vectashield medium (H1200; Vector Laboratories, Burlingame, CA) and analyzed with a Leica DMRBE microscope (Leica, Rijswijk, The Netherlands). Pictures were taken at 40 × (oil, 1.00 NA) or 100 × (oil, 1.30 N) magnification with a Leica DC350F Camera using Leica FW400 software and processed with Adobe Photoshop (Adobe Systems, San Jose, CA). For detection of endogenous Triad1 by multicolor flow cytometry (Cytomics FC500; Becton Dickinson, Franklin Lakes, NJ), cells were stained with a Triad1 antibody using fix-and-perm approaches as described27 followed by blocking of cells with rabbit serum and subsequent staining for relevant surface markers. For Triad1 Northern blot hybridization 10 μg total RNA isolated from NB4 cells treated with ATRA (1 × 10-6 M) for various time points or 1.0 μg mRNA from hematopoietic cells was hybridized using Triad1 clone 14-5 as a probe. For GST-captures, 10 μg immobilized GST or GST-Triad1 was incubated with cell lysates from 1 × 107 NB4 cells. For capture of His-tagged proteins, lysates from transfected 293T cells were incubated with His-select beads (Sigma) under denaturing conditions (6 M guanidiumchloride). For immunoprecipitations, lysates from transfected COS cells were incubated with indicated antibodies. Bound proteins were washed, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotted, and stained with relevant antibodies.
Yeast 2-hybrid analysis
For yeast 2-hybrid analyses UbcH bait28 and Triad1 prey constructs were transfected to the yeast strain AH109 (Clontech). This strain contains 2 selectable markers (histidine and adenine) and a lacZ reporter. A positive interaction in yeast was defined by growth of blue colonies 4 days after plating cells on leucine, tryptophane, adenine, and histidine-deficient synthetic defined (SD) medium containing X-α-gal.
Proteasome activity measurements and in vitro ubiquitination assays
Proteasome activity was determined in primary murine bone marrow cells after treatment with 1 × 10-8 M MG132. For measurements, 67.5 μg protein lysates (concentration determined using Bradford assay) were incubated with the proteasome substrate Suc-Leu-Leu-Val-Tyr-AMC (Boston Biochem, Cambridge, MA) for 2 hours at 37°C as described.29 7-Amino-4-methylcoumarin (AMC) liberated from the substrate by proteasome activity was determined by measuring fluorescence (Fluorstar, BMG; Isogen Life Sciences, Maarssen, The Netherlands).29 For in vitro ubiquitination assays His-Triad1 was isolated from Hi5 cells (Baculovirus Bac-to-Bac system; Invitrogen) using His-select spin columns followed by elution using 250 nM imidazole. Imidazole was removed from His-Triad1 protein eluates using a Microcon YM-10 filter (Amicon, Bedford, MA), and protein concentrations were determined with the Bradford assay. Ubiquitination assays were performed using 1.6 μg reticulocyte fraction IIA (Boston Biochem), bovine Ub (Sigma), or recombinant UbK48-only (Boston Biochem) with freshly prepared His-Triad1 for 60 minutes at 37°C followed by staining of ubiquitinated proteins in Western blot analysis using a Ub-specific antibody (C61; Sigma) as described.30
Clonogenic assays (CFU-GM) and myeloid cell proliferation
Retroviral vectors (pLZRS) were transfected to retroviral producer phoenix cells (kindly provided by Dr G. Nolan). Primary murine bone marrow cells isolated from femurs from 6-week-old female CBA/CA mice were prestimulated for 2 days with stem cell factor (SCF), interleukin 12 (IL-12), IL-3, Flt-3, thrombopoietin (TPO), and α transforming growth factor-β (α-TGFβ) and subsequently infected with retroviruses on retronectin-coated dishes. After 24 hours GFP-positive cells were sorted by FACS and seeded in semisolid medium (5000 cells/well) containing pokeweed mitogen-stimulated mouse spleen conditioned medium, horse serum, and SCF. Colonies were counted 5 days later yielding between 83 and 125 colonies after granulocyte-macrophage colony-forming unit (CFU-GM) of empty vector transduced cells and morphologically analyzed after May-Grünwald-Giemsa staining. To determine proliferation in liquid assays, 5000 cells were seeded in 96-well plates using the above-mentioned medium without agar. Relative cell numbers were determined by FACS using fluorescent beads. Apoptosis and DNA histograms were determined by flow cytometry using AnnexinV and propidium iodide staining, respectively. Percentages of cells in various phases of cell cycle were determined using ModFit software (Verity Software House, Topsham, ME). These studies were approved by the DEC Committee, RUNMC, Nijmegen, The Netherlands.
Results
Identification of genes induced during granulocytic differentiation
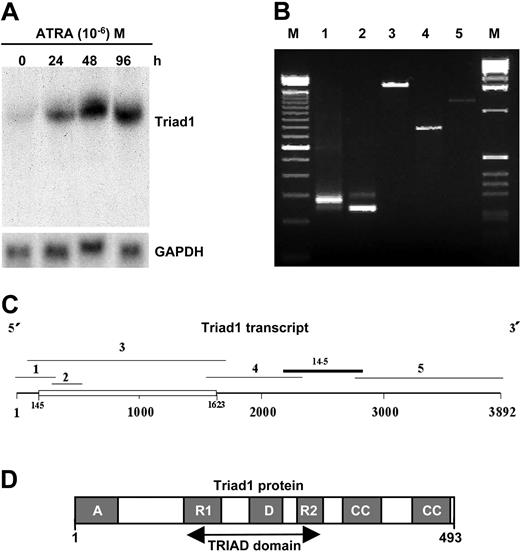
Acute promyelocytic leukemia cells can be forced to differentiate along the granulocytic lineage by the vitamin A derivative ATRA. To identify genes involved in granulocytic differentiation we performed a subtractive PCR screen using RNA isolated from promyelocytic leukemia (NB4) cells before and 18 hours after ATRA treatment. Northern blotting using one of the subtracted clones (clone 14-5, GenBank accession number AF099153) showed the induction of a 4-kilobase (Kb) mRNA in NB4 cells 24 hours after ATRA treatment to reach maximum levels at 48 hours (Figure 1A). The subtracted clone appeared identical to human randomly cloned expressed cDNA sequences identified in database searches (not shown). By using these sequences in database walks a 3.9-Kb cDNA contig was constructed. This contig was confirmed by amplification of overlapping RT-PCR fragments and 5′- and 3′-RACE (Figure 1B-C). Sequence analysis of the 3.9-Kb transcript identified a poly-adenylation signal (ATAAA) 24 base pair (bp) upstream of the 3′ end of the transcript and a translational start codon at cDNA position 145. This start codon is accompanied by a consensus Kozak sequence and is followed by a 1479-bp ORF (Figure S1). The coding sequences were confirmed by sequence analysis of 12 independent clones (GenBank accession number AF099149). In line with multiple bands observed in RT-PCR (Figure 1B), 3 Triad1 clones contained different insertions of extra sequences present at cDNA position 399 possibly representing splice variants (GenBank accession numbers AF099150, AF099151, and AF099152).
Identification of Triad1. (A) Northern blot hybridization using subtracted clone 14-5 detecting the ATRA-mediated induction of a 4-Kb transcript in NB4 cells (time of ATRA incubation is given at the top in hours). Rehybridization with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe indicates equal loading. (B) Triad1 5′- and 3′-RACE and RT-PCR fragments as depicted in panel 1C (lane numbers correspond to fragment numbers in panel 1C). Alternative splicing results in the detection of 2 RT-PCR fragments (lane 2). RACE and PCR fragments 1 to 5 were amplified using primer combinations indicated in Table 1. (C) Schematic representation of the 3.9-Kb ATRA-induced Triad1 transcript (see Figure S1 for sequences). Indicated are the 1479 open reading frame (ORF) (open box) and overlapping cDNA and 5′- and 3′-RACE fragments as well as the localization of fragment 14-5 representing the clone identified in the subtractive PCR screen. (D) Schematic representation of Triad1 protein domains. A indicates acidic domain; R1 and R2, RING finger structures; D, DRIL domain; CC, coiled coil domain.
Identification of Triad1. (A) Northern blot hybridization using subtracted clone 14-5 detecting the ATRA-mediated induction of a 4-Kb transcript in NB4 cells (time of ATRA incubation is given at the top in hours). Rehybridization with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe indicates equal loading. (B) Triad1 5′- and 3′-RACE and RT-PCR fragments as depicted in panel 1C (lane numbers correspond to fragment numbers in panel 1C). Alternative splicing results in the detection of 2 RT-PCR fragments (lane 2). RACE and PCR fragments 1 to 5 were amplified using primer combinations indicated in Table 1. (C) Schematic representation of the 3.9-Kb ATRA-induced Triad1 transcript (see Figure S1 for sequences). Indicated are the 1479 open reading frame (ORF) (open box) and overlapping cDNA and 5′- and 3′-RACE fragments as well as the localization of fragment 14-5 representing the clone identified in the subtractive PCR screen. (D) Schematic representation of Triad1 protein domains. A indicates acidic domain; R1 and R2, RING finger structures; D, DRIL domain; CC, coiled coil domain.
Identification of Triad1
The 1479-bp ORF in the 3.9-Kb transcript predicts the formation of a 493-aa protein (Figure 1D). We have called this protein Triad1 referring to its cysteine-histidine rich TRIAD motif.31 The TRIAD motif consists of 2 RING fingers that flank the DRIL domain. The DRIL (also known as IBR [in between RING fingers]32 ) domain resembles other cysteine-rich motifs as the LAP (leukemia-associated protein)/PHD, LIM, and RING finger domains.31 Recent data indicate that the C-terminal RING finger (RING2) of the TRIAD motif has a distinct topology from the classic RING finger structure and that it binds only 1 zinc atom instead of 2.33 Further Triad1 protein analysis identified an acidic N-terminal region, 2 C-terminal coiled coil regions, and 2 putative nuclear localization signals. To determine whether Triad1 is conserved during evolution we performed database searches and identified full-length murine Drosophila melanogaster and Caenorhabditis elegans orthologs (Figure S1). The conservation in the last 231 aa C-terminal part of Triad1 between D melanogaster and C elegans compared with human Triad1 is very high, being 80% and 64% identical, respectively. Comparison of complete murine and human Triad1 shows that the proteins are 98% identical. The strong evolutionary conservation of Triad1 suggests that it exerts important biologic functions.
Triad1 is induced during normal granulocytic/monocytic differentiation
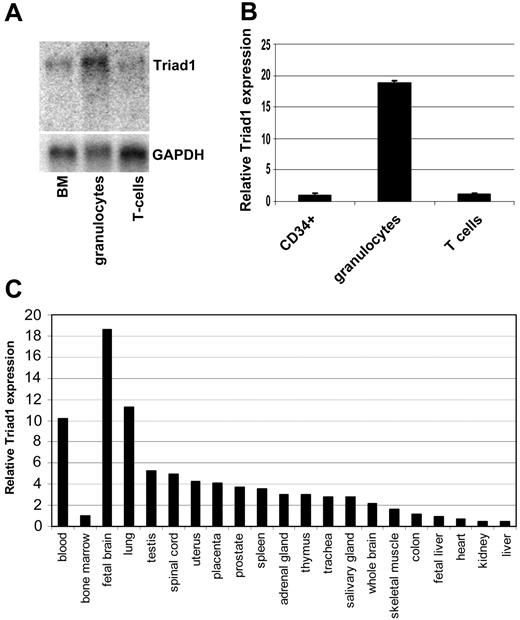
TRIAD1 gene expression is induced during forced granulocytic differentiation of promyelocytic leukemia (NB4) cells. To determine the relevance of this observation we analyzed TRIAD1 expression in normal human hematopoiesis. TRIAD1 Northern blot hybridization using RNA isolated from total bone marrow, mature granulocytes, and T cells revealed strongest TRIAD1 expression in granulocytes (Figure 2A). In agreement, TRIAD1 quantitative RT-PCR detected high expression in mature granulocytes and low expression in T cells and immature CD34+ cells, indicating that TRIAD1 is induced during maturation of immature CD34+ cells toward mature granulocytes (Figure 2B). Further expression studies showed detectable TRIAD1 expression by real-time PCR in a large set of human tissues tested, indicating that the function of Triad1 is probably not limited to hematopoiesis (Figure 2C).
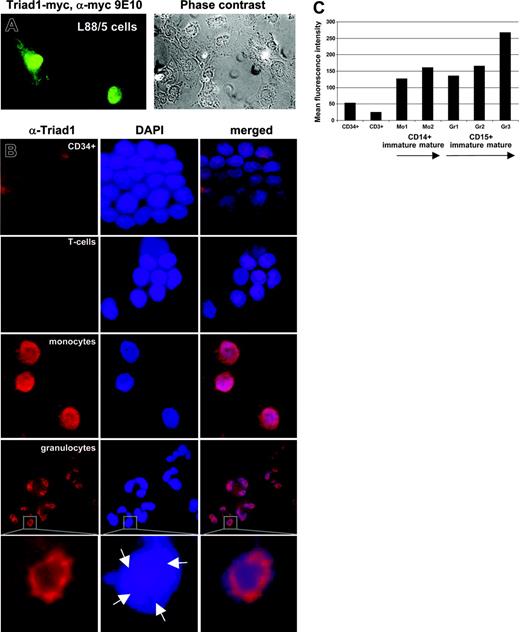
To determine the subcellular localization of Triad1 a C-terminally myc epitope-tagged Triad1 (Triad1-myc) expression vector was transfected to L88/5 stromal cells. Indirect immunofluorescence (IF) using the myc epitope recognizing antibody 9E10 revealed a clear nuclear diffuse staining with exclusion of the nucleoli (Figure 3A). In cells with highest IF levels, some cytoplasmic staining was also observed. To determine the endogenous Triad1 expression in primary hematopoietic cells an affinity-purified Triad1 antibody was used in indirect IF. Analysis of immature CD34+ and T cells revealed a weak nuclear staining (Figure 3B). In contrast, a strong nuclear localization was observed in both granulocytes and monocytes. Although the localization in monocytes was nuclear diffuse, a ring-shaped nuclear organization was observed in granulocytes (Figure 3B). To determine at what stage Triad1 was induced during myelopoiesis we stained fresh bone marrow and leukapheresis samples with a Triad1 antibody and analyzed the cells by flow cytometry. Costaining with CD34 (immature progenitors), CD3 (T cells), CD14 (monocytes), or CD15 (granulocytes) showed that Triad1 is lowly expressed in immature CD34+ and T cells and that the expression gradually increases on maturation toward committed myelomonocytic progenitors to reach highest levels in mature monocytes and granulocytes (Figure 3C; Figure S2). Together, these data indicate that Triad1 is induced at the mRNA and protein level during differentiation of immature blood cells toward both monocytes and granulocytes and that it localizes predominantly to the nucleus in primary human hematopoietic cells.
Triad1 is highly expressed in granulocytes. (A) Northern blot hybridization using Triad1 clone 14-5 as a probe detects high Triad1 expression in mature granulocytes compared with complete bone marrow and T cells. (B) Triad1 quantitative RT-PCR detects high expression in granulocytes and low expression in immature CD34+ and T cells. Data are from 2 samples from unrelated healthy volunteers. Expression in CD34+ cells was set at 1.0, and values were normalized for porphobilinogen deaminase (PBGD) expression as described.26 (C) Triad1 real-time PCR detects high expression in peripheral blood compared with bone marrow (expression in total bone marrow was set at 1.0). Among nonhematopoietic tissues highest expression was observed in fetal brain.
Triad1 is highly expressed in granulocytes. (A) Northern blot hybridization using Triad1 clone 14-5 as a probe detects high Triad1 expression in mature granulocytes compared with complete bone marrow and T cells. (B) Triad1 quantitative RT-PCR detects high expression in granulocytes and low expression in immature CD34+ and T cells. Data are from 2 samples from unrelated healthy volunteers. Expression in CD34+ cells was set at 1.0, and values were normalized for porphobilinogen deaminase (PBGD) expression as described.26 (C) Triad1 real-time PCR detects high expression in peripheral blood compared with bone marrow (expression in total bone marrow was set at 1.0). Among nonhematopoietic tissues highest expression was observed in fetal brain.
Triad1 is an E3 Ub ligase
The RING finger structure provides an interface for interaction with E2 Ubcs, and all RING finger proteins that bind Ubcs exhibit Ub ligase activity.20 Because Triad1 harbors 2 RING fingers we tested whether it binds to Ubcs using yeast 2-hybrid assays. For these experiments 2 Triad1 prey-fragments were used, 1 coding for amino acids 1 to 119, Triad1[1-119], lacking the TRIAD structure and 1 for amino acids 109 to 493, Triad1[109-493], including the TRIAD structure. No interaction was observed between Triad1[1-119] and a panel of Ubcs28 tested, including CDC34, UbcH5, UbcH6, UbcH7, UbcH8, UbcH9, UbcH10, and UbcH13 (not shown). In contrast, a clear interaction between Triad1[109-493] and UbcH7 was observed as indicated by growth of yeast cells on medium lacking both histidine and adenine (Figure 4A). When less stringent yeast 2-hybrid conditions were used (histidine selection only) an interaction between Triad1[109-493] and UbcH6 and UbcH13 (also known as Bendless) was also found (data not shown). Because endogenous UbcH7 is readily detectable in NB4 cells, we incubated cell lysates from these cells with GST-Triad1 to confirm the UbcH7 interaction as observed in yeast 2-hybrid assays. Bound proteins were stained with an UbcH7 antibody in Western blot analysis, showing that Triad1 efficiently binds endogenous UbcH7 from NB4 cells (Figure 4B). Staining for UbcH9 in these experiments was negative, whereas UbcH9 is also easily detectable in NB4 cells by Western blotting (not shown). To test whether the N-terminal RING finger was involved in UbcH7 binding we constructed 2 independent Triad1 RING finger point mutants in which 1 of the predicted zinc-coordinating amino acids were changed into alanine (H158A, C161A). Mutations were introduced in the Triad1 prey-construct (Triad1[109-493]) and used in yeast 2-hybrid assays. Although normal Triad1[109-493] interacts with UbcH7 in yeast 2-hybrid assays, both point mutants did not interact with UbcH7, indicating that the N-terminal RING finger of Triad1 is essential for UbcH7 binding (Figure 4A). To further test whether Triad1 might function as Ub ligase, we determined whether Triad1 binds ubiquitinated proteins. To this end, Triad1-myc and Flag-tagged Ub (Flag-Ub) were coexpressed in the presence or absence of proteasome inhibitors followed by myc IP. Triad1 staining of myc-immunoprecipitated proteins in Western blot analysis revealed that proteasome inhibition resulted in stabilization of Triad1. Flag staining of proteins bound to Triad1 detected a smear of ubiquitinated proteins that appeared more intense when transfected cells were treated with a proteasome inhibitor, suggesting that Triad1 binds ubiquitinated proteins (Figure 4C). Because the observed smear might represent exclusively ubiquitinated Triad1 species instead of ubiquitinated proteins bound to Triad1, we repeated the experiment using GFP-Triad1 and Flag-Ub. Subsequent GFP IP followed by Triad1 staining in Western blot analysis revealed the presence of unmodified Triad1, whereas Flag staining revealed a smear of ubiquitinated proteins, including proteins with sizes smaller compared with Triad1. Because the smaller proteins do not represent Triad1, we concluded that Triad1 binds ubiquitinated proteins (Figure 4D). To further test whether Triad1 functions as E3 Ub ligase, we performed an in vitro ubiquitination assay using reticulocyte fraction IIA (enriched in E1 and E2 activity and devoid of Ub and proteasome activity), with or without Ub and His-Triad1. Staining of reaction mixtures with a Ub-specific antibody in Western blot analysis revealed the presence of Triad1-dependent ubiquitinated proteins (Figure 4E). Triad1 binds to UbcH7. In concert with E3 Ub ligases, UbcH7 supports the formation of Ub chains linked through lysine 48 (K48) that are recognized by the proteasome. To test whether Triad1 can catalyze the formation of Ub chains linked through K48, we also used recombinant Ub carrying only 1 lysine residue at position 48 (UbK48-only), showing that Triad1 catalyzes the formation of Ub chains linked through K48. The interaction between Triad1 and UbcH7 as well as ubiquitinated proteins and the in vitro ubiquitination assay strongly suggest that Triad1 functions as E3 Ub ligase that may target proteins for proteasomal degradation.
Triad1 is a predominantly nuclear protein and is highly expressed in mature granulocytes and monocytes. (A) IF of Triad1-myc reveals a nuclear diffuse localization with exclusion of nucleoli in L88/5 cells.53 In cells with strong nuclear staining some cytoplasmic staining is also observed. Phase-contrast image (phase contrast) indicating cellular structures are given at the right. Immunofluorescence of empty vector-transduced cells did not reveal significant staining (not shown). (B) A Triad1-specific affinity-purified antibody detects low endogenous Triad1 expression in CD34+ and T cells and high nuclear expression in mature granulocytes and monocytes. DNA was stained with 4′-6-diamino-2-phenylindole (DAPI) and merged pictures are indicated. Representative results from 1 of 3 unrelated healthy volunteers are indicated. Triad1 IF detects in granulocytes a ring-shaped nuclear sublocalization. Strongest Triad1 expression is observed in the DAPI dull region (arrows) within the lobes of the nuclei. As negative control, cells were stained with preimmune serum taken prior to peptide vaccination, revealing no significant signals (not shown). (C) Intracellular Triad1 was stained on fresh bone marrow (n = 1) or leukapheresis material (n = 1) and analyzed by flow cytometer yielding similar results. The figure shows the mean fluorescence of indicated compartments of the leukapheresis sample. Lowest expression was observed in CD3+ and CD34+ cells. Within the CD14+ fraction lowest expression was observed in the more immature fraction (Mo1) compared with mature monocytes (Mo2). Likewise, in immature myelocytic fractions (Gr1 including promyelocytes and Gr2 including myelocytes/metamyelocytes) lower expression was observed compared with mature granulocytes (Gr3). Cells were costained for Triad1 and CD45 in combination with CD34, CD3, CD14, or CD15. Within the CD14+ and CD15+ populations immature and mature compartments were defined based on CD45 staining and side scatter as described (see also Figure S2).54 Staining with preimmune serum resulted in 30- to 100-fold lower signals (not shown).
Triad1 is a predominantly nuclear protein and is highly expressed in mature granulocytes and monocytes. (A) IF of Triad1-myc reveals a nuclear diffuse localization with exclusion of nucleoli in L88/5 cells.53 In cells with strong nuclear staining some cytoplasmic staining is also observed. Phase-contrast image (phase contrast) indicating cellular structures are given at the right. Immunofluorescence of empty vector-transduced cells did not reveal significant staining (not shown). (B) A Triad1-specific affinity-purified antibody detects low endogenous Triad1 expression in CD34+ and T cells and high nuclear expression in mature granulocytes and monocytes. DNA was stained with 4′-6-diamino-2-phenylindole (DAPI) and merged pictures are indicated. Representative results from 1 of 3 unrelated healthy volunteers are indicated. Triad1 IF detects in granulocytes a ring-shaped nuclear sublocalization. Strongest Triad1 expression is observed in the DAPI dull region (arrows) within the lobes of the nuclei. As negative control, cells were stained with preimmune serum taken prior to peptide vaccination, revealing no significant signals (not shown). (C) Intracellular Triad1 was stained on fresh bone marrow (n = 1) or leukapheresis material (n = 1) and analyzed by flow cytometer yielding similar results. The figure shows the mean fluorescence of indicated compartments of the leukapheresis sample. Lowest expression was observed in CD3+ and CD34+ cells. Within the CD14+ fraction lowest expression was observed in the more immature fraction (Mo1) compared with mature monocytes (Mo2). Likewise, in immature myelocytic fractions (Gr1 including promyelocytes and Gr2 including myelocytes/metamyelocytes) lower expression was observed compared with mature granulocytes (Gr3). Cells were costained for Triad1 and CD45 in combination with CD34, CD3, CD14, or CD15. Within the CD14+ and CD15+ populations immature and mature compartments were defined based on CD45 staining and side scatter as described (see also Figure S2).54 Staining with preimmune serum resulted in 30- to 100-fold lower signals (not shown).
Triad1 is a Ub ligase. (A) Human Triad1 interacts with UbcH7 in yeast 2-hybrid assays as shown by growth on double histidine and adenine selection (left). The Triad1 mutants H158A and C161A fail to bind to UbcH7 (right). Used bait (left) and prey-constructs (right) are indicated. EV indicates empty vector control. (B) GST-Triad1 efficiently captures UbcH7 from human NB4 cell lysates, whereas GST alone does not. (C) Cotransfection of Triad1-myc and Flag-Ub followed by myc immunoprecipitation (IP) and Flag staining detects a smear of ubiquitinated proteins [X-(ub)n] bound to Triad1. The amount of ubiquitinated proteins bound to Triad1 was more intense when cells were treated with a proteasome inhibitor (12.5 μM MG132). (D) Cotransfection of GFP-Triad1 and Flag-Ub followed by GFP IP. Immunoprecipitates were split in 2 parts and run on the same gel as indicated and independently stained for Triad1 and Flag, respectively, revealing GFP-Triad1 (arrowhead) and a smear of ubiquitinated proteins ([X-(ub)n], including proteins with sizes smaller compared with Triad1. Asterisk represents immunoglobulin H (IgH). (E) In vitro Ub ligase assay. Reticulocyte fraction IIA, Triad1 (0.03 μg) or 10*Triad1 (0.3 μg) without Ub (-), with wild-type Ub (WT) or UbK48-only (K48) were incubated followed by staining of ubiquitinated proteins. Note the marked increase of both wild-type and K48-only ubiquitinated species after addition of Triad1. Increased dose of Triad1 correlates with increase in ubiquitinated proteins. Equal protein transfer between the tested conditions is indicated by the Ub band (bottom).
Triad1 is a Ub ligase. (A) Human Triad1 interacts with UbcH7 in yeast 2-hybrid assays as shown by growth on double histidine and adenine selection (left). The Triad1 mutants H158A and C161A fail to bind to UbcH7 (right). Used bait (left) and prey-constructs (right) are indicated. EV indicates empty vector control. (B) GST-Triad1 efficiently captures UbcH7 from human NB4 cell lysates, whereas GST alone does not. (C) Cotransfection of Triad1-myc and Flag-Ub followed by myc immunoprecipitation (IP) and Flag staining detects a smear of ubiquitinated proteins [X-(ub)n] bound to Triad1. The amount of ubiquitinated proteins bound to Triad1 was more intense when cells were treated with a proteasome inhibitor (12.5 μM MG132). (D) Cotransfection of GFP-Triad1 and Flag-Ub followed by GFP IP. Immunoprecipitates were split in 2 parts and run on the same gel as indicated and independently stained for Triad1 and Flag, respectively, revealing GFP-Triad1 (arrowhead) and a smear of ubiquitinated proteins ([X-(ub)n], including proteins with sizes smaller compared with Triad1. Asterisk represents immunoglobulin H (IgH). (E) In vitro Ub ligase assay. Reticulocyte fraction IIA, Triad1 (0.03 μg) or 10*Triad1 (0.3 μg) without Ub (-), with wild-type Ub (WT) or UbK48-only (K48) were incubated followed by staining of ubiquitinated proteins. Note the marked increase of both wild-type and K48-only ubiquitinated species after addition of Triad1. Increased dose of Triad1 correlates with increase in ubiquitinated proteins. Equal protein transfer between the tested conditions is indicated by the Ub band (bottom).
Triad1 is targeted by the Ub-proteasome pathway
In addition to their substrates, many Ub ligases are ubiquitinated themselves.13 Ubiquitination of Triad1 was studied by coexpressing Triad1 and His-tagged Ub (His-Ub) followed by selection of His-tagged proteins under denaturing conditions. Staining with a Triad1-specific antibody detected discrete bands with higher molecular weight compared with unmodified Triad1, indicating that Triad1 is ubiquitinated (Figure 5A). Proteasome inhibition resulted in an increase in ubiquitinated Triad1 forms, indicating that ubiquitinated Triad1 is subject to proteasomal degradation. Similar results were obtained when Triad1 and Flag-Ub were coexpressed followed by Flag IP under nondenaturing conditions (Figure 5B). The discrete Triad1 Ub-modified forms observed in these IPs combined with the smear of ubiquitinated proteins detected on Triad1 IP when cotransfected with Ub as shown in Figure 4C-D further indicate that, apart from being ubiquitinated itself, Triad1 also binds ubiquitinated proteins.
Triad1 is ubiquitinated and targeted for degradation by the proteasome. (A) Cotransfection of Triad1 and His-Ub followed by selection of ubiquitinated proteins using His-select beads under denaturing conditions followed by Triad1 staining detects multiple discrete ubiquitinated Triad1 forms [Triad1-(ub)n]. Proteasome inhibition (12.5 μM MG132) results in an increase in Triad1 ubiquitination. Left lane contains positive control containing lysate from Triad1 (arrowhead) transduced 293T cells. Asterisk indicates an a-specific band. (B) Cotransfection of Flag-Ub and Triad1 followed by Flag IP and staining for Triad1 detects multiple ubiquitinated Triad1 species (Triad1-(ub)n) that were more intense when cells were treated with a proteasome inhibitor prior to cell lysis. Note that unmodified Triad1 (arrow) coimmunoprecipitates with ubiquitinated Triad1.
Triad1 is ubiquitinated and targeted for degradation by the proteasome. (A) Cotransfection of Triad1 and His-Ub followed by selection of ubiquitinated proteins using His-select beads under denaturing conditions followed by Triad1 staining detects multiple discrete ubiquitinated Triad1 forms [Triad1-(ub)n]. Proteasome inhibition (12.5 μM MG132) results in an increase in Triad1 ubiquitination. Left lane contains positive control containing lysate from Triad1 (arrowhead) transduced 293T cells. Asterisk indicates an a-specific band. (B) Cotransfection of Flag-Ub and Triad1 followed by Flag IP and staining for Triad1 detects multiple ubiquitinated Triad1 species (Triad1-(ub)n) that were more intense when cells were treated with a proteasome inhibitor prior to cell lysis. Note that unmodified Triad1 (arrow) coimmunoprecipitates with ubiquitinated Triad1.
Ub ligase activity of Triad1 inhibits clonogenic growth of primary murine bone marrow cells. (A) Empty vector (EV) or Triad1 retrovirally transduced murine bone marrow cells with increasing expression (Triad1 I, II, and III, respectively) were used in CFU-GM, revealing a Triad1 concentration-dependent inhibition of clonogenic growth (n = 3). Triad1 staining using lysates from 100 000 transduced bone marrow cells revealed no detectable levels in EV-transduced cells and increasing expression in cells with increasing GFP positivity (fractions II and III, respectively). NC indicates negative control. (B) Morphologic analysis of cells grown in CFU-GM shows a modest increase in the percentage of granulocytes and decrease in erythroblasts on Triad1 expression. (C) Triad1 RING mutants H158A and C161A do not inhibit clonogenic growth, showing that the N-terminal RING finger is essential for the clonogenic inhibition exerted by wild-type Triad1 (n = 2). Comparable expression of wild-type Triad1 and mutants was shown by Western blotting using lysates from 100 000 transduced bone marrow cell (equal loading was checked by Ponceau staining, not shown). (D) Treatment of murine bone marrow cells grown in liquid medium with MG132 indicates maximal tolerated dose with limited toxicity of 1 × 10-8 M. Cells (5000) were seeded in 96 wells and cultured for 3 days. Relative cell numbers were determined by flow cytometry using fluorescent beads (Beckman Coulter). (E) Treatment of primary murine bone marrow cells with 1 × 10-8 M MG132 for 2 days resulted in an increase in ubiquitinated proteins (x-(ub)n). (F) Proteasome activity measurements (indicated as fluorescence units [FUs]) on lysates taken from primary murine bone marrow cells that were treated with 1 × 10-8 M MG132 for 1 or 2 days showed clear proteasomal inhibition. (G) Proteasome inhibition (MG132) relieves the suppressive effect of Triad1 in CFU-GM (n = 3). MG132 was added to cells prior to cell seeding. For all tested conditions observed numbers of colonies were indicated relative to number of empty vector (EV) obtained colonies (set at 100). (H) Treatment of Triad1-transduced cells with independent proteasome inhibitors counteracted the suppressive effects of Triad1 on colony formation. The number of obtained colonies of mock-treated Triad1 transduced cells was set at 100, and relative colony numbers obtained for cells treated with indicated proteasome inhibitors are indicated (1 × 10-8 M MG132 [n = 3], 1 × 10-9 M PS341 [n = 2], 5 × 10-10 M epoxomycin [epox; n = 1], and 1 × 10-8 M lactacystin [lacta; n = 1]). Error bars indicate standard deviations.
Ub ligase activity of Triad1 inhibits clonogenic growth of primary murine bone marrow cells. (A) Empty vector (EV) or Triad1 retrovirally transduced murine bone marrow cells with increasing expression (Triad1 I, II, and III, respectively) were used in CFU-GM, revealing a Triad1 concentration-dependent inhibition of clonogenic growth (n = 3). Triad1 staining using lysates from 100 000 transduced bone marrow cells revealed no detectable levels in EV-transduced cells and increasing expression in cells with increasing GFP positivity (fractions II and III, respectively). NC indicates negative control. (B) Morphologic analysis of cells grown in CFU-GM shows a modest increase in the percentage of granulocytes and decrease in erythroblasts on Triad1 expression. (C) Triad1 RING mutants H158A and C161A do not inhibit clonogenic growth, showing that the N-terminal RING finger is essential for the clonogenic inhibition exerted by wild-type Triad1 (n = 2). Comparable expression of wild-type Triad1 and mutants was shown by Western blotting using lysates from 100 000 transduced bone marrow cell (equal loading was checked by Ponceau staining, not shown). (D) Treatment of murine bone marrow cells grown in liquid medium with MG132 indicates maximal tolerated dose with limited toxicity of 1 × 10-8 M. Cells (5000) were seeded in 96 wells and cultured for 3 days. Relative cell numbers were determined by flow cytometry using fluorescent beads (Beckman Coulter). (E) Treatment of primary murine bone marrow cells with 1 × 10-8 M MG132 for 2 days resulted in an increase in ubiquitinated proteins (x-(ub)n). (F) Proteasome activity measurements (indicated as fluorescence units [FUs]) on lysates taken from primary murine bone marrow cells that were treated with 1 × 10-8 M MG132 for 1 or 2 days showed clear proteasomal inhibition. (G) Proteasome inhibition (MG132) relieves the suppressive effect of Triad1 in CFU-GM (n = 3). MG132 was added to cells prior to cell seeding. For all tested conditions observed numbers of colonies were indicated relative to number of empty vector (EV) obtained colonies (set at 100). (H) Treatment of Triad1-transduced cells with independent proteasome inhibitors counteracted the suppressive effects of Triad1 on colony formation. The number of obtained colonies of mock-treated Triad1 transduced cells was set at 100, and relative colony numbers obtained for cells treated with indicated proteasome inhibitors are indicated (1 × 10-8 M MG132 [n = 3], 1 × 10-9 M PS341 [n = 2], 5 × 10-10 M epoxomycin [epox; n = 1], and 1 × 10-8 M lactacystin [lacta; n = 1]). Error bars indicate standard deviations.
Ub ligase activity of Triad1 regulates myelopoiesis
To determine the biologic relevance of the induction of Triad1 during myeloid differentiation, we tested whether Triad1 expression affected the clonogenic growth of primary immature murine bone marrow cells in CFU-GM assays. Primary murine bone marrow cells were transduced with pLZRS-derived viruses containing a GFP-IRES (internal ribosome entry site)-Triad1 sequence allowing for separate GFP and Triad1 expression. Fractions with low, intermediate and high GFP expression were sorted. GFP expression correlated with Triad1 expression as shown by Western blot analysis (Figure 6A; Figure S3A). Compared with empty vector controls (GFP alone), Triad1 expression resulted in a concentration-dependent inhibition of clonogenic growth. Although low Triad1 expression did not inhibit clonogenic growth, intermediate Triad1 expression resulted in an inhibition of greater than 60%, whereas highest Triad1 expression resulted in a strong inhibition of clonogenic growth of greater than 80% (Figure 6A). In addition, colonies derived from high and intermediate Triad1-expressing cells were significantly smaller compared with empty vector controls (Figure S3B). To determine whether Triad1 affects myeloid differentiation, colonies were recovered from semisolid medium and morphologically analyzed. This revealed a modest Triad1-dependent increase in the percentage of granulocytes and decrease in erythroblasts compared with empty vector transduced cells (Figure 6B).
The observed inhibition of clonogenic growth suggests a role for Triad1 in the development of immature-committed murine bone marrow cells toward mature myeloid cells. In many Ub ligases the RING finger is essential for Ubc binding as well as Ub ligase activity.20 Mutations in RING fingers abrogating the interaction with Ubcs result in impaired ubiquitination. To test whether the Ub ligase activity of Triad1 was involved in the clonogenic inhibition of myeloid progenitor cells, we introduced the H158A and C161A RING finger mutations that abrogate UbcH7 binding in full-length Triad1-coding sequences in the retroviral vectors and used these constructs in CFU-GM colony assays. Strikingly, in contrast to normal Triad1, expression of the Triad1 point mutants did not affect the clonogenic growth of primary immature cells (Figure 6C). Because the mutant proteins were expressed at levels comparable to wild-type Triad1 as shown by Western blot analysis, this indicates that the N-terminal RING finger of Triad1 plays an important role in the regulation of proliferation and/or differentiation of myeloid progenitor cells. In E3 Ub ligases, the RING finger structure has been shown to be essential for Ub ligase activity.20 Because UbcH7 can catalyze the formation of Ub chains resulting in protein degradation and the Triad1 RING finger mutations abrogate the inhibitory effect on clonogenic growth, our data suggest that the ubiquitination and subsequent proteasomal degradation of Triad1 target proteins results in clonogenic inhibition. To test this, we determined whether the inhibition of clonogenic growth exerted by Triad1 could be relieved by proteasome inhibition. For these experiments, 1 × 10-8 M of the proteasome inhibitor MG132 was used because at this concentration limited toxicity on empty vector-transduced cells was observed in liquid assays (Figure 6D). At this concentration, the bone marrow cells showed an increase in ubiquitinated species and decreased proteasomal activity (Figures 6E-F, respectively). Interestingly, proteasome inhibition partially reversed the Triad1-induced suppression of colony formation (Figure 6G). When corrected for the suppressive effects of MG132 on empty vector-transduced cells, a greater than 2-fold increase in Triad1-positive colonies was observed when the proteasome was inhibited (Figure 6H). Moreover, the colonies derived from MG132-treated Triad1-transduced cells were significantly larger as compared with nontreated cells (Figure S3B). In addition to the proteasome, MG132 inhibits other proteases. To rule out the possibility that the increase in colony numbers of Triad1-transduced cells after treatment with MG132 depended on other mechanisms rather then inhibition of the 26S proteasome, we repeated the experiments using other more specific proteasome inhibitors, including PS-341, epoxomycin, and lactacystin. This yielded similar results as observed for MG132 (Figure 6H). Thus, the growth inhibitory effect exerted by Triad1 relies on the N-terminal RING finger and can be counteracted by proteasome inhibition, indicating that degradation of Triad1 targets are implicated in myelopoiesis.
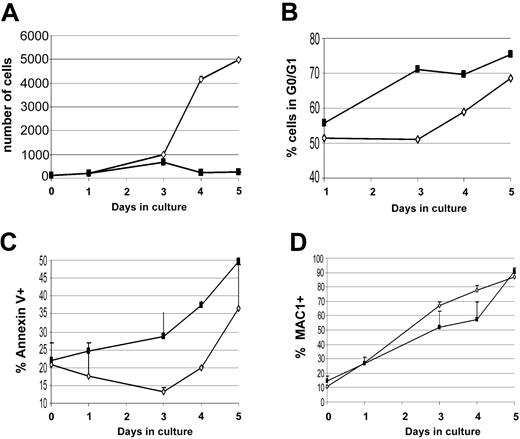
To further investigate how Triad1 affected CFU-GM colony formation, we cultured Triad1 retrovirally transduced bone marrow cells in liquid medium and measured proliferation, cell cycle progression (DNA histograms), differentiation, and apoptosis. Compared with empty vector control, Triad1 expression resulted in a dramatic inhibition of proliferation (Figure 7A). This was accompanied by a marked increase in the percentage of cells in G0/G1 phase, and an increase in annexin V positivity (Figures 7B-C). In these experiments no significant changes in myeloid differentiation as indicated by Mac1 positivity was observed (Figure 7D). Thus, the inhibition of clonogenic growth by Triad1 is probably caused by a cell-cycle arrest associated with increased apoptosis. From these findings we conclude that the ubiquitination and degradation of Triad1 target proteins contributes to the development of early committed myeloid progenitors toward mature monocytes and granulocytes.
Triad1 inhibits proliferation of committed myeloid progenitors in liquid cultures. (A) Proliferation of transduced bone marrow cells in liquid medium, indicating dramatic growth inhibition of Triad1-transduced cells compared with empty vector-transduced cells. Cell number at start of culture was set at 100, and relative numbers during culture time (days) were plotted. ⋄ and ▪ represent, respectively, empty vector- and Triad1-transduced cells. (B) Reduced growth rate of Triad1-transduced cells is accompanied by an increase in the percentage of cells in G0/G1 phase (DNA histograms were determined by flow cytometry using propidium iodide staining; Figure S3C). (C) Triad1-transduced cells grown in liquid medium show enhanced annexin V positivity compared with empty vector-transduced cells. (D) Triad1 expression does not result in alterations in the percentage of Mac1-positive cells compared with empty vector-transduced cells. Error bars indicate standard deviations.
Triad1 inhibits proliferation of committed myeloid progenitors in liquid cultures. (A) Proliferation of transduced bone marrow cells in liquid medium, indicating dramatic growth inhibition of Triad1-transduced cells compared with empty vector-transduced cells. Cell number at start of culture was set at 100, and relative numbers during culture time (days) were plotted. ⋄ and ▪ represent, respectively, empty vector- and Triad1-transduced cells. (B) Reduced growth rate of Triad1-transduced cells is accompanied by an increase in the percentage of cells in G0/G1 phase (DNA histograms were determined by flow cytometry using propidium iodide staining; Figure S3C). (C) Triad1-transduced cells grown in liquid medium show enhanced annexin V positivity compared with empty vector-transduced cells. (D) Triad1 expression does not result in alterations in the percentage of Mac1-positive cells compared with empty vector-transduced cells. Error bars indicate standard deviations.
Discussion
The modification of proteins with ubiquitin plays an important role in a variety of basic cellular processes, including cell division, differentiation, and apoptosis. Protein poly-ubiquitination linked through lysine 48 of Ub results in degradation of tagged proteins by the 26S proteasome. Proteins are ubiquitinated by E2-E3 Ub ligase complexes. The E3 Ub ligases play a key role in this process because these enzymes determine which cellular proteins are ubiquitinated. We report here that the E3 Ub ligase Triad1 is induced during both granulocytic and monocytic differentiation (Figures 1, 2, 3). Triad1 contains 2 RING finger structures. By providing an interface for interaction with E2 Ub-conjugating enzymes, the RING finger constitutes an essential domain in many E3 Ub ligases. In line with this, we found that Triad1 physically interacts with the E2 Ub-conjugating enzyme UbcH7 through its N-terminal RING finger and to ubiquitinated proteins (Figure 4A-D). Moreover, in vitro ubiquitination experiments showed that Triad1 supports ubiquitination of both wild-type and UbK48-only (Figure 4E). Ub chains linked through lysine 48 are efficiently recognized by the proteasome, resulting in substrate degradation. This indicates that Triad1 may mark proteins for proteasomal degradation.
Triad1 exhibits a predominantly nuclear staining in all cell types tested (Figure 3). In stromal cells (L88/5) and in primary monocytes the nuclear staining is diffuse, whereas a ring-shaped localization is observed within the nuclear lobes of primary granulocytes (Figure 3). Although these data may suggest that Triad1 substrates are primarily nuclear proteins, a protein-interaction map of the Drosophila proteome34 revealed that Drosophila Triad1 (CG5709) binds in addition to Ubc7 (CG12799) to both cytoplasmic and nuclear proteins. These include cytoplasmic orthologs of signaling molecule Traf3 (TNF [tumor necrosis factor] receptor-associated factor-3) involved in TNF-receptor-mediated signal transduction and E3 ub ligase Hakai involved in ubiquitination of E-cadherin (respectively, CG4394 and CG10263), but also nuclear proteins, including the ortholog of the transcription factor HOX11L2 (CG7937) implicated in acute lymphoblastic leukemia development and the ortholog of the helix-loop-helix transcription factors NHLH1 and NHLH2 (CG3052) implicated in neurogenesis.35-38
Forced Triad1 expression in primary murine bone marrow cells inhibited colony formation in CFU-GM assays. Two N-terminal RING finger point mutants that are unable to bind the E2 UbcH7 did not inhibit clonogenic growth, indicating that the N-terminal RING finger of Triad1 is involved in the inhibition of clonogenic growth (Figure 6A,C). As the RING finger is involved in the catalytic activity (ie, Ub conjugation)13,20 in virtually all E3 RING Ub ligases, this suggested that the ubiquitination of Triad1 target proteins caused the inhibition of clonogenic growth. In conjunction with E3 Ub ligases, UbcH7 is known to catalyze the formation of Ub chains that result in proteasomal degradation of tagged proteins. To further study the clonogenic inhibition of Triad1 in CFU-GM assays we repeated the experiments in the presence of proteasome inhibitors and found that this partially reversed the effects of Triad1 (Figure 6G-H). We conclude that proteasomal degradation of Triad1 targets plays an important role in myelopoiesis. In liquid cultures of transduced bone marrow cells, Triad1 expression resulted in a strong inhibition of proliferation whereas differentiation remained unaltered. The inhibition of proliferation was associated with a G0/G1 accumulation and increased apoptosis (Figure 7).
Triad1 is a member of the class of TRIAD (or RING-IBR-RING) proteins.31,32 Since the description of this class of proteins, several TRIAD proteins have been reported to function as E3 Ub ligase, including Triad3, Parkin, Parc (pulmonary and activation-regulated chemokine), and p53RFP (p53-associated Parkin-like cytoplasmic protein).39-42 These Ub ligases are involved in various biologic processes, including immunity, neuronal development, and p53 functioning. Triad3 targets Toll-like receptors 4 and 9 for ubiquitination.40 By sensing antigens, Toll-like receptors play a role in both innate and adapted immunity. Ligand binding of these receptors result in nuclear factor[kappa]B (NF-κB) activation and Triad3 regulates the ubiquitination and concomitant turnover of these receptors, thereby controlling the intensity and duration of Toll-like receptor function.40 Another well-studied TRIAD protein is Parkin, which is, like Triad1, highly expressed in fetal brain and involved in neuronal development.43 Congenital mutations in Parkin cause juvenile parkinsonism, and acquired loss-of-heterozygosity of Parkin has been described in various human malignancies.44-48 The TRIAD proteins Parc and p53RFP are involved in p53 function.41,42 Parc functions as E3 Ub ligase that binds to p53. Although Parc does not ubiquitinate p53, it sequesters this protein in the cytoplasm. The other TRIAD protein, p53RFP, is a p53 target gene. p53RFP binds to the cyclin-dependent kinase inhibitor p21waf1. Because p53RFP overexpression results in reduced p21waf1 levels, p53RFP might function as Ub ligase for p21waf1.41 The majority of the reported TRIAD proteins have been shown to interact with UbcH7, which is in line with the observation that Triad1 also interacts with UbcH7. By using less stringent conditions in yeast 2-hybrid assays, we also observed an interaction between Triad1 and UbcH13 (not shown). Interestingly, UbcH13 catalyzes the formation of Ub chains (linked through lysine 63) that are not recognized by the proteasome but that alter the activity of proteins.49-51 Two recent studies have shown that the TRIAD protein Parkin also binds to UbcH13 in addition to UbcH7 and that it can catalyze the formation of both lysine 48- and 63-linked Ub chains.30,52 As Triad1 may bind UbcH13 it will be important to test whether it also exhibits activity that supports ubiquitination linked through lysine 63 of Ub. Nevertheless, the inhibition of clonogenic growth exerted by Triad1 described here depends at least for a part on targeting proteins for proteasomal degradation, because proteasome inhibition partially reversed the effects of Triad1. To further understand how the Ub-proteasome pathway contributes to myelopoiesis, it will be of great interest to identify which cellular proteins are targeted for ubiquitination by Triad1.
Prepublished online as Blood First Edition Paper, August 23, 2005; DOI 10.1182/blood-2005-04-1450.
Supported by grants from the Dutch Cancer Society (J.A.F.M., L.V.E., C.A.J.E.-V., and G.N.), the Royal Netherlands Academy of Sciences KNAW (J.H.J.), and the Vanderes Foundation (B.A.V.D.R.).
C.A.J.E.-V. performed the subtractive polymerase chain reaction (PCR) and Northern blot analysis; B.A.V.D.R. cloned Triad1 and performed database analyses; J.A.F.M., G.N., and A.M. performed granulocyte-macrophage colony-forming unit (CFU-GM) assays; J.A.F.M. and B.A.V.D.R. cloned and sequenced Triad1 constructs; B.A.V.D.R., J.A.F.M., and L.V.E. generated the Triad1 antibody and performed Triad1 immunofluorescence and real-time PCR; J.A.F.M. performed yeast 2-hybrid studies, Western blot analyses, in vitro ubiquitination, proteasome activity, and liquid culture studies; J.A.F.M., T.D.W., B.L., J.H.J., and B.A.V.D.R. designed the studies; and B.A.V.D.R. wrote the paper.
J.H.J. and B.A.V.D.R. contributed equally to this work.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Martin Scheffner for valuable discussions and help in setting up ubiquitination assays and for providing us with pcDNA3.1-His-Ub, Herman Swarts for generating Triad1 Baculovirus, Dr Philip James for 2 hybrid vectors, Dr Yukio Okano for Ubc yeast 2-hybrid bait constructs, Dr Toshiaki Suzuki for pcDNA3.1-Flag-Ub, Dr G. Nolan for pLZRS and the phoenix cell line ϕ-NX-A.

![Figure 4. Triad1 is a Ub ligase. (A) Human Triad1 interacts with UbcH7 in yeast 2-hybrid assays as shown by growth on double histidine and adenine selection (left). The Triad1 mutants H158A and C161A fail to bind to UbcH7 (right). Used bait (left) and prey-constructs (right) are indicated. EV indicates empty vector control. (B) GST-Triad1 efficiently captures UbcH7 from human NB4 cell lysates, whereas GST alone does not. (C) Cotransfection of Triad1-myc and Flag-Ub followed by myc immunoprecipitation (IP) and Flag staining detects a smear of ubiquitinated proteins [X-(ub)n] bound to Triad1. The amount of ubiquitinated proteins bound to Triad1 was more intense when cells were treated with a proteasome inhibitor (12.5 μM MG132). (D) Cotransfection of GFP-Triad1 and Flag-Ub followed by GFP IP. Immunoprecipitates were split in 2 parts and run on the same gel as indicated and independently stained for Triad1 and Flag, respectively, revealing GFP-Triad1 (arrowhead) and a smear of ubiquitinated proteins ([X-(ub)n], including proteins with sizes smaller compared with Triad1. Asterisk represents immunoglobulin H (IgH). (E) In vitro Ub ligase assay. Reticulocyte fraction IIA, Triad1 (0.03 μg) or 10*Triad1 (0.3 μg) without Ub (-), with wild-type Ub (WT) or UbK48-only (K48) were incubated followed by staining of ubiquitinated proteins. Note the marked increase of both wild-type and K48-only ubiquitinated species after addition of Triad1. Increased dose of Triad1 correlates with increase in ubiquitinated proteins. Equal protein transfer between the tested conditions is indicated by the Ub band (bottom).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/13/10.1182_blood-2005-04-1450/2/m_zh80240588130004.jpeg?Expires=1763495772&Signature=Ai4ULoZqJ7FjjURpUSkrYqS3dfTNR6R4Zv1U5wkTMsk6jEbkUpU~HOI~P1974SbI1VGndOkFqsq72TlUs9Iq01kw2t7Y8m7Qn7ffYf9NUzMG5WkNpKxYNohxuKc7jeLJaWCfZDLLCjO5Crx~u7blal5enyjw4vSJKci9eTyiiBzXlgimmyK18sqzcdMg7gv3-CF3niedKCX9DwYcivAqGpv2ifhaDBPdNgStvPSrF5w52Lw4ni8jkWTFHcK2IFR0UimrUfQ8CrLs7UOb9hdEkNHGYfcwxm5WK~w7XdCApQ1FCwZTNdD2NlPo4WoJXuOFkgNI0bUQy2UnGJZ7eD95Uw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Triad1 is ubiquitinated and targeted for degradation by the proteasome. (A) Cotransfection of Triad1 and His-Ub followed by selection of ubiquitinated proteins using His-select beads under denaturing conditions followed by Triad1 staining detects multiple discrete ubiquitinated Triad1 forms [Triad1-(ub)n]. Proteasome inhibition (12.5 μM MG132) results in an increase in Triad1 ubiquitination. Left lane contains positive control containing lysate from Triad1 (arrowhead) transduced 293T cells. Asterisk indicates an a-specific band. (B) Cotransfection of Flag-Ub and Triad1 followed by Flag IP and staining for Triad1 detects multiple ubiquitinated Triad1 species (Triad1-(ub)n) that were more intense when cells were treated with a proteasome inhibitor prior to cell lysis. Note that unmodified Triad1 (arrow) coimmunoprecipitates with ubiquitinated Triad1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/13/10.1182_blood-2005-04-1450/2/m_zh80240588130005.jpeg?Expires=1763495772&Signature=catidnyduJ~j0MD247OSNXt7jx5cP6KwSJqNCRBVPYGlgV-SBDwJ26F9mlWoBSX3GPVzX4Bw~z2uC1ZJ9KJc8kfPdom6ckJE9OKEVbu1ctSHlI5GFDC5LiG3M8MxzZzILhISbyyF2Dk4K2vusB3MxV66~4YdSVDnBd1TGHJvMxgX-RqgrbzCz32cGGa9nEImBi~aQBSK~zqkp2XjP2trbjZEXlO7yS0OBrJt30S3zyuSWkI1Sp2bhM5cDeQTFstAE8Oc91qRYSLcu5lz-~U83E98wEb5wSEOSWpvFrSRIGq2ZQ2iCmZeBL9xpAHTR3NNDa8YR~HFjMCev3ALI5-z8Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Ub ligase activity of Triad1 inhibits clonogenic growth of primary murine bone marrow cells. (A) Empty vector (EV) or Triad1 retrovirally transduced murine bone marrow cells with increasing expression (Triad1 I, II, and III, respectively) were used in CFU-GM, revealing a Triad1 concentration-dependent inhibition of clonogenic growth (n = 3). Triad1 staining using lysates from 100 000 transduced bone marrow cells revealed no detectable levels in EV-transduced cells and increasing expression in cells with increasing GFP positivity (fractions II and III, respectively). NC indicates negative control. (B) Morphologic analysis of cells grown in CFU-GM shows a modest increase in the percentage of granulocytes and decrease in erythroblasts on Triad1 expression. (C) Triad1 RING mutants H158A and C161A do not inhibit clonogenic growth, showing that the N-terminal RING finger is essential for the clonogenic inhibition exerted by wild-type Triad1 (n = 2). Comparable expression of wild-type Triad1 and mutants was shown by Western blotting using lysates from 100 000 transduced bone marrow cell (equal loading was checked by Ponceau staining, not shown). (D) Treatment of murine bone marrow cells grown in liquid medium with MG132 indicates maximal tolerated dose with limited toxicity of 1 × 10-8 M. Cells (5000) were seeded in 96 wells and cultured for 3 days. Relative cell numbers were determined by flow cytometry using fluorescent beads (Beckman Coulter). (E) Treatment of primary murine bone marrow cells with 1 × 10-8 M MG132 for 2 days resulted in an increase in ubiquitinated proteins (x-(ub)n). (F) Proteasome activity measurements (indicated as fluorescence units [FUs]) on lysates taken from primary murine bone marrow cells that were treated with 1 × 10-8 M MG132 for 1 or 2 days showed clear proteasomal inhibition. (G) Proteasome inhibition (MG132) relieves the suppressive effect of Triad1 in CFU-GM (n = 3). MG132 was added to cells prior to cell seeding. For all tested conditions observed numbers of colonies were indicated relative to number of empty vector (EV) obtained colonies (set at 100). (H) Treatment of Triad1-transduced cells with independent proteasome inhibitors counteracted the suppressive effects of Triad1 on colony formation. The number of obtained colonies of mock-treated Triad1 transduced cells was set at 100, and relative colony numbers obtained for cells treated with indicated proteasome inhibitors are indicated (1 × 10-8 M MG132 [n = 3], 1 × 10-9 M PS341 [n = 2], 5 × 10-10 M epoxomycin [epox; n = 1], and 1 × 10-8 M lactacystin [lacta; n = 1]). Error bars indicate standard deviations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/13/10.1182_blood-2005-04-1450/2/m_zh80240588130006.jpeg?Expires=1763495772&Signature=uA7UaA-JzDRQOWTyTzbQK0gEEFLWkEb7iXbUybkGFWd2xSogsmBeh3QjZ5VTiuF5T5Oy6qmBiO1Jt8Aid69OHJykCWmuwgTNmKTMcsbxMyXXydToXDPK60NmvZ5Btfee2dakyQ~qc~M3NWnLeb5m2-ZYh1OQpI465qTSUlv1YPo2VknV65Nc6v~9mYxx36cnKMn-g3DnkHjTG2CluQFvBSAZGkGGXNejGB9q0loi97hED0xLXtQfrky8pQJ6oHhNHhKjMNUuT8-CtQcdky8JtjJwXqOFZ6BHpEjv3SKDpGpVofkYYKoooI4aplGMKE-A2AZSbN~cx7o~5gNCuhGIsg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal