Abstract
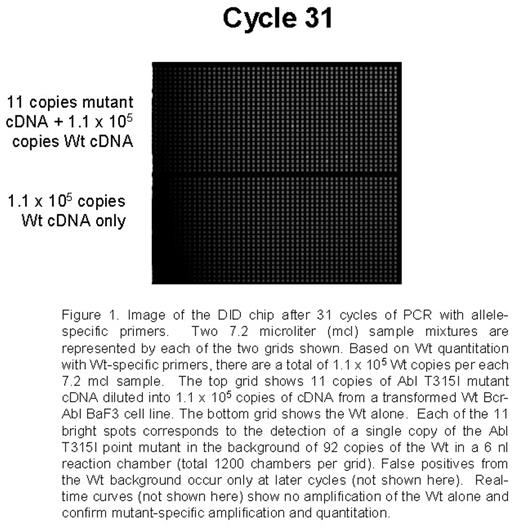
Point mutations in the Abl tyrosine kinase domain are the main mechanism of secondary resistance to imatinib mesylate (IM) therapy in chronic myeloid leukemia (CML) patients. Current mutation detection approaches including direct sequencing, DHPLC, and SSCP range in sensitivity from 5% to 20%. Detection of mutations in early treatment and possibly pre-treatment samples could allow patient therapy to be altered before resistance is detected cytogenetically or disease progression. A sensitive assay using a novel nanofluidic chip designed for digital isolation and detection (DID) by mutant allele-specific amplification was used to detect rare mutant copies of Abl in a very high background of wild-type (Wt) Abl cDNA. The DID chip partitions a nucleic acid sample into thousands of isolated reaction chambers, thereby enriching rare targets and enabling absolute quantitative, specific and sensitive detection of rare sequences and mutations. Each sample mix of 7.2 mcl containing the necessary reagents for a quantitative PCR 5′-nuclease assay is filled into a nanofluidic network. The sample is partitioned into 1,200 chambers using microfluidic valves fabricated by multilayer-soft-lithography. Each 6 nl reaction chamber contains a fraction of the original nucleic acid sample and reagents for detecting the target of interest. Since the mutant target is rare, the majority of partitions will not contain the mutant, while a small number of partitions will contain just one mutant copy. Single copy detection using the DID chip is reproducible and reliable. For initial proof of concept experiments a set of assays able to detect a group of p-loop mutations and the T315I mutation were optimized using model mixtures. Studies by French centers and Australian groups suggest that these mutations confer worse prognosis and the T315I mutation is refractory to other targeted therapies. Mutation-specific PCR assays were developed for the following mutations: G250E, Q252H (2 substitutions), Y253F, E255K, E255V, and T315I. Initial experiments utilized mutated Bcr-Abl cDNA and plasmids serially diluted into the background of 105 equivalents of Wt Bcr-Abl cDNA from a transformed BaF3 cell line. As few as 9-88 copies were specifically and reliably detected in each of the mutation-specific assays in the background of 105 Wt copies. A non-extendable blocker primer for the Wt and streptavidin beads are also being tested to enrich for mutants prior to PCR. Wt Abl is also quantified to allow for the determination of percent Abl mutated in the assays applied to clinical samples.
Conclusion: This method utilizes a novel platform to provide highly sensitive and specific detection of known point mutations and can be applied to early clinical samples. The assay is unique in that it can be extended to a large group of mutations as detection is achieved in very small amounts of rare samples partitioned into thousands of wells prior to PCR.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal