In the thymus, 2 types of Lin–Sca-1+ (lineage-negative stem cell antigen-1–positive) progenitors can generate T-lineage cells: c-Kithi interleukin-7 receptor α–negative (c-KithiIL-7Rα–) and c-KitloIL-7Rα+. While c-KithiIL-7Rα– progenitors are absent, c-KitloIL-7Rα+ progenitors are abundant in the lymph nodes (LNs). c-KitloIL-7Rα+ progenitors undergo abortive T-cell commitment in the LNs and become arrested in the G1 phase of the cell cycle because they fail both to up-regulate c-myb, c-myc, and cyclin D2 and to repress junB, p16INK4a, and p21Cip1/WAF. As a result, development of LN c-KitloIL-7Rα+ progenitors is blocked at an intermediate CD44+CD25lo development stage in vivo, and LN-derived progenitors fail to generate mature T cells when cultured with OP9-DL1 stromal cells. LN stroma can provide key signals for T-cell development including IL-7, Kit ligand, and Delta-like–1 but lacks Wnt4 and Wnt7b transcripts. LN c-KitloIL-7Rα+ progenitors are able to generate mature T cells when cultured with stromal cells producing wingless-related MMTV integration site 4 (Wnt4) or upon in vivo exposure to oncostatin M whose signaling pathway intersects with Wnt. Thus, supplying Wnt signals to c-KitloIL-7Rα+ progenitors may be sufficient to transform the LN into a primary T-lymphoid organ. These data provide unique insights into the essence of a primary T-lymphoid organ and into how a cryptic extrathymic T-cell development pathway can be amplified.
Introduction
In all animals with an adaptive immune system, the thymus is the primary lymphoid organ for T-cell development and no other organ can compensate for defective thymic function.1 This is problematic considering that progressive thymus atrophy ultimately affects all aging subjects and can even impinge on younger subjects affected by several serious illnesses.2-4 A key question that has baffled immunologists for 40 years is the nature of the signals provided by thymic stromal cells that are necessary and sufficient for T-cell development.5
Strikingly, recent studies have shown that a bone marrow stromal cell line ectopically expressing the Notch ligand Delta-like–1 (OP9-DL1) acquired the capacity to induce the differentiation of hematopoietic progenitors into functional T cells in vitro.6,7 A startling and important implication is that the 3-dimensional thymic microenvironment and the presence of thymic epithelial cells are not essential for T-cell development.6 Thymus-independent T-cell development can also take place in vivo.8-10 Thus, using transgenic mice bearing a green fluorescent protein (GFP) gene placed under the control of the recombination-activating gene 2 (RAG2) promoter, Guy-Grand et al9 showed that T lymphopoiesis occurred in lymph nodes (LNs) and less in the Peyer patches of athymic mice. This cryptic T-cell development pathway however generates only limited numbers of mature T cells.9 Unexpectedly, signals transmitted by the leukemia inhibitory factor (LIF) receptor following prolonged exposure to mouse LIF or bovine oncostatin M (OM) amplify the cryptic LN T-lymphopoietic pathway and transform the mouse LNs into primary T-lymphoid organs.11-16 Thus, about 215 × 106 Thy1+CD4+CD8+ cells are present in the mesenteric LNs of 12-week-old lckOM transgenic mice that express the OM transgene under the control of the proximal lymphocyte protein tyrosine kinase (lck) promoter.15 Studies of OM-transgenic mice showed that this extrathymic pathway is thymus independent, generates functional T lymphocytes, and is regulated by a cyclooxygenase-2–dependent proliferation of high endothelial venules.16-19 The goal of our work was to determine why LNs are normally unable to support T-cell development and how OM can alleviate this defect. We surmised that such knowledge would provide invaluable information on the essence of a primary T-lymphoid organ, that is, how stromal cells regulate crucial early steps in T-cell development.
Materials and methods
Mice
Flow cytometry analysis and cell sorting
The following antibodies were used: biotin and phycoerythrin–cyanin 7 (PE-Cy7) anti-CD8α (53-6.7); biotin anti-CD8β (53-5.8); allophycocyanin (APC)–Cy7 anti-CD4; biotin anti-NK1.1 (PK136); biotin, APC, and fluorescein isothiocyanate (FITC) anti–T-cell receptor β (anti-TCRβ; H57); biotin anti-TCRγδ (GL-3); FITC and PE anti-CD44 (IM7); biotin, APC-Cy7, PE, and APC anti-CD25 (PC61); biotin mouse lineage panel (CD3ϵ, CD11b, CD45R/B220, Ly6C, Ly6G [GR-1], TER-119/erythroid cells [Ly-76]); purified anti-CD127 (interleukin-7 receptor α [IL-7Rα], A7R34) detected with goat anti–rat FITC; APC anti-CD117 (c-Kit, 2B8); FITC anti-CD24 (heat-stable antigen [HSA]); PE-Cy5 and PE anti–stem cell antigen-1 (anti–Sca-1; E13-161.7); FITC anti–bromodeoxyuridine (anti-BrdU; 3D4) with its isotype control (MOPC-21); and FITC anti–B-cell lymphoma 2 (anti–Bcl-2; 3F11) with its isotype control (A19-3). Biotinylated antibodies were detected with streptavidin peridinin chlorophyll protein or PE-Cy7. Anti-CD127 was purchased from eBioscience (San Diego, CA); the other antibodies mentioned earlier in this paragraph, as well as annexin V–FITC, were purchased from BD Pharmingen (San Diego, CA) and Cedarlane Laboratories (Hornby, ON, Canada). Polyclonal purified anti–phospho–signal transducers and activators of transcription 3 (anti–phospho-Stat3; Tyr705; Signaling Technology, Beverly, MA) was detected with FITC goat anti–rabbit immunoglobulin G F(ab)2 (Abcam; Cambridge, MA). Intracellular staining was done as previously described for BrdU,20 TCRβ, Bcl-2,21 and phospho-Stat3.22 Cells were analyzed on a FACSCalibur flow cytometer using CellQuest software and sorted on a FACSVantage SE system with FACSDiVa option (BD Biosciences, San Jose, CA). To preserve cell integrity and avoid loss of rare cell subsets, electronic sorting was done without preliminary cell depletion. The purity of sorted cell populations was above 98%.
RT-PCR analysis
RNA was prepared from cells sorted in trizol reagent (Invitrogen, Burlington, ON, Canada) followed by chloroform extraction and RNA precipitation following the manufacturer's instructions. We performed reverse transcriptase–polymerase chain reaction (RT-PCR) with Qiagen OneStep RT-PCR Kit (Valencia, CA). Previously described RT-PCR conditions and primers were used for Hprt, Rag1, γc, Ptrca, and Cd3e.23 Primers and annealing temperatures were as follows for Hes1 (forward) 5′-GCCAGTGTCAACACGACACCGG-3′, (reverse) 5′-TCACCTCGTTCATGCACTCG3′ (66°C); and for Deltex1 (forward) 5′-CACTGGCCCTGTCCACCCAGCCTTGGCAGG-3′, (reverse) 5′-GGGAAGGCGGGCAACTCAGGCCTCAGG-3′ (55°C). Negative controls were performed in all assays (water and no RT).
Quantitative RT-PCR
Lymphoid cells were separated from stromal cells by mechanical mashing of lymphoid organs through a cell strainer as described.18,24 The mRNA was extracted in trizol reagent and reverse transcription was carried out using SuperScript II RNaseH Reverse Transcriptase (Invitrogen). Real-Time RT-PCR was performed with an ABI Prism Sequence Detection System 7700 (Applied Biosystems, Foster City, CA) using TaqMan Universal PCR Master Mix (Applied Biosystems). Triplicate wells were averaged and the target gene values were normalized for Hprt content. We used specific primers and probes (TaqMan gene expression assays) from Applied Biosystems.
T-cell progenitors and OP9 cell cocultures
Double-negative 1 (DN1) and DN4 lineage-negative (Lin–) progenitors were sorted according to surface expression of CD44, c-kit, and Sca-1. Unless stated otherwise, sorted cells were seeded at 4 × 104 cells/well onto 24-well tissue plates containing a confluent monolayer of OP9 cells transfected with constructs encoding (i) GFP alone; (ii) GFP and DL1; or (iii) GFP, DL1, and wingless-related MMTV integration site 4 (Wnt4). Wnt4 plasmid (Upstate Biotechnology, Lake Placid, NY) transfection of OP9-DL1 cells was carried out using FuGene 6 (Roche Biochemicals, Rotkreuz, Switzerland) according to manufacturer's instructions. All cocultures were performed in the presence of IL-7 and fms-like tyrosine kinase-3 ligand (Flt3L; Peprotech, Rocky Hill, NJ).6 Cocultures were harvested by forceful pipetting at the indicated time points and stained for flow cytometry analysis.
Results
Lymphoid progenitors committed to the T lineage are present in the LNs
The least mature thymocytes are termed DN1 cells and express a Lin–CD44+CD25– surface phenotype. Two subsets of DN1 thymocytes can generate T lymphocytes: c-KithiIL-7Rα– early thymic progenitors (ETPs) and c-KitloIL-7Rα+ common lymphoid progenitors (CLPs).25-30 Thymocytes subsequently go through DN2 (CD44+CD25+), DN3 (CD44–CD25+), and DN4 (CD44–CD25–) stages before giving rise to CD4+CD8+ double-positive T cells.
LNs can support in situ generation of mature single-positive (SP) T cells following intravenous injection of DN thymocytes but not of hematopoietic stem cells into athymic hosts.31 This means that the lack of T-cell development in the LNs under normal conditions is due to the failure of LNs to attract T-cell progenitors or to support some early event at the DN stage. To discover the early step in T-cell development that occurs in the thymus and the OM-transgenic LNs but not the wild-type (wt) LNs, we first analyzed populations of Lin– cells in these organs. All analyses of LN cells in this work were performed on mesenteric LNs. We discriminated 3 subsets of DN1 phenotype cells according to the level of c-Kit expression (negative, low, or high) because previous reports showed that this marker identifies cell subsets with different T-cell progenitor potential.26-28,32 Overall, DN1 phenotype cells were present in similar numbers in the thymus and wt LNs and were more abundant in the OM+ LNs (Figure 1A-B). However, notable discrepancies were found among DN1 cell subsets in the 3 organs. Strikingly, c-Kithi DN1 cells were present exclusively in the thymus (Figure 1C-D). In contrast, c-Kit– and c-Kitlo DN1 phenotype cells were more abundant in the wt LNs than the thymus and even more so in the OM+ LNs (Figure 1C). Furthermore, among c-Kitlo DN1 cells, the percentage of Sca-1+ elements was lower in the wt and OM+ LNs (∼19%) than in the thymus (∼ 38%; Figure 1D). The vast majority of c-Kit– and c-Kitlo thymic and LN DN1 cells were IL-7Rα+, whereas thymic c-Kithi cells were IL-7Rα– (data not shown).
Relative to the thymus, wt and OM+ LNs showed an increased proportion of cells bearing a pre-DN2 phenotype (CD44+CD25lo; Figure 1A). Detection of Hes1, Deltex1, Rag1, and Cd3e transcripts indicates that DN1 and pre-DN2 subsets in wt and OM+ LNs contain cells committed to the T lineage (Figure 2A-B). DN2 and DN3 cells were practically undetectable in wt LNs, yet cells with a DN4 phenotype were present (Figure 1A-B). At the population level, the transcriptome of “illegitimate” wt LN DN4 phenotype cells was not identical to that of genuine thymic DN4 cells, as shown by differences in levels of Rag1, Ptcra, and Hes1 transcripts (Figure 2E). However, at least some of the illegitimate DN4 phenotype cells in the wt LNs were committed to the T lineage: (i) they contained Cd3e transcripts (Figure 2E); (ii) about 19% expressed intracytoplasmic TCRβ chains (Figure 2F); and (iii) when cultured for 7 days in the presence of OP9-DL1 stromal cells, which can support all stages of T-cell development, wt LN–derived DN4 cells generated CD4+CD8+ and SP TCRβ+ T cells (Figure 2G). In contrast to the wt LNs, the numbers of cells with DN2, DN3, and DN4 phenotype were similar in OM+ LNs and thymus (Figure 1B). Furthermore, DN2, DN3, and DN4 cells in the OM+ LNs were similar to those in the thymus with regard to the levels of several transcripts and expression of the c-Kit protein (Figure 2C-E). The sole difference between thymic and OM+ LN DN cells was the lower proportion of DN4 cells with rearranged TCRβ chains in the OM+ LNs (Figure 2F).
LNs contain heterogeneous subsets of Lin– DN cells. (A) To estimate the proportion of cells with DN1-DN4 phenotype, lymphoid cells from the thymus, wt LNs, and OM+ LNs were stained for CD25, CD44, and lineage markers (CD3ϵ, CD8α, CD8β, CD11b, CD45R/B220, Ly6C, Ly6G, NK1.1, TER-119, TCRβ, and TCRγ). Numbers in the various quadrants correspond to percentage of Lin– cells stained with CD25 and CD44. (B) Number of cells with DN1-DN4 phenotype in the 3 lymphoid organs (mean ± SD; n = 3). (C) Number of c-Kit–, c-Kitlo, and c-Kithi DN1 phenotype cells per 106 lymphoid cells. Gated Lin–CD44+CD25– cells were stained for c-Kit (mean ± SD; n = 3). No DN1 phenotype c-Kithi cells were detected in the wt and OM+ LNs (*). (D) Expression of c-Kit on DN1 phenotype cells (Lin–CD44+CD25–) and of Sca-1 on both c-Kitlo and c-Kithi subsets are shown for each organ. Numbers correspond to percentages of cells in each quadrant. Data in panels A and D are representative of 1 experiment out of 3.
LNs contain heterogeneous subsets of Lin– DN cells. (A) To estimate the proportion of cells with DN1-DN4 phenotype, lymphoid cells from the thymus, wt LNs, and OM+ LNs were stained for CD25, CD44, and lineage markers (CD3ϵ, CD8α, CD8β, CD11b, CD45R/B220, Ly6C, Ly6G, NK1.1, TER-119, TCRβ, and TCRγ). Numbers in the various quadrants correspond to percentage of Lin– cells stained with CD25 and CD44. (B) Number of cells with DN1-DN4 phenotype in the 3 lymphoid organs (mean ± SD; n = 3). (C) Number of c-Kit–, c-Kitlo, and c-Kithi DN1 phenotype cells per 106 lymphoid cells. Gated Lin–CD44+CD25– cells were stained for c-Kit (mean ± SD; n = 3). No DN1 phenotype c-Kithi cells were detected in the wt and OM+ LNs (*). (D) Expression of c-Kit on DN1 phenotype cells (Lin–CD44+CD25–) and of Sca-1 on both c-Kitlo and c-Kithi subsets are shown for each organ. Numbers correspond to percentages of cells in each quadrant. Data in panels A and D are representative of 1 experiment out of 3.
Two major points can be made from these data. First, Lin–c-KithiIL-7Rα– DN1 cells, whose phenotype corresponds to that of ETPs,26 are present exclusively in the thymus. A corollary is that, at least in the OM+ LNs, mature T cells can be produced in the absence of c-KithiIL-7Rα– DN1 cells. This extrathymic pathway is truly thymus independent, as shown in athymic mice reconstituted with OM transgenic fetal liver or injected with OM.14,15,18 Second, accumulation of pre-DN2 cells in wt and OM+ LNs and emergence of DN2 and DN3 cells in OM+ but not wt LNs suggest that failure of wt LNs to support T-cell development is due to a blockade of the DN1-to-DN2 transition that is alleviated in the OM+ LNs.
Proliferation of DN cells
In the thymus, DN cells proliferate extensively, particularly at the DN2 and DN4 stages.20 To analyze the proliferation of DN cell subsets in the thymus and LNs, BrdU was injected intraperitoneally, mice were killed 40 minutes later, and cell cycle status was determined by staining with anti-BrdU antibody and 7-aminoactinomycin D (7AAD).33 In addition, the proportion of apoptotic cells was estimated by annexin V labeling. As opposed to their thymic counterparts, all DN phenotype cells in the wt LNs were arrested at the G1 phase of the cell cycle with virtually no cells in S phase (Figure 3A). In the OM+ LNs, the percentage of cells in S phase was similar to thymocytes for DN1, DN2, and DN3 cells but significantly lower for the pre-DN2 and DN4 subsets (Figure 3A). Among DN cells in the wt LNs, lack of proliferation was correlated with higher proportion of apoptotic cells compared with the thymus and the OM+ LNs (Figure 3C).
Since DN1 cells found in lymphoid organs are heterogeneous (Figure 1D), we sought to provide a more accurate estimation of their mitotic behavior by assessing BrdU incorporation in cell subsets expressing different levels of c-Kit (Figure 3B). In the thymus, BrdU+ DN1 cells were mainly found in the c-Kitlo and c-Kithi cell subsets (Figure 3B). In contrast, BrdU incorporation by DN1 cells in LNs was independent of c-Kit level, being of similar and relatively modest magnitude among c-Kitneg and c-Kitlo cells, and increased about 2-fold in OM+ relative to wt LNs (Figure 3B). Thus, cell cycle status of DN1 cells was correlated with c-Kit expression in the thymus but not wt or OM+ LNs. The low level of BrdU incorporation among c-Kitlo DN1 cells in the LNs relative to the thymus suggests that the LN stroma fails to provide either Kit ligand or another signal that promotes proliferation of c-Kitlo DN1 cells in the thymus.
Key differences between thymus and LN stroma involve Wnt proteins
The data in Figure 1 show that failure of wt LNs to support T-cell development is due to an inability to complete the DN1-to-DN2 transition. This defect is largely alleviated in OM+ LNs. T-cell development is however not entirely thymuslike in the OM+ LNs where accumulation of pre-DN2 cells and relatively low proliferation of DN4 phenotype cells were found. Signals required for the development of thymocytes at the DN1-DN2 stage are initiated by key ligands that control proliferation and survival (IL-7, kit ligand, and possibly Wnt proteins)34-39 and T-cell lineage commitment (Delta-like Notch-1 ligands).40,41 Expansion of the DN4 cell subset depends on expression of the pre-TCR (at the DN3 stage), which has no ligand, and is probably enhanced by Wnt signals.33,39,42-45 We therefore performed quantitative real-time RT-PCR on the stroma of lymphoid organs to evaluate the expression profile of IL-7, Kit ligand, Delta-like proteins, and 6 Wnt proteins that are normally present in the thymus.46,47 We also assessed expression of the Flt3L cytokine gene because, although it is not essential for T-cell development, it may influence the proliferation and survival of lymphoid progenitors.48,49
We found no deficit of the following transcripts in the wt LNs relative to the thymus: Il7, Kitl, Flt3L, Delta-like–1 and –4, Wnt1, Wnt7a, Wnt10a, and Wnt10b (Figure 4). Furthermore, none of these transcripts was more abundant in the OM+ compared with wt LNs. However, 2 salient differences were observed between the thymus and LNs: Wnt4 and Wnt7b transcripts were present in the thymus (as previously reported46,47 ) but absent in the LNs (P < .001 and P < .005, respectively; Figure 4). Although we cannot formally exclude that lack of Wnt7b in the LNs may be biologically relevant, we elected to focus our attention (and culture experiments described in the last paragraph of “Results”) on Wnt4 for the following reasons: (i) Wnt4, which regulates FoxN1 expression, is the most abundantly expressed Wnt family member in both embryonic thymic epithelium as well as mature thymic cortical epithelium46,47 ; (ii) aside from Wnt1, Wnt4 is the only Wnt protein for which a role in thymopoiesis is supported by experimental evidence36,50 ; and (iii) OP9-DL1 stromal cells that can support all steps of T-cell development express Wnt4 but not Wnt7b (Figure 4). Stromal fractions may be contaminated by a few adherent lymphoid cells.18,24 Evaluation by quantitative RT-PCR of Wnt4 transcripts in thymus lymphoid and stromal fractions confirmed that stromal cells were the main if not the sole site of Wnt4 transcription in the thymus (data not shown). Lack of Wnt4 in the LNs could be pivotal since T-cell development in the LNs (Figures 1A and 3A) is impaired at 2 stages where Wnt signals have been proposed to influence thymocyte development39,42,43 : expansion of the DN2 and of the DN4 compartments. Lack of Wnt4 protein in the LNs could provide a parsimonious explanation for both defects.
LNs contain lymphoid progenitors committed to the T lineage. RT-PCR analysis of DN1 (A), pre-DN2 (B), and DN4 (E) cells sorted from wt thymus, wt LNs, and OM+ LNs. One-step RT-PCR was done on the same mRNA samples for transcripts of interest and for Hprt. (C) c-Kit expression on DN2 and DN3 subsets from wt thymus (dotted line) and OM+ LNs (solid line). Intracellular TCRβ (icTCRβ) chain expression in DN3 (D) and DN4 (F) subsets. (G) Sorted DN4 cells (105; Lin–CD8–CD44–CD25–) harvested from wt LNs were cocultured on OP9-DL1 cells and analyzed for T-cell development after 7 days of in vitro culture. Numbers indicate cell population percentages. In panels F and G, dotted lines represent staining with isotype control antibodies.
LNs contain lymphoid progenitors committed to the T lineage. RT-PCR analysis of DN1 (A), pre-DN2 (B), and DN4 (E) cells sorted from wt thymus, wt LNs, and OM+ LNs. One-step RT-PCR was done on the same mRNA samples for transcripts of interest and for Hprt. (C) c-Kit expression on DN2 and DN3 subsets from wt thymus (dotted line) and OM+ LNs (solid line). Intracellular TCRβ (icTCRβ) chain expression in DN3 (D) and DN4 (F) subsets. (G) Sorted DN4 cells (105; Lin–CD8–CD44–CD25–) harvested from wt LNs were cocultured on OP9-DL1 cells and analyzed for T-cell development after 7 days of in vitro culture. Numbers indicate cell population percentages. In panels F and G, dotted lines represent staining with isotype control antibodies.
Survival and proliferation of DN cells are impaired in wt LNs. (A) Analysis of cell cycle status of DN phenotype cells. Forty minutes after injection of 1 mg BrdU intraperitoneally, mice were killed and cells were stained with 7AAD and antibodies against BrdU, CD25, CD44, and lineage markers. Numbers correspond to the percentages of cells in the G1/0, S, and G2+M phase of the cell cycle. One representative experiment out of 3. (B) Lin– CD44+c-Kit subsets in cycling thymocytes. Mice received 2 injections (1 mg each) of BrdU at 2-hour intervals. Twenty-four hours later, prepared cells were stained with antibodies against BrdU, CD44, c-Kit, and lineage markers (which included CD25). Numbers indicate the percentage of BrdU+ cells. One representative experiment out of 3. (C) Percentages of annexin V+ cells in wt thymus, wt LNs, and OM+ LNs are indicated in the graphs.
Survival and proliferation of DN cells are impaired in wt LNs. (A) Analysis of cell cycle status of DN phenotype cells. Forty minutes after injection of 1 mg BrdU intraperitoneally, mice were killed and cells were stained with 7AAD and antibodies against BrdU, CD25, CD44, and lineage markers. Numbers correspond to the percentages of cells in the G1/0, S, and G2+M phase of the cell cycle. One representative experiment out of 3. (B) Lin– CD44+c-Kit subsets in cycling thymocytes. Mice received 2 injections (1 mg each) of BrdU at 2-hour intervals. Twenty-four hours later, prepared cells were stained with antibodies against BrdU, CD44, c-Kit, and lineage markers (which included CD25). Numbers indicate the percentage of BrdU+ cells. One representative experiment out of 3. (C) Percentages of annexin V+ cells in wt thymus, wt LNs, and OM+ LNs are indicated in the graphs.
LN stroma lacks Wnt4 and Wnt7b transcripts. The mRNA expression profile of selected genes in the stroma of wt thymus and LNs, OM+ LNs, and OP9-DL1 cells. The mRNA values were normalized according to Hprt and thymic stroma mRNA levels were set as 1. Data are mean ± SD from 3 or 4 independent experiments (*no detectable mRNA after 50 amplification cycles). Differences between groups were evaluated with Student t test. Levels of statistical significance for comparison of wt thymus versus wt LNs are ¥P < .04, §P < .005, and ‡P <.001. †Levels of DL1 transcripts for OP9-DL1 cells (715 ± 55) are not shown on the graph.
LN stroma lacks Wnt4 and Wnt7b transcripts. The mRNA expression profile of selected genes in the stroma of wt thymus and LNs, OM+ LNs, and OP9-DL1 cells. The mRNA values were normalized according to Hprt and thymic stroma mRNA levels were set as 1. Data are mean ± SD from 3 or 4 independent experiments (*no detectable mRNA after 50 amplification cycles). Differences between groups were evaluated with Student t test. Levels of statistical significance for comparison of wt thymus versus wt LNs are ¥P < .04, §P < .005, and ‡P <.001. †Levels of DL1 transcripts for OP9-DL1 cells (715 ± 55) are not shown on the graph.
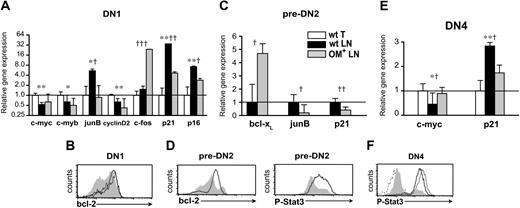
LN DN cells show distinct gene expression profiles. Real-time RT-PCR assays and FACS analysis measuring gene and protein expression in DN1 (A-B), pre-DN2 (C-D), and DN4 (E-F) cells sorted from lymphoid organs. The mRNA levels of the wt thymus for DN1 (A) and DN4 (E) cells and of wt LN for pre-DN2 (C) cells were set as 1. Hprt mRNA levels were used to normalize cDNA content among subpopulations. (A,C,E) Data are mean ± SD from 3 independent experiments. Differences between groups were evaluated with Student t test. Levels of statistical significance for comparison of wt thymus versus wt LNs are *P < .02 and **P < .003 and for comparison of wt LN versus OM+ LNs, †P < .04, ††P < .008, and †††P < .001. Intracellular staining for Bcl-2 and phospho-Stat3 (P-Stat3) proteins was done (B,D,F) on wt thymus (solid black line), wt LNs (gray shaded histograms), and OM+ LNs (solid gray line) DN cells. Secondary antibody was used as a negative control for P-Stat3 staining (dotted line).
LN DN cells show distinct gene expression profiles. Real-time RT-PCR assays and FACS analysis measuring gene and protein expression in DN1 (A-B), pre-DN2 (C-D), and DN4 (E-F) cells sorted from lymphoid organs. The mRNA levels of the wt thymus for DN1 (A) and DN4 (E) cells and of wt LN for pre-DN2 (C) cells were set as 1. Hprt mRNA levels were used to normalize cDNA content among subpopulations. (A,C,E) Data are mean ± SD from 3 independent experiments. Differences between groups were evaluated with Student t test. Levels of statistical significance for comparison of wt thymus versus wt LNs are *P < .02 and **P < .003 and for comparison of wt LN versus OM+ LNs, †P < .04, ††P < .008, and †††P < .001. Intracellular staining for Bcl-2 and phospho-Stat3 (P-Stat3) proteins was done (B,D,F) on wt thymus (solid black line), wt LNs (gray shaded histograms), and OM+ LNs (solid gray line) DN cells. Secondary antibody was used as a negative control for P-Stat3 staining (dotted line).
Wnt and LIF/OM signaling pathways in DN phenotype cells
Wnt signaling is complex: there are 18 Wnt proteins in the mouse and their target genes differ among various cell types.51,52 The transcriptional response specifically elicited by discrete Wnt proteins, particularly Wnt4, has not been fully characterized in immature T cells. To discover whether and how lack of Wnt signals could hamper T-cell development, we used quantitative PCR to study the expression of genes that have been both implicated in thymocyte development37,53-55 and shown to be regulated by Wnt signals in various cell types.37,56-58 We performed these studies in the 2 subsets of DN phenotype cells that are present in significant numbers in both the thymus and wt LNs, that is, DN1 and DN4 cells (Figure 1A-B). Wnt signaling promotes cell proliferation by increasing transcription of c-myb, c-myc, and c-fos and decreasing that of junB. Key downstream events include induction of cyclin D2 by c-myc59 and repression of 2 cyclin-dependent kinase inhibitors (p16INK4a and p21Cip1/WAF1) that are induced by junB and repressed by c-fos.60,61 In line with this, transcript levels of c-myb, c-myc, and cyclin D2 were lower whereas those of junB, p16INK4a, and p21Cip1/WAF1 were higher in wt LNs compared with thymus DN cells (Figure 5A,E). However, c-fos levels were not deficient in the wt LNs relative to thymus DN1 cells (Figure 5A). Thus, aside from c-fos levels, transcript profiles provide consistent albeit indirect evidence for a dearth of Wnt signals in DN cells from the wt LNs relative to the thymus. This suggests that in DN cells, Wnt4 (and possibly Wnt7b; Figure 4) signals may have a nonredundant effect on genes such as c-myb, c-myc, and junB but are not essential for induction of c-fos.
Bovine OM binds only to the LIF receptor in mouse.11-13 While extrathymic T-cell development in OM-transgenic mice must therefore be induced by OM binding to the LIF receptor, it has not been determined whether this interaction occurs specifically in immature T cells. To address this, we studied the 3 subsets of DN phenotype cells present in both the wt and OM+ LNs (DN1, pre-DN2, and DN4; Figure 1A-B). Signals from the LIF receptor partially overlap with those induced by Wnt signaling62 and have a similar impact on transcription of c-fos, junB, p16INK4a, p21Cip1/WAF1, and c-myc.63-65 Comparison of transcript levels in the OM+ relative to wt LNs supports the idea that OM signals in DN cells from the OM+ LNs compensate for the lack of Wnt signaling: levels of c-fos and c-myc were higher whereas those of junB, p16INK4a, and p21Cip1/WAF1 were decreased in DN cells from the OM+ relative to the wt LNs (Figure 5A,C,E). Supplementary evidence for OM signaling66,67 in DN cells from the OM+ LNs included up-regulation of Bcl-2 in DN1 and pre-DN2 cells (Figure 5B,D), of Bcl-xL in pre-DN2 cells (Figure 5C), and of phospho-Stat3 in pre-DN2 and DN4 cells (Figure 5D,F).
In vitro differentiation of c-Kitlo and c-Kithi progenitors
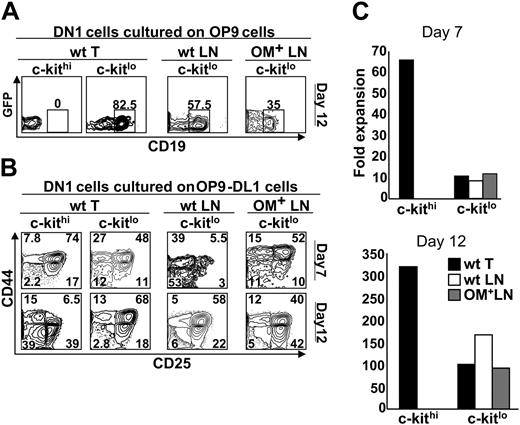
We next asked whether culture with stromal cells expressing Wnt4 could allow DN1 phenotype cells from the LNs to undergo T-lineage differentiation. OP9-DL1 cells express Wnt4, albeit at lower levels than thymic stromal cells (Figure 4). Thus, we cultured the following subsets of DN1 phenotype cells in the presence of OP9-DL1 stromal cells: c-Kithi (Sca-1+) cells from the thymus, as well as the Sca-1– and Sca-1+ subsets of c-Kit– and c-Kitlo cells from the thymus, wt LNs, and OM+ LNs. As expected, for all cell subsets tested, no development toward the T lineage was observed in presence of OP9 cells, that is, in the absence of the Notch ligand Delta-like–1 (data not shown). In the presence of OP9-DL1 cells, T-cell differentiation was observed with thymic c-Kithi cells and c-KitloSca-1+ cells from the 3 lymphoid organs (Figure 6B). In contrast, no T-cell differentiation (appearance of DN2 phenotype cells) was observed with c-Kit–Sca-1–, c-Kit–Sca-1+, and c-KitloSca-1– subsets (data not shown). Interestingly, the behavior in culture of c-Kit– and c-Kitlo cell subsets was not influenced by their site of origin (thymus, wt LNs, or OM+ LNs; Figure 6). Consistent with previous studies,28 c-Kithi (thymic) DN1 cells cultured with OP9-DL1 cells proliferated extensively and generated DN4 cells after 12 days (Figure 6B) and CD4+CD8+ as well as SP T cells after 18 days (data not shown). In comparison with c-Kithi DN1 cells, c-KitloSca-1+DN1 cells (from the thymus or LNs) showed 2 deficits: (i) in terms of absolute numbers, they accumulated to lower levels on days 7 and 12 (Figure 6C); and (ii) their progeny showed a very low proportion of DN4 cells on day 12 (Figure 6B). Furthermore, c-KitloSca-1+ differed from c-Kithi DN1 thymic cells in that only the former generated substantial numbers of CD19+ B cells when cultured on OP9 cells (Figure 6A). Thus, when cultured with OP9-DL1 cells, c-KitloSca-1+ DN1 cells from the thymus and LNs progress well up to the DN3 stage, but expansion of their DN4 cell progeny is limited.
c-Kitlo and c-Kithi progenitors display different differentiation potential when grown on OP9 and OP9-DL1 cells. The following subsets of Lin–CD44+CD25–DN1 cells were sorted: c-KitloSca-1+ cells from the thymus, wt LNs, and OM+ LNs, and c-KithiSca-1+ cells from the thymus. These DN1 cell populations were plated on a confluent monolayer of (A) OP9 cells or (B) OP9-DL1 cells and analyzed by flow cytometry at the indicated time points. (C) Fold expansion of progenitors plated on OP9-DL1 cells was measured by dividing the number of cells harvested by the number of cells initially plated. The analysis is of 1 representative experiment out of 3.
c-Kitlo and c-Kithi progenitors display different differentiation potential when grown on OP9 and OP9-DL1 cells. The following subsets of Lin–CD44+CD25–DN1 cells were sorted: c-KitloSca-1+ cells from the thymus, wt LNs, and OM+ LNs, and c-KithiSca-1+ cells from the thymus. These DN1 cell populations were plated on a confluent monolayer of (A) OP9 cells or (B) OP9-DL1 cells and analyzed by flow cytometry at the indicated time points. (C) Fold expansion of progenitors plated on OP9-DL1 cells was measured by dividing the number of cells harvested by the number of cells initially plated. The analysis is of 1 representative experiment out of 3.
The OM transgene is under the control of the lck proximal promoter in lckOM mice. Expression of the lck proximal promoter is up-regulated in DN3 cells and remains substantial in DN4, CD4+CD8+, and SP T cells.68 Thus, a plethora of T-lineage cells produces OM in the lckOM LNs.15,17 In contrast, when c-KitloSca-1+ DN1 cells from lckOM LNs were plated on OP9-DL1 cells, their initial development took place in the absence of OM-producing cells. Thus, they did not fare better than c-KitloSca-1+ DN1 cells derived from wt LNs (Figure 6B).
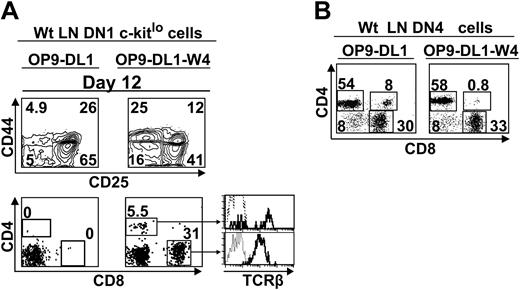
OP9-DL1 stromal cells express only low levels of Wnt4, about 15% those of the thymus stroma (Figure 4). We therefore engineered OP9-DL1 cells expressing levels of Wnt4 transcripts similar to the thymus (data not shown) and tested their ability to support the development of c-KitloSca-1+ LN DN1 phenotype cells. Provision of thymuslike amounts of Wnt4 by OP9-DL1 stromal cells (OP9-DL1–W4 cells) increased by 3-fold the percentage of DN4 cells generated from c-KitloSca-1+ LN DN1 cells on day 12 (Figure 7A). Moreover, overexpression of Wnt4 on stromal cells allowed c-KitloSca-1+ LN DN1 cells to generate TCRαβ SP T cells as early as day 12 of culture (Figure 7A). By contrast, no SP T cells were produced when c-KitloSca-1+ LN DN1 cells were cultured for up to 25 days with OP9-DL1 stromal cells (data not shown). In addition to enhancement of DN4 cell expansion, Wnt4 may regulate differentiation events downstream of the DN4 stage since it induced a modest but reproducible shortening of the time required for transition from the DN4 to SP phenotype (Figure 7B). Thus, increased expression of Wnt4 on OP9-DL1–W4 cells was sufficient to allow LN c-KitloSca-1+ cells to generate mature T cells. DN1 cells plated on OP9-DL1–W4 cells generated more CD8 than CD4 T cells (Figure 7A) because OP9 cells express major histocompatibility complex (MHC) class I but do not express MHC class II.6 That CD8 T cells were not favored when DN4 cells were cultured with stromal cells (Figure 7B) probably reflects the fact that some DN4 cells had initiated positive selection on MHC class II in vivo.69
Discussion
It has previously been shown that 2 types of Lin–Sca-1+ progenitors can generate T cells in the thymus: c-KithiIL-7Rα– and c-KitloIL-7Rα+.25-27 We report herein that these progenitors fail to generate T cells in the wt LNs for different reasons: c-KithiIL-7Rα– are absent in the LNs whereas c-KitloIL-7Rα+ are present but cell cycle arrested (Figure 1C). Considering that the relation between c-KithiIL-7Rα– and c-KitloIL-7Rα+ DN1 cells is unclear,26,27 the absence of c-KithiIL-7Rα– DN1 cells in the wt and OM+ LNs means that LNs either fail to attract these progenitors or fail to support their in situ generation from c-Kit– or c-Kitlo cells. On the contrary, c-KitloIL-7Rα+ are more abundant in the wt LNs than the thymus and even more so in the OM+ LNs (Figure 1C). Nevertheless, full differentiation of c-KitloIL-7Rα+ progenitors cannot take place in the wt LNs because of the absence of signals that may be provided by Wnt4 (and possibly Wnt7b) in the thymus and by OM in the OM-transgenic LNs. Accumulation of Lin–c-KitloIL-7Rα+ cells in OM+ LNs appears to be a local phenomenon because we found no increase of Lin–c-KitloIL-7Rα+ cell numbers in the bone marrow of OM+ mice (data not shown). It is tempting to speculate that accumulation of Lin–c-KitloIL-7Rα+ cells may be related to the proliferation of high endothelial venules found in OM+ LNs.18 A corollary warranting further investigation is that LN high endothelial venules might express a unique ligand important for homing of Lin–c-KitloIL-7Rα+ progenitors. All analyses of LN cells in this work were performed on mesenteric LNs. However, our data can probably be generalized to other LNs inasmuch as extrathymic T-cell development in OM-transgenic mice was found not only in mesenteric but also in cervical and axillary LNs.15
This work strongly suggests that in the wt LNs the T-lineage differentiative potential of Lin–c-KitloIL-7Rα+ cells is thwarted by the absence of Wnt signaling. First, Lin– DN cell subsets in the wt LNs are characterized by 3 features reported in the thymus of mutant mice with disruption of the Wnt signaling pathway: blockade of DN1→DN2 transition with accumulation of pre-DN2 cells (Figure 1), presence of illegitimate DN4 cells in absence of DN2 and DN3 cells (Figure 1), and cell cycle arrest of DN4 cells (Figure 3). Indeed, transcription factor Tcf-1 (T-cell transcription factor 1) is essential for DN1 thymocytes to reach the DN2 stage, and accumulation of pre-DN2 cells has been reported in the thymus of mice with impaired guanosine triphosphatase (GTPase) Rho or c-Kit signaling (growth factor independent-1 [Gfi1–/–]).70,71 Moreover, illegitimate DN4 cells were found in the thymus of c-Kit–deficient (Vickid) and FoxN1-mutant mice, and noncycling DN4 cells were reported in Brahma-related gene (Brg)–deficient mice.23,33,72 The common link among Tcf-1, GTPase Rho, c-Kit, Brg, and FoxN1 is that they are involved in Wnt signaling.46,52,57,73 Second, among key ligands that are produced by stromal cells and that can regulate early steps of T-cell development, Wnt4 and Wnt7b were the sole transcripts that were deficient in the LNs relative to the thymus (Figure 4). Third, expression profiling (Figure 5) incriminates deficient Wnt signaling as a plausible explanation for the proliferative defect of wt LNs relative to thymus DN cells (Figure 3). Fourth, when cultured with OP9-DL1 stromal cells expressing thymuslike levels of Wnt4, wt LN Lin–c-KitloSca-1+IL-7Rα+ progenitors generate SP T cells (Figure 7). Nevertheless, generation of Wnt transgenic mice will be essential to directly evaluate whether T-cell development in the LN is normally hampered solely by the lack of Wnt signals.
LN c-Kitlo progenitors can complete T-cell development when grown on stromal cells transfected with Wnt4 (OP9-DL1-W4). (A) Sorted DN1 cells (4 × 103; Lin–CD44+CD25–Sca1+c-kitlo) from wt LNs were plated on 6-well tissue culture plates containing a confluent monolayer of OP9-DL1 cells transfected or not with Wnt4 and analyzed by flow cytometry after 12 days in culture. Dotted line represents isotype control. (B) Sorted DN4 cells (105; Lin–CD44–CD25–) harvested from wt LNs were cocultured on OP9-DL1 or OP9-DL1-W4 cells and analyzed on day 7. Numbers indicate cell population percentages. The analysis is of 1 representative experiment out of 2.
LN c-Kitlo progenitors can complete T-cell development when grown on stromal cells transfected with Wnt4 (OP9-DL1-W4). (A) Sorted DN1 cells (4 × 103; Lin–CD44+CD25–Sca1+c-kitlo) from wt LNs were plated on 6-well tissue culture plates containing a confluent monolayer of OP9-DL1 cells transfected or not with Wnt4 and analyzed by flow cytometry after 12 days in culture. Dotted line represents isotype control. (B) Sorted DN4 cells (105; Lin–CD44–CD25–) harvested from wt LNs were cocultured on OP9-DL1 or OP9-DL1-W4 cells and analyzed on day 7. Numbers indicate cell population percentages. The analysis is of 1 representative experiment out of 2.
In wt mice, LNs fail to support T-cell development primarily because they are unable to sustain the DN1-to-DN2 transition. This defect is largely but not completely alleviated in OM+ LNs (Figures 1A and 3A). Additional studies are therefore needed to define more precisely events downstream of Wnt and OM/LIF signaling that influence the development of early lymphoid progenitors. Studies in other cell types suggest that OM/LIF signals partially overlap with Wnt signals and have a similar impact on transcription of c-fos, junB, p16INK4a, p21Cip1/WAF1, and c-myc.63-65 However, only Wnt may sustain expression of the transcription factors Oct-3/4, Rex-1, and Nanog.62
In contrast to c-KitloIL-7Rα+ cells, thymic c-KithiIL-7Rα– cells did not require high levels of Wnt4 to generate T cells in vitro (Figure 6). Two possibilities could explain this unexpected discrepancy between the 2 types of progenitors: c-KithiIL-7Rα– cells do not require Wnt signals, or they can use Wnt ligands other than Wnt4. Considering the great controversy over the involvement of Wnt signaling in T-cell development,38,39,50,52 both scenarios must be considered perfectly plausible. Notably, our in vitro culture data support the growing consensus that the proliferation rate and T-cell generation potential of thymic c-KithiIL-7Rα– are unmatched by other DN1 cell subsets.26,28 Major questions are why c-KithiIL-7Rα– cells are found only in the thymus and whether these cells could generate T cells as efficiently in the LNs as in the thymus. Nonetheless, one concept emerging from our work is that in stark contrast with what is seen in the thymus, T-cell production in the LNs occurs in the absence of c-KithiIL-7Rα– cells. This raises the interesting question of whether progenitor type (c-KithiIL-7Rα– vs c-KitloIL-7Rα+) influences the behavior and function of their T-cell progeny.
Whether they were from the thymus or LNs, c-KitloSca-1+IL-7Rα+ populations had the same behavior when cultured in vitro (Figure 6). Their phenotype and their ability to generate T and B cells suggest that they are closely related to bone marrow CLPs,25,27 though clonogenic assays would be required to formally demonstrate that these populations contain bipotent precursors of T and B cells. Of note, CLPs injected into athymic hosts were recently shown to rapidly generate functional CD8 T cells in elusive extrathymic sites.74 The present study suggests that the LN is a primary (if not the sole) site where CLPs can generate T cells. Furthermore, coupled to the present work, demonstration that high numbers of extrathymic CD4 SP cells are produced in the OM+ LNs14-16 indicates that CLP phenotype cells can generate not only CD8 but also CD4 T cells in a thymus-independent fashion. Demonstration that the T-cell generation potential of CLP phenotype cells is dramatically amplified by Wnt4 could be relevant for treatment of subjects with T-cell lymphopenia, considering the recent demonstration that CLP-derived T cells can protect against lethal murine cytomegalovirus infection.74 Caution is always warranted when extrapolating data from mouse hematolymphoid precursors to human.75 Nevertheless, the high level of conservation of the Wnt pathway and the 98% amino acid identity between mouse and human Wnt4 suggest that our data on mouse Wnt4 may apply to human. The use of Wnt4 protein could therefore have a valuable role in developing ex vivo culture systems for generating therapeutically relevant numbers of T lymphocytes from extrathymic lymphoid progenitors.
Prepublished online as Blood First Edition Paper, March 3, 2005; DOI 10.1182/blood-2004-12-4886.
Supported by Canadian Institutes of Health Research (CIHR) grant MOP-42384 and the Fonds de la Recherche en Santé du Québec. I.L. is supported by a training grant from the CIHR. J.C.Z-.P. is supported by an Investigator Award from the CIHR. C.P. holds a Canada Research Chair in Immunobiology.
R.T. and I.L. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to PRO-DNA Diagnostic for help with quantitative PCR and J. A. Kashul for editorial assistance.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal