Abstract
Human parvovirus B19 (B19) infects human erythroid cells expressing P antigen. However, some cell lines that were positive for P antigen failed to bind B19, whereas some cell lines had an ability to bind B19 despite undetectable expression of P antigen. We here demonstrate that B19 specifically binds with Ku80 autoantigen on the cell surface. Furthermore, transfection of HeLa cells with the gene of Ku80 enabled the binding of B19 and allowed its entry into cells. Moreover, reduction of cell-surface expression of Ku80 in KU812Ep6 cells, which was a high-sensitive cell line for B19 infection, by short interfering RNA for Ku80 resulted in the marked inhibition of B19 binding in KU812Ep6 cells. Although Ku80 originally has been described as a nuclear protein, human bone marrow erythroid cells with glycophorin A or CD36, B cells with CD20, or T cells with CD3 were all positive for cell-surface expression of Ku80. B19 infection of KU812Ep6 cells and bone marrow cells was inhibited in the presence of anti-Ku80 antibody. Our data suggest that Ku80 functions as a novel coreceptor for B19 infection, and this finding may provide an explanation for the pathologic immunity associated with B19 infection.
Introduction
Human parvovirus B19 (B19) infects erythroid-lineage cells through P antigen and causes various clinical symptoms such as erythema infectiosum, anemia, polyarthritis, or fetal hydrops in humans.1,2 The cellular receptor for B19 infection has been regarded as blood group P antigen based on the failure of B19 infection in a patient with an hereditary defect of P antigen.3 However, the target cells of B19 may be not be exclusively P-antigen–positive erythroid-lineage cells, as illustrated by the poor relationship between P antigen expression levels and the efficiency of B19 infection4 or the failure of B19 binding to globoside.5 Recently, Weigel-Kelley et al described the role of α5β1 integrin as the cellular coreceptor for B19 infection.6 The notion that B19 receptor is not solely P antigen may be compatible with clinical findings that B19 has been detected in mononuclear cells of blood or tonsils with acute or prolonged B19 infection.7,8 Also, following B19 infection, the numbers of peripheral blood lymphocytes may decrease despite undetectable levels of P antigen on their cell surface.9,10 Finally, autoimmune-like phenomena including antinuclear antibodies, rheumatoid factors, or antiphospholipid antibodies are often associated with B19 infection,8,11 and the levels of tumor necrosis factor α (TNF-α) and interferon γ (IFN-γ) secreted from macrophages or T cells are elevated during acute or prolonged B19 infection.12 Clinical studies have shown that B19 DNA can be amplified from joint samples by polymerase chain reaction (PCR),13,14 and infective B19 was detected in the articular lesions of patients with rheumatoid arthritis. B19 transcripts and B19 protein viral protein 1 (VP1) were also present in T cells, B cells, macrophages, and follicular dendritic cells.14 The cellular mechanism that may allow B19 binding and its entry into nonerythroid cells has not been elucidated. In the present study, we explored a putative receptor for B19 that was distinct from P antigen.
Materials and methods
Cells
Macrophage cell lines U937, urinary bladder carcinoma cell line T24, colon cancer cell line SW620, renal adenocarcinoma cell line ACHN, and HeLa cells were provided by the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University (Sendai, Japan). Human erythroid cell line KU812Ep615 was provided by E. Miyagawa (Institute of Fuji Rebio, Tokyo, Japan). T-cell line H9 was purchased from American Type Culture Collection (Manassas, VA). Bone marrow samples were obtained from the volunteers who gave informed consent for the use of their samples for our study. Informed consent was provided in accordance with the Declaration of Helsinki.
Human parvovirus B19
Serum from patient 1 with acute B19 infection was used as the source for B19 in the in vitro infection study. The serum contained 2.5 × 1014 copies of B19-DNA per milliliter but was negative for IgM and IgG anti-B19 antibodies.16 B19 was purified using cellulose hollow fiber, was provided by Dr. K. Yamaguchi,17 and used as an antigen for enzyme-linked immunosorbent assay (ELISA).
Recombinant human parvovirus B19 empty capsid protein
Recombinant human parvovirus B19 empty capsid protein (rB19ECP), prepared as described previously,18 was kindly provided by K. Kamata at Denka Seiken (Tokyo, Japan). rB19ECP was composed of VP1 and VP2 at a ratio of 5:95, respectively.
Antibodies
Monoclonal anti-Ku80 antibodies that recognized N-terminus (amino acids 3-22) or C-terminus of Ku80 (amino acids 610-705) were purchased from Oncogene (Boston, MA) and BD Biosciences (San Jose, CA), respectively. PAR3 is a mouse monoclonal antibody recognizing VP2, which shared with VP1 of B19.19,20 1F5 is a mouse monoclonal antibody with anti-idiotypic activity to an anti-DNA antibody,21 which was used as a control for flow cytometry analysis. GL4, a rabbit polyclonal antigloboside antibody (IgG and IgM), was purchased from Matreya (State College, PA). Monoclonal anti–α5 and anti–β1 integrin antibodies were purchased from Chemicon (Temecula. CA). Other monoclonal antibodies were purchased from BD Biosciences.
In vitro infection of B19
Cells (2 × 106) in 0.5 mL RPMI were infected with B19 containing serum from 1 (diluted at 2 × 1011 copies of B19 DNA/mL) for 30 minutes on ice and washed extensively 3 times with phosphate-buffered saline (PBS), pH 7.2, for evaluation of B19 adsorption. To study B19 replication, the prepared cells in 3 mL RPMI containing 10% fetal bovine serum (FBS) were further incubated at 37°C for 48 hours in a 5% CO2 humidified atmosphere, followed by 3 extensive washes with PBS and then evaluated for B19 protein and B19 DNA.
Protein precipitation and purification of precipitated protein
Cell-surface protein of H9 cells was labeled with sulfo-NHS esters of biotin (Pierce, Rockford, IL), followed by protein precipitation. H9 cells were treated with Nonidet P-40 lysis buffer (1% Nonidet P-40, 140 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM EDTA [ethylenediaminetetraacetic acid], 50 mM Tris [tris(hydroxymethyl)aminomethane)–HCl, pH 7.4) and immunoprecipitated with rB19ECP- or bovine serum albumin (BSA)–conjugated cyanogen bromide (CNBr)–Sepharose (Amersham Bioscience, Piscataway, NJ). The precipitated samples were separated under denaturing conditions in a 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel, followed by electrotransfer to a polyvinylidene difluoride (PVDF) membrane. Protein was detected with enhanced chemiluminescence (ECL) Western blotting detection system (Amersham Biosciences) and visualized by LAS-1000 (Fujifilm, Tokyo, Japan). A crude membrane fraction of H9 cells (1 × 1011) was prepared and solubilized in 1% Nonidet P-40 lysis buffer. The fraction was then precipitated with rB19ECP-conjugated CNBr-Sepharose for 16 hours at 4°C, and the precipitated proteins were separated by SDS-PAGE followed by Coomassie blue staining. Protein sequencing was carried out by Toray Research Center (Kamakura, Japan).
Flow cytometry analysis
Cells were suspended in 100 μL 1% BSA-PBS and incubated with 5 μg/mL test antibodies on ice for 30 minutes. Cells were then washed 3 times with PBS. Cells that required secondary antibodies for detection were further incubated with fluorescein isothiocyanate (FITC)–conjugated goat anti–mouse IgG (or FITC-conjugated anti–rabbit IgG for the GL4 primary antibody; Sigma, St Louis, MO) at 1:200 on ice for 30 minutes. Cells were washed 3 times with PBS before flow cytometry analysis (Becton Dickinson, San Jose, CA). For the detection of Ku80 on the cell surface of bone marrow cells, cells were first reacted with 5 μg/mL anti-Ku80 antibody followed by an incubation with FITC-conjugated anti–mouse IgG antibody. After being washed, cells were reacted with the cell-lineage–specific antibodies (anti–glycophorin A, anti-CD3, anti-CD20, anti-CD56, anti-CD14, or anti-CD36 antibodies) conjugated with phycoerythrin (PE; BD Biosciences) according to the manufacturer's instruction.
In the in vitro infection study, B19-infected cells were fixed with 4% paraformaldehyde followed by permeabilization with SAP buffer (0.1% saponin, 0.05% NaN3 in Hanks balanced salt solution). Next, cells were incubated with PAR3 at a concentration of 5 mg/mL on ice for 30 minutes, followed by the same procedure as described.
ELISA and quantitative PCR
Preparation of cell fraction from B19-infected cells
Cells (6 × 105) were infected with B19 for 30 minutes on ice. Following three washes with PBS, pH 7.2, DNA was extracted from 2 × 105 cells to measure adsorbed B19. The remaining 4 × 105 cells were further incubated for 30 minutes at 37°C, followed by 3 washes with PBS, pH 4.5. To obtain cytoplasm fractions of B19-infected cells, cells were treated with lysis buffer A (100 mM Tris-HCl, pH 7.5, 1% Triton X-100, 5 mM EDTA, 50 mM NaCl, and 100 μM PMSF), and centrifuged. Then, DNA was extracted from the supernatant to measure B19-DNA in cytoplasm. The pellets were washed with lysis buffer A 3 times and treated with lysis buffer B (100 mM Tris-HCl, pH 7.5, 1% Triton X-100, 5 mM EDTA, 500 mM NaCl, and 100 μM PMSF). Following centrifugation, DNA was extracted from the supernatant to measure B19-DNA in nuclei.
Transfection
Five micrograms of expression vector pcD22 containing pKu80 was used for the transfection of 1 × 105 HeLa cells, and pcD was used as a vector-only control. Transfection was done using the lipofectin method (Invitrogen, Carlsbad, CA). Transfected HeLa cells were infected with B19 for 30 minutes at 37°C. After being washed 3 times with PBS, pH 7.2, cells were collected with 5 mM EDTA-PBS, and B19 was detected by confocal microscopy analysis. Transfected HeLa cells were incubated with 1 μg/mL biotinylated rB19ECP in the presence of 5 μg/mL inhibitor antibodies for 30 minutes at 37°C. After being washed 3 times with PBS, pH 7.2, cells were collected with 5 mM EDTA-PBS, and rB19ECP or Ku80 was detected by confocal microscopy analysis.
RNA interference of K80 in KU812Ep6
The short interfering RNA (siRNA) for Ku80 was synthesized targeting the sequence between nucleotide numbers 130 and 148: 5′-CAAGCAAAGAAGGUGAUAAdTdT-3′ (sense), 3′-dTdTGUUCGUUUCUUCCACUAUU-5′ (antisense). Disordered siRNA of scrambled nucleotide sequence, used as negative control, was 5′-GCGCGCUUUGUAGGAUUCGdTdT-3′ (sense), 3′-dTdTCGCGCGAAACAUCCUAAGC-5′ (antisense). Synthesized siRNA (200 nM) was transfected to 1 × 106 KU812Ep6 cells by Cell Line Nucleofector Kit V (Amaxa, Gaithersburg, MD) according to the manufacturer's instructions. Transfected cells were subjected to flow cytometry analysis and in vitro infection study of B19, after 48 hours of incubation.
Detection of B19 in HeLa cells by confocal laser microscopy
Cells were grown on glass microslides and fixed with 4% paraformaldehyde in PBS for 10 minutes at room temperature. Cells were blocked with PBS containing 10% FBS for 30 minutes at 4°C, followed by incubation with mouse monoclonal anti-B19 antibody PAR3 (10 μg/mL) for 30 minutes at 4°C, then washed with PBS twice, and incubated with FITC-conjugated goat anti–mouse IgG (1:100; Sigma) for 30 minutes at 4°C. To detect localization of Ku80 and rB19ECP, cells on glass microslides were incubated with mouse monoclonal anti-Ku80 antibody (5 μg/mL) for 30 minutes at 4°C, washed with PBS twice, and incubated with tetramethyl isothiocyanate (TRITC)–conjugated goat anti–mouse IgG (1:50; Sigma) for 30 minutes at 4°C; cells were further incubated with avidin-FITC (1:100; Gibco, Carlsbad, CA) for 30 minutes at 4°C for the detection of labeled rB19ECP. Confocal miscroscopy analysis was performed with a D-ECLIPSE CI (Nikon, Kawasaki, Japan) mounted with 20×/0.50 or 40×/0.75 Plan Fluor dry objective lenses. Excitation at 488 nm from an argon laser and at 543 nm from a helium-neon laser was used. Images were acquired with E2-Cl 2.00 software (Nikon) and processed with Adobe Photoshop 7.0.1 (Adobe Systems, San Jose, CA).
Results
Identification of B19-binding protein on the cell surface of nonerythroid cells
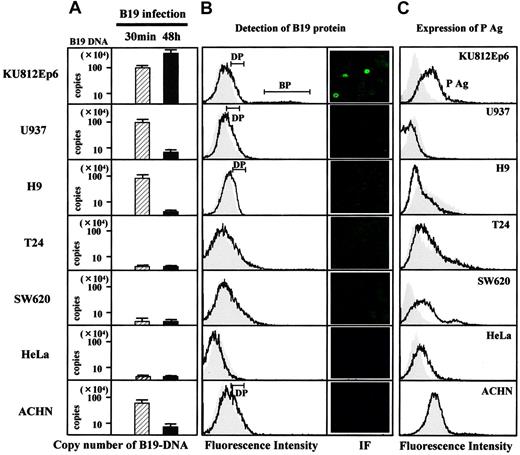
To identify a putative receptor for B19, we first checked the expression of P antigen (Figure 1). Flow cytometry analysis revealed that α5β1 integrin6 was also positive on the surface of all cell lines tested (data not shown). We then studied the binding and replication of B19 in association with the expression of P antigen and α5β1 integrin. Quantitative study for cell-surface binding, B19 DNA replication, and fluorescence-activating cell sorting (FACS) analysis using anti–B19 protein (VP2) antibody PAR3 revealed that B19 binds not only to a P antigen–expressing erythroid cell line KU812Ep6 but also to a macrophage cell line, U937, to a T-cell line, H9, and a renal carcinoma cell line, ACHN, in which P antigen was undetectable on the cell surface. None of the cell lines, T24, SW620, and HeLa, bound B19 despite surface P antigen expression (left column in Figure 1A). FACS analysis at 48 hours after B19 infection revealed 2 types of staining patterns for B19 protein following immunohistochemistry using PAR3: (1) intense staining in KU812Ep6 and (2) weak staining in Ku812Ep6, U937, H9, and ACHN (left column in Figure 1B). Replication of B19 DNA and the synthesis of B19 protein was observed in KU812Ep6, but not in any of the other cells, irrespective of the presence of P antigen (right column in Figure 1A and right column in Figure 1B) or α5β1 integrin.
B19 infectivity and expression of P antigen. Each cell line (2 × 106) was inoculated with B19 (1 × 1011 copies of B19 DNA) for 30 minutes at 4°C and washed with PBS, pH 7.2, 3 times. Half of the cells in each group were used for evaluation of B19 adsorption (left column in panel A), and remaining cells in 3 mL RPMI containing 10% FBS were further incubated at 37°C for 48 hours to measure B19 DNA replication (right column in panel A) or to detect B19 protein (B). (A) B19 binding and replication of B19 in various cell lines. B19-infected cells were quantified for B19 DNA as described in “Materials and methods.” The left column (▨) is regarded as B19 adsorption, and the right column (▪) as B19 replication. The scale for B19 DNA is shown in logarithm. (B) Detection of B19 protein in B19-infected cells. After a 48-hour incubation with B19, the cells were washed 3 times with PBS and they were fixed with 4% paraformaldehyde followed by permeabilization with SAP buffer (0.1% saponin, 0.05% NaN3 in Hanks balanced salt solution). Then, cells were incubated with PAR3 at a concentration of 5 μg/mL on ice for 30 minutes, followed by an incubation with FITC-conjugated goat anti–mouse IgG. The expression of B19 protein in cytoplasm was analyzed by flow cytometry with PAR3 (line) or isotype-matched antibody 1F5 (shadow; left panel), or by immunofluorescence (IF) staining with PAR3 (right panel). Two types of positive patterns were observed in flow cytometry: dull positive (DP) pattern in KU812Ep6, U937, H9, and ACHN; bright positive (BP) pattern in KU812Ep6. (C) Flow cytometry analysis of P antigen expression on the cell surface. Indicated cells were incubated with antigloboside antibody, GL4, followed by PE-labeled anti–rabbit IgG. Shadow represents staining using rabbit IgG as a negative control.
B19 infectivity and expression of P antigen. Each cell line (2 × 106) was inoculated with B19 (1 × 1011 copies of B19 DNA) for 30 minutes at 4°C and washed with PBS, pH 7.2, 3 times. Half of the cells in each group were used for evaluation of B19 adsorption (left column in panel A), and remaining cells in 3 mL RPMI containing 10% FBS were further incubated at 37°C for 48 hours to measure B19 DNA replication (right column in panel A) or to detect B19 protein (B). (A) B19 binding and replication of B19 in various cell lines. B19-infected cells were quantified for B19 DNA as described in “Materials and methods.” The left column (▨) is regarded as B19 adsorption, and the right column (▪) as B19 replication. The scale for B19 DNA is shown in logarithm. (B) Detection of B19 protein in B19-infected cells. After a 48-hour incubation with B19, the cells were washed 3 times with PBS and they were fixed with 4% paraformaldehyde followed by permeabilization with SAP buffer (0.1% saponin, 0.05% NaN3 in Hanks balanced salt solution). Then, cells were incubated with PAR3 at a concentration of 5 μg/mL on ice for 30 minutes, followed by an incubation with FITC-conjugated goat anti–mouse IgG. The expression of B19 protein in cytoplasm was analyzed by flow cytometry with PAR3 (line) or isotype-matched antibody 1F5 (shadow; left panel), or by immunofluorescence (IF) staining with PAR3 (right panel). Two types of positive patterns were observed in flow cytometry: dull positive (DP) pattern in KU812Ep6, U937, H9, and ACHN; bright positive (BP) pattern in KU812Ep6. (C) Flow cytometry analysis of P antigen expression on the cell surface. Indicated cells were incubated with antigloboside antibody, GL4, followed by PE-labeled anti–rabbit IgG. Shadow represents staining using rabbit IgG as a negative control.
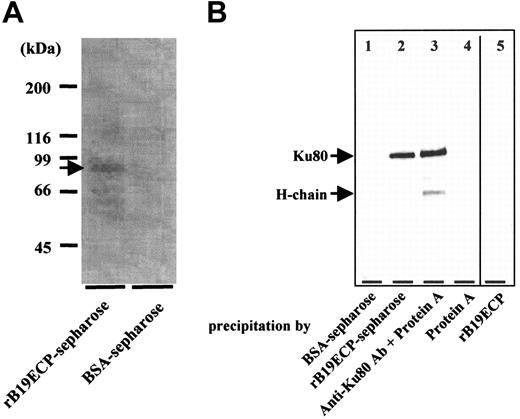
Determination of B19-binding protein on surface of T cell line H9. (A) Isolation of B19-binding protein from H9 surface. Surface proteins H9 of cells were biotinylated. Cell lysate from 1 × 1011 biotinylated H9 cells was mixed with rB19ECP-conjugated Sepharose or with BSA-conjugated Sepharose. Precipitated protein was isolated and reacted with streptavidin–horseradish-peroxidase conjugate on PVDF membranes, followed by the chemiluminescence detection. (B) Western blotting of protein from H9 surface with anti-Ku80 antibody. Lanes 1-4 show cell lysate precipitated with indicated protein or protein-conjugated Sepharose. Lane 5 shows the rB19ECP (1 μg) resolved by electrophoresis under denaturing conditions.
Determination of B19-binding protein on surface of T cell line H9. (A) Isolation of B19-binding protein from H9 surface. Surface proteins H9 of cells were biotinylated. Cell lysate from 1 × 1011 biotinylated H9 cells was mixed with rB19ECP-conjugated Sepharose or with BSA-conjugated Sepharose. Precipitated protein was isolated and reacted with streptavidin–horseradish-peroxidase conjugate on PVDF membranes, followed by the chemiluminescence detection. (B) Western blotting of protein from H9 surface with anti-Ku80 antibody. Lanes 1-4 show cell lysate precipitated with indicated protein or protein-conjugated Sepharose. Lane 5 shows the rB19ECP (1 μg) resolved by electrophoresis under denaturing conditions.
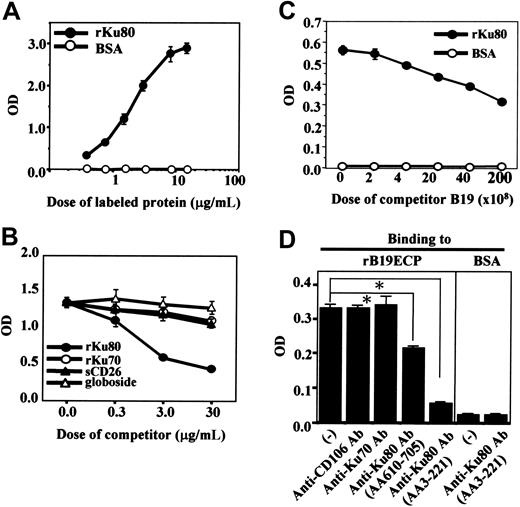
To determine the cell-surface molecule responsible for B19 binding to H9 cells, a recombinant empty capsid protein of B19 (rB19ECP) was used. Biotinylated rB19ECP bound H9 in a dose-dependent manner (data not shown). We then purified the rB19ECP-binding molecule from the cell surface of H9 using rB19ECP-conjugated Sepharose (rB19ECP-Sepharose). The precipitated 80-kDa protein (Figure 2A) was analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry. The obtained data were collated and submitted for homology search using the Swiss Prot and NCB Inr databases. The Ku80 autoantigen was identified as the gene product with the highest homology in both databases. As a confirmation, the rB19ECP-binding 80-kDa protein reacted with anti-Ku80 antibody (Figure 2B). Competitive ELISA further confirmed the specific binding between Ku80 and B19. Biotinylated recombinant Ku80 (rKu80) reacted with rB19ECP fixed to microwells (Figure 3A); the binding was selectively inhibited by unlabeled rKu80 but not by recombinant Ku70 (rKu70), globoside, or recombinant soluble CD26 (sCD26)23 (Figure 3B). This binding was also inhibited in the presence of native B19 particles from infected patients (Figure 3C). Two anti-Ku80 antibodies significantly inhibited the binding of biotinylated rKu80 and rB19ECP, whereas anti-Ku70 antibody or anti-CD106 antibody failed to inhibit the binding (Figure 3D).
Ku80 participates in B19 binding and subsequent entry
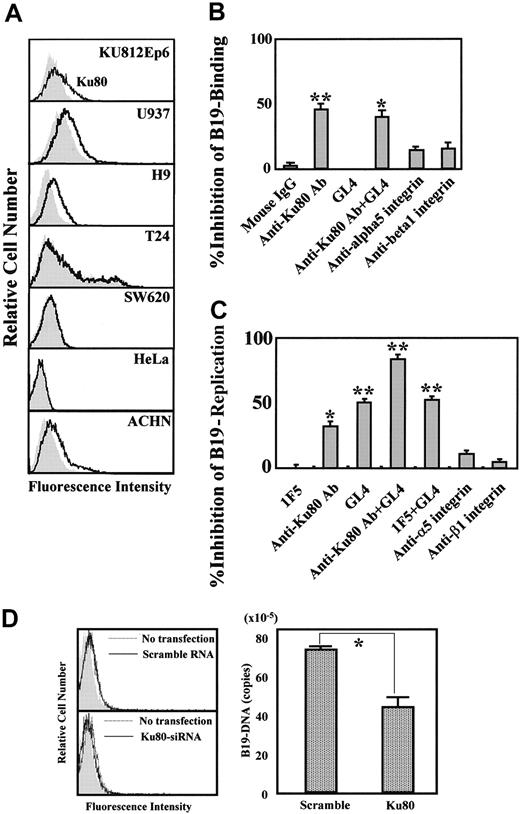
We next investigated whether Ku80 would participate in B19 binding on the cell surface and facilitate B19 entry. KU812Ep6, U937, H9, and ACHN cells efficiently bound B19 (Figure 1A) and all of these cells clearly expressed Ku80 on their surface (Figure 4A). On the other hand, Ku80 was undetectable on T24, SW620, and HeLa cells, and no binding of B19 occurred (Figures 4A and 1A). An in vitro infection experiment demonstrated efficient replication of B19 DNA in KU812Ep6 cells that expressed both Ku80 and P antigen. B19 failed to amplify itself in U937, H9, and ACHN cells, which express Ku80 but no detectable levels of P antigen on the cell surface (Figures 1 and 4A). T24, SW620, and HeLa cells were nonpermissive for B19 infection although they expressed P antigen (Figure 1) and α5β1 integrin.
Specific binding of rB19ECP to Ku80. (A) Specific binding of Ku80 to rB19ECP. Indicated concentration of biotinylated rKu80 or biotinylated BSA was reacted with rB19ECP fixed to 96 microwells and detected by ELISA. (B) Competitive ELISA for rB19ECP binding to rKu80. Biotinylated rKu80 (2 μg/mL) was reacted with rB19ECP fixed to wells in the presence of indicated doses of unlabeled rKu80, rKu70, sCD26, or globoside. (C) Inhibition of rB19ECP binding to rKu80 by purified B19. Biotinylated rKu80 (1 μg/mL) was added to rB19ECP fixed to wells in the presence of B19 that was purified from B19+ serum with repeated microfiltration. Doses of B19 are expressed as copy numbers of B19 DNA. (D) Inhibition of rB19ECP binding to rKu80 by anti-Ku80 antibodies. Binding of biotinylated rKu80 or biotinylated BSA to rB19ECP fixed to wells was measured in the presence of isotype-matched mouse monoclonal antibodies as indicated.
Specific binding of rB19ECP to Ku80. (A) Specific binding of Ku80 to rB19ECP. Indicated concentration of biotinylated rKu80 or biotinylated BSA was reacted with rB19ECP fixed to 96 microwells and detected by ELISA. (B) Competitive ELISA for rB19ECP binding to rKu80. Biotinylated rKu80 (2 μg/mL) was reacted with rB19ECP fixed to wells in the presence of indicated doses of unlabeled rKu80, rKu70, sCD26, or globoside. (C) Inhibition of rB19ECP binding to rKu80 by purified B19. Biotinylated rKu80 (1 μg/mL) was added to rB19ECP fixed to wells in the presence of B19 that was purified from B19+ serum with repeated microfiltration. Doses of B19 are expressed as copy numbers of B19 DNA. (D) Inhibition of rB19ECP binding to rKu80 by anti-Ku80 antibodies. Binding of biotinylated rKu80 or biotinylated BSA to rB19ECP fixed to wells was measured in the presence of isotype-matched mouse monoclonal antibodies as indicated.
Role of Ku80 in B19 infection in vitro. (A) Ku80 expression on cell surface. The indicated cell lines were reacted with 5 μg/mL mouse monoclonal anti-Ku80 antibody (line) or 5 μg/mL isotype-matched mouse monoclonal antibody 1F5 (shadow), followed by FITC-labeled anti–mouse IgG antibodies. Cells were washed with PBS, and cell-surface expression of Ku80 was analyzed by flow cytometry. (B) Blocking of B19 adsorption by anti-Ku80 antibody or antigloboside antibody. KU812Ep6 cells (2 × 106) were infected with B19 (2 × 1011 copies of B19 DNA) on ice for 30 minutes in the presence of the indicated antibodies (5 μg/mL) and extensively washed with PBS 3 times. To activate α5β1 integrin, anti-integrin antibodies were used in the presence of divalent ions (1 mM Mn2+, 1 mM Mg2+). B19 DNA in each group was quantified by quantitative PCR. The blocking ability of B19 binding by each antibody was expressed as percent decrease of B19-DNA in each group compared to that in antibody-untreated cells. **P < .01, *P < .05 by Student t test. (C) Blocking of B19 replication by anti-Ku80 antibody or antigloboside antibody. KU812Ep6 cells were infected with B19 and washed as described. Cells were further incubated for 48 hours at 37°C and washed with PBS 3 times before the quantitative study of B19 DNA. To activate α5β1 integrin, anti-integrin antibodies were used in the presence of divalent ions (1 mM Mn2+, 1 mM Mg2+). The blocking ability of B19 replication by each antibody was expressed as described. **P < .01, *P < .05 by Student t test. (D) RNA interference of Ku80 in KU812Ep6 cells. Cell-surface expression of Ku80 was examined by flow cytometry in scramble RNA or siRNA of Ku80-transfected KU812Ep6 cells (left panel). KU812Ep6 cells treated with indicated RNA were reacted with 5 μg/mL mouse monoclonal anti-Ku80 antibody or 5 μg/mL isotype-matched mouse monoclonal antibody 1F5 (shadow), followed by FITC-labeled anti–mouse IgG antibodies. B19 association of siRNA-transfected KU812Ep6 cells was evaluated by quantitative PCR (right panel). Sample DNA was prepared from extensively washed scramble RNA or siRNA of Ku80-transfected KU812Ep6 cells after 2 hours of incubation with B19. *P < .01 by Student t test.
Role of Ku80 in B19 infection in vitro. (A) Ku80 expression on cell surface. The indicated cell lines were reacted with 5 μg/mL mouse monoclonal anti-Ku80 antibody (line) or 5 μg/mL isotype-matched mouse monoclonal antibody 1F5 (shadow), followed by FITC-labeled anti–mouse IgG antibodies. Cells were washed with PBS, and cell-surface expression of Ku80 was analyzed by flow cytometry. (B) Blocking of B19 adsorption by anti-Ku80 antibody or antigloboside antibody. KU812Ep6 cells (2 × 106) were infected with B19 (2 × 1011 copies of B19 DNA) on ice for 30 minutes in the presence of the indicated antibodies (5 μg/mL) and extensively washed with PBS 3 times. To activate α5β1 integrin, anti-integrin antibodies were used in the presence of divalent ions (1 mM Mn2+, 1 mM Mg2+). B19 DNA in each group was quantified by quantitative PCR. The blocking ability of B19 binding by each antibody was expressed as percent decrease of B19-DNA in each group compared to that in antibody-untreated cells. **P < .01, *P < .05 by Student t test. (C) Blocking of B19 replication by anti-Ku80 antibody or antigloboside antibody. KU812Ep6 cells were infected with B19 and washed as described. Cells were further incubated for 48 hours at 37°C and washed with PBS 3 times before the quantitative study of B19 DNA. To activate α5β1 integrin, anti-integrin antibodies were used in the presence of divalent ions (1 mM Mn2+, 1 mM Mg2+). The blocking ability of B19 replication by each antibody was expressed as described. **P < .01, *P < .05 by Student t test. (D) RNA interference of Ku80 in KU812Ep6 cells. Cell-surface expression of Ku80 was examined by flow cytometry in scramble RNA or siRNA of Ku80-transfected KU812Ep6 cells (left panel). KU812Ep6 cells treated with indicated RNA were reacted with 5 μg/mL mouse monoclonal anti-Ku80 antibody or 5 μg/mL isotype-matched mouse monoclonal antibody 1F5 (shadow), followed by FITC-labeled anti–mouse IgG antibodies. B19 association of siRNA-transfected KU812Ep6 cells was evaluated by quantitative PCR (right panel). Sample DNA was prepared from extensively washed scramble RNA or siRNA of Ku80-transfected KU812Ep6 cells after 2 hours of incubation with B19. *P < .01 by Student t test.
Ku80 functions as a coreceptor for B19 infection together with P antigen
We then performed an inhibition test for B19 infection of KU812Ep6 cells using antibodies against Ku80, P antigen, α5β1 integrin. Anti-Ku80 antibody inhibited B19 binding, whereas anti-P antibody, GL4, did not inhibit B19 binding. Anti–α5 and anti–β1 integrin antibodies caused a slight inhibition of B19 binding (Figure 4B). Both anti-Ku80 antibody and GL4 also inhibited B19 replication in KU812Ep6 cells. The simultaneous presence of both antibodies more strongly inhibited the replication of B19 DNA (Figure 4C). Presence of anti–α5 and anti–β1 integrin antibodies caused only a slight inhibition of B19 replication (Figure 4C). In other experiments, KU812Ep6 cells were treated with siRNA against Ku80 and then tested for the replication of B19 at B19 infection study. The results revealed the suppression of B19 binding to the KU812Ep6 cells with reduced expression of Ku80 (Figure 4D).
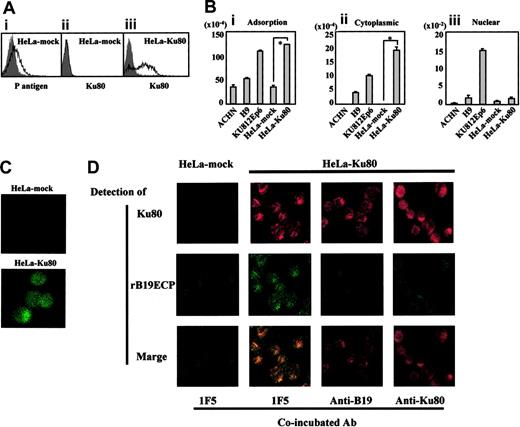
Transfection of Ku80 to HeLa cells. (A) Expression of Ku80 on HeLa cells transfected with pKu80. Ku80 cDNA was inserted to expression plasmid pcD and the resulted pKu80 was transfected to HeLa cells (HeLa-Ku80) using lipofectin. Empty pcD was used for a mock transfection (HeLa-mock). The transfected cells (2 × 105) were incubated with 5 μg/mL of each antibody, anti-Ku80 antibody (ii,iii), GL4 (A1), or isotype-matched mouse monoclonal antibodies or rabbit serum (shadow), and then analyzed for the expression of P antigen or Ku80 on the cell surface. Figures show HeLa-mock expressed P antigen but not Ku antigen on the surface (i,ii), whereas HeLa-Ku80 expressed Ku80 (iii). (B) Increased binding and viral entry of B19 in HeLa-Ku80. The indicated cells (6 × 105) were infected with B19 (2 × 1011 copies of B19 DNA) for 30 minutes on ice. After washing cells 3 times with PBS, pH 7.2, DNA was extracted from 2 × 105 cells. Remaining cells were further incubated for 30 minutes at 37°C. After washing cells 3 times with PBS, pH 4.5, a cytoplasmic and nuclear fraction was prepared, and then DNA was extracted from each fraction. Prepared DNA was subjected to a quantitative PCR to quantify B19 DNA. *P < .01 by Student t test. (C) B19 infection to HeLa-Ku80. HeLa-mock or HeLa-Ku80 cells (2 × 105) were infected with B19 (2 × 1011 copies of B19 DNA) for 30 minutes at 37°C. After being washed 3 times with PBS, cells were collected with 5 mM EDTA-PBS, pH 7.2, fixed with 4% paraformaldehyde and reacted with PAR3, followed by FITC-labeled anti–mouse IgG antibody as a secondary antibody. Thus prepared cells were then subjected to a confocal microscope analysis. The panel represents B19 entered into HeLa-Ku80. (D) Colocalization of rB19ECP and Ku80. HeLa-mock or HeLa-Ku80 (2 × 105) cells were incubated with biotinylated rB19ECP (1 μg/mL) in the presence of 5 μg/mL inhibitor antibody indicated for 30 minutes at 37°C. After being washed 3 times with PBS, pH 7.2, cells were collected with 5 mM EDTA-PBS, and rB19ECP or Ku80 was detected by confocal microscopy analysis. Ku80 was detected by anti-Ku80 antibody followed by TRITC-labeled anti–mouse IgG antibody as a secondary antibody. Detection of biotinylated rB19ECP was done by avidin-FITC as described in “Materials and methods.”
Transfection of Ku80 to HeLa cells. (A) Expression of Ku80 on HeLa cells transfected with pKu80. Ku80 cDNA was inserted to expression plasmid pcD and the resulted pKu80 was transfected to HeLa cells (HeLa-Ku80) using lipofectin. Empty pcD was used for a mock transfection (HeLa-mock). The transfected cells (2 × 105) were incubated with 5 μg/mL of each antibody, anti-Ku80 antibody (ii,iii), GL4 (A1), or isotype-matched mouse monoclonal antibodies or rabbit serum (shadow), and then analyzed for the expression of P antigen or Ku80 on the cell surface. Figures show HeLa-mock expressed P antigen but not Ku antigen on the surface (i,ii), whereas HeLa-Ku80 expressed Ku80 (iii). (B) Increased binding and viral entry of B19 in HeLa-Ku80. The indicated cells (6 × 105) were infected with B19 (2 × 1011 copies of B19 DNA) for 30 minutes on ice. After washing cells 3 times with PBS, pH 7.2, DNA was extracted from 2 × 105 cells. Remaining cells were further incubated for 30 minutes at 37°C. After washing cells 3 times with PBS, pH 4.5, a cytoplasmic and nuclear fraction was prepared, and then DNA was extracted from each fraction. Prepared DNA was subjected to a quantitative PCR to quantify B19 DNA. *P < .01 by Student t test. (C) B19 infection to HeLa-Ku80. HeLa-mock or HeLa-Ku80 cells (2 × 105) were infected with B19 (2 × 1011 copies of B19 DNA) for 30 minutes at 37°C. After being washed 3 times with PBS, cells were collected with 5 mM EDTA-PBS, pH 7.2, fixed with 4% paraformaldehyde and reacted with PAR3, followed by FITC-labeled anti–mouse IgG antibody as a secondary antibody. Thus prepared cells were then subjected to a confocal microscope analysis. The panel represents B19 entered into HeLa-Ku80. (D) Colocalization of rB19ECP and Ku80. HeLa-mock or HeLa-Ku80 (2 × 105) cells were incubated with biotinylated rB19ECP (1 μg/mL) in the presence of 5 μg/mL inhibitor antibody indicated for 30 minutes at 37°C. After being washed 3 times with PBS, pH 7.2, cells were collected with 5 mM EDTA-PBS, and rB19ECP or Ku80 was detected by confocal microscopy analysis. Ku80 was detected by anti-Ku80 antibody followed by TRITC-labeled anti–mouse IgG antibody as a secondary antibody. Detection of biotinylated rB19ECP was done by avidin-FITC as described in “Materials and methods.”
The role of Ku80 as a coreceptor for B19 infection was also supported by a transfection experiment using HeLa cells that were nonpermissive for B19 infection. Figure 5A shows that the surface of Ku80-transfected HeLa cells (HeLa-Ku80) became positive for Ku80 expression and binding of B19 to the cells was significantly enhanced (Figure 5B). Quantitative analysis of B19 DNA (Figure 5B) and confocal laser microscopy (Figure 5C) confirmed that B19 DNA and B19 protein were present in the cytoplasmic fraction of HeLa-Ku80 cells 30 minutes after infection, similar to Ku812Ep6. Furthermore, a coincubation experiment of rB19ECP and HeLa-Ku80 revealed the colocalization of rB19ECP and Ku80 in the cytoplasm or membrane (or both) of HeLa-Ku80 (Figure 5D). Moreover, association of rB19ECP and HeLa-Ku80 was apparently inhibited by the presence of anti-B19 antibody or anti-Ku80 antibody (Figure 5D).
Ku80 is expressed on the surface of bone marrow cells
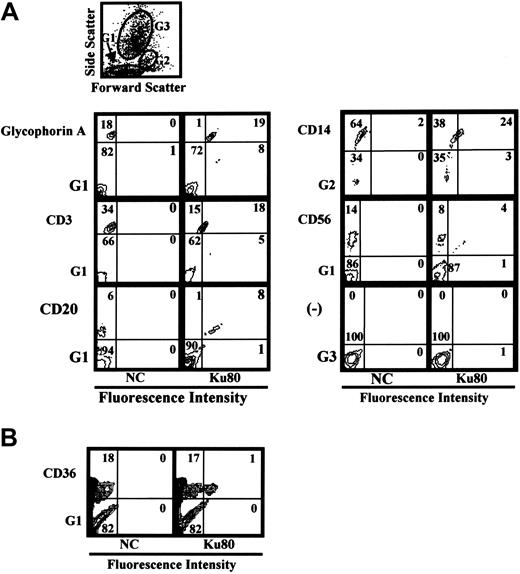
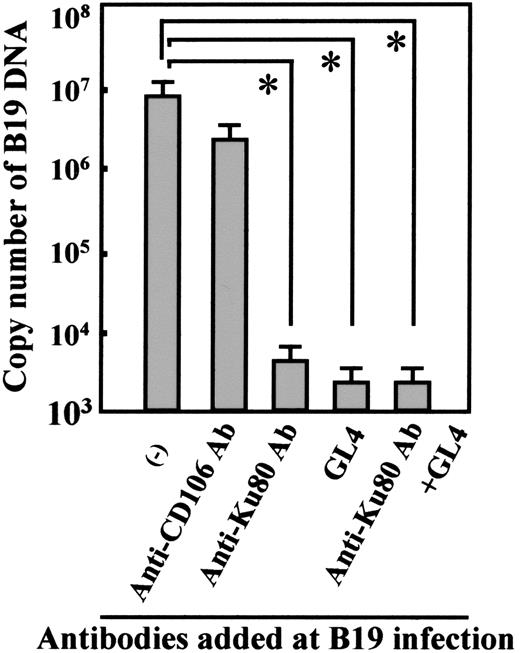
Because Ku80 is known as a nuclear protein, it is important to determine whether or not Ku80 is expressed on the cell-surface in vivo. Ku80 was not detected on the cell surface of peripheral blood mononuclear cells (data not shown). We then examined cell-surface expression of Ku80 in bone marrow cells because bone marrow cells are potential targets of B19 infection. Flow cytometry analysis of bone marrow cells demonstrated that Ku80 was highly expressed on the cell surface of erythroid progenitor cells expressing glycophorin A as well as on the surface of immune cells such as CD20+, CD3+, or CD14+ cells in bone marrow (Figure 6A). A small portion (5.6%) of CD36+ bone marrow cells, which may be permissive to B19 infection,23 were also positive for the expression of Ku80 on the cell surface (Figure 6B). B19 binding to bone marrow cells was inhibited in the presence of anti-Ku80 antibody at B19 infection in vitro (data not shown). Figure 7 shows that the replication of B19 in bone marrow cells was significantly inhibited in the presence of anti-Ku80 antibody or GL4. The inhibition rate of B19 replication in the presence of both anti-Ku80 antibody and GL4 was similar to that in the presence of GL4.
Cell-surface expression of Ku80 in human bone marrow cells. Flow cytometry analysis of Ku80 expression on the cell surface. Bone marrow cells were reacted with indicated antibodies and anti-Ku80 antibody as described in “Materials and methods,” and then the expression of surface molecules was analyzed. Prior to the study, each sample had been analyzed by the scattered plot. The results showed that the glycophorin A+, CD3+, CD20+, CD56+, or CD36+ cells were scattered in gate 1 (G1), and CD14+ cells in gate 2 (G2), and that there were no glycophorin A+, CD3+, CD20+, CD56+, or CD36+ cells in gate 3 (G3). Then the expression of Ku80 on cell surface in gated cells was analyzed. The gate used in each experiment is shown at left-lower side of each plot. (A) Gates used in the experiment and detection of Ku80 on the surface of various cell lineages. (B) Detection of Ku80 on the surface of CD36+ bone marrow cells.
Cell-surface expression of Ku80 in human bone marrow cells. Flow cytometry analysis of Ku80 expression on the cell surface. Bone marrow cells were reacted with indicated antibodies and anti-Ku80 antibody as described in “Materials and methods,” and then the expression of surface molecules was analyzed. Prior to the study, each sample had been analyzed by the scattered plot. The results showed that the glycophorin A+, CD3+, CD20+, CD56+, or CD36+ cells were scattered in gate 1 (G1), and CD14+ cells in gate 2 (G2), and that there were no glycophorin A+, CD3+, CD20+, CD56+, or CD36+ cells in gate 3 (G3). Then the expression of Ku80 on cell surface in gated cells was analyzed. The gate used in each experiment is shown at left-lower side of each plot. (A) Gates used in the experiment and detection of Ku80 on the surface of various cell lineages. (B) Detection of Ku80 on the surface of CD36+ bone marrow cells.
Discussion
The presented data implicate Ku80 as a coreceptor involved in B19 infection. U937, H9, and ACHN cells expressing Ku80 showed B19 binding, but some cells with P antigen failed to bind B19 unless these cells expressed Ku80 on their surface. A marked increase in B19 binding in Ku80-transfected HeLa cells and the inhibition of B19 infectivity by anti-Ku80 antibody or siRNA to Ku80 suggests a Ku80-dependent B19 interaction with the targeted cells. Specific inhibition of B19 binding by anti-Ku80 antibody that recognized the N-terminus of the Ku80 protein suggests that B19 interacts with specific sites of Ku80 on the cell surface. Further, Epstein-Barr virus or hepatitis virus C failed to bind either to Ku80-expressing HeLa or U937 cells (data not shown). These results suggest that Ku80 is one of the specific receptors for B19 infection.
Ku is a heterodimeric DNA-binding protein consisting of a 70-kDa (Ku70) and an 80-kDa (Ku80) subunit and was originally identified as a nuclear antigen recognized by autoantibodies in patients with systemic lupus erythematosus and scleroderma.25 Ku has a central role in multiple nuclear processes, including DNA repair, chromosome maintenance, transcription regulation, and V(D)J recombination. Ku is abundant in the nucleus, consistent with its function as a DNA-protein kinase (DNA-PK).26,27 However, recent studies have shown cytoplasm or surface localization of Ku in various types of cells, including of leukemia, multiple myeloma, and tumor cell lines. Ku is a component of the DNA-PK complex in membrane rafts of mammalian cells.26 Although the role of surface Ku80 has not been well clarified,28 signal transduction and Ku80 are coupled in both B and T cells,25,28,29 and localization of the DNA-PK complex in lipid rafts suggests a putative role in the signal transduction pathway following ionizing radiation.26 It was recently reported that Ku interacts with metalloproteinase 9 at the cell surface of highly invasive hematopoietic cells of normal and tumor cell origin, and Ku80/MMP-9 interaction at the cell membrane may result in contribution to the invasion of tumor cells through regulation of extracellular matrix remodelling.30 Further, the membrane form of Ku, whose expression is induced at hypoxia, mediates cell adhesion of plasma cells,30-32 indicating a role for Ku as an adhesion receptor for fibronectin.33 The present study showed that Ku80 is positive on the surface of CD3+ cells, CD20+ cells, CD14+ cells, glycophorin A+ cells, and CD36+ cells from bone marrow where B19 infection is permissive.
We have discovered a novel role of Ku80 as a cellular receptor in B19 infection. Anti-Ku80 antibody, however, did not cause complete inhibition of B19 infection, whereas pretreatment with anti-Ku80 antibody together with GL4 strongly inhibited B19 infectivity in KU812Ep6 cells and human bone marrow cells, showing the necessity of P antigen as a receptor. A recent report showed that α5β1 integrin has a role in B19 entry into host cells,6 and KU812Ep6, U937, H9, ACHN, and HeLa cells all expressed α5β1 integrin on their surface (data not shown). However, B19 entry into U937- and H9-expressing Ku80 and α5β1 integrin or HeLa cells with P antigen and α5β1 integrin was insufficient or negative (Figures 1 and 5B). B19 entry was marked in KU812Ep6 cells or Ku80-HeLa cells that expressed Ku80, P antigen, and α5β1 integrin on their surface, showing the necessity of P antigen for efficient binding and the virus entry afterward. Anti–α5 and anti–β1 integrin antibodies, which inhibited the entry of B19 into K562 cells,6 caused a slight inhibition of B19 binding as well as B19 replication in KU812Ep6, supporting the participation of α5β1 integrin in B19 infection. We are currently investigating the precise mechanism of the interaction among B19-related receptors such as P antigen, Ku80, and α5β1 integrin in association with the following signal transduction in B19-infected cells.
The use of multiple receptors for entry into cells has been observed frequently in virus infection, such as by α herpesviruses, HHV-8 or HIV.34,35 We have shown that B19 uses at least 2 receptors, Ku80 and P antigen, in the process of infection. Ku80 may function as an efficient B19-capturing molecule on the cell surface and may also contribute to B19 entry into cells; markedly enhanced entry of B19 in Ku80-HeLa cells (Figure 5C-D) suggests that Ku80 mediates efficient B19 entry in cooperation with P antigen and probably with α5β1 integrin.6 Although Ku80 can interact with Epstein-Barr virus protein in the nucleus,36 this study is the first to show the use of Ku80 antigen as a cellular receptor for virus infection. Despite marked entry of B19, synthesis of B19 protein was unsuccessful in Ku80-HeLa cells, but was possible only in erythroid cell lines, indicating that unknown intracellular factors may be required for B19 replication in the targeted cells.37,38
Blocking of B19 infection of bone marrow cells by anti-Ku80 antibody or antigloboside antibody. Bone marrow cells (2 × 106) were infected with B19 (2 × 1011 copies of B19 DNA) with the indicated antibodies and evaluated for quantity of B19 DNA as described. Anti-CD106 antibody was a mouse monoclonal antibody used as a negative control. The differences in the results between control (–) and other samples were statistically analyzed. *P < .01 by Student t test.
Blocking of B19 infection of bone marrow cells by anti-Ku80 antibody or antigloboside antibody. Bone marrow cells (2 × 106) were infected with B19 (2 × 1011 copies of B19 DNA) with the indicated antibodies and evaluated for quantity of B19 DNA as described. Anti-CD106 antibody was a mouse monoclonal antibody used as a negative control. The differences in the results between control (–) and other samples were statistically analyzed. *P < .01 by Student t test.
Ku80 is not found in circulating mononuclear cells from healthy volunteers but is positive on the surface of B19-binding cells in vivo, such as immune cells in tonsils, erythroblasts, T cells, B cells, macrophages in bone marrow, and immune cells including follicular dendritic cells in rheumatoid joints, indicating the surface expression of Ku antigen may be restricted by environmental conditions. Of interest is that the oxygen levels are markedly low in bone marrow and joints39-41 compared with that in blood, and surface Ku80 is inducible with hypoxia.31,32 A recent study suggests the efficiency of B19 infection increases with hypoxia.42 These studies suggest that surface Ku80 induced with hypoxia may participate in the process of B19 infection of joints and bone marrow.
Ku80 expression on the surface of immune cells in bone marrow in vivo may explain clinical findings associated with B19 infection to nonerythroid cells. Namely, B19 infection often causes a decreased number of leukocytes or lymphocytes in blood during acute B19 infection, as well as increased levels of TNF-α and IFN-γ in blood or rheumatoid joints, and the detection of B19 on T cells, B cells, or macrophages in tonsils, bone marrow, or rheumatoid joints. B19 may infect immune cells in bone marrow or the synovium and persist to lead to secrete an inflammatory cytokine through the activation of AP1 and AP2 by B19 NS1.43 Stimulation of cellular receptors with B19 may trigger activation of signal cascades in host cells, which may explain why immune cells in acute and prolonged B19 infection or in the joints of rheumatoid arthritis are functionally altered.
Prepublished online as Blood First Edition Paper, August 2, 2005; DOI 10.1182/blood-2005-02-0536.
Supported by a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science, Sports and Culture in Japan.
Y.M. and T.S.-I. designed with research, performed research, and wrote the paper; K.K.-I., J.H., T.K., and T.I. performed research; Y.H. analyzed data; and Y.K. and T.S. designed research.
Y.M. and T.S.-I. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to E. Miyagawa for KU812Ep6, K. Kamata for rB19ECP, T. Mimori for rKu80 and rKu70, C. Morimoto for sCD26, K. Yamaguchi for purified B19, and S. Shibahara for the pKu80.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal