Abstract
KIT exon 8 mutations are located in the extracellular portion of the receptor and are strongly associated with core-binding factor (CBF)-acute myeloid leukemia (AML). To characterize the functional role of these mutants, we analyzed the proproliferative and antiapoptotic potential of 3 KIT exon 8 mutations in interleukin 3 (IL-3)-dependent Ba/F3 cells. All KIT exon 8 mutants induced receptor hyperactivation in response to stem cell factor (SCF) stimulation in terms of proliferation and resistance toward apoptotic cell death. A representative KIT exon 8 mutant showed spontaneous receptor dimerization, phosphorylation of mitogen-activated protein kinase (MAPK), and conferred IL-3-independent growth to Ba/F3 cells. MAPK and phosphatidylinositol 3-kinase (PI3-kinase) activation was essential for the phenotype of this mutant. Additionally, imatinib inhibited proliferation of KIT exon 8 mutant-expressing Ba/F3 cells. Our data show that KIT exon 8 mutations represent gain-of-function mutations and might represent a new molecular target for treatment of CBF leukemias. (Blood. 2005;105:3319-3321)
Introduction
KIT is a member of the type III receptor tyrosine kinase family1 and plays a crucial role in normal hematopoiesis and acute myeloid leukemia (AML).2,3
Exon 8 deletion plus insertion mutations of KIT have been described in patients with core-binding factor (CBF)-AMLs, that is, AML M4eo with inv(16) or AML M2 with t(8;21).4-6 These mutations are localized in the extracellular domain of KIT, whereas other known mutations in AML are located at Asp816 in the tyrosine kinase domain of the receptor.3 The occurrence of KIT exon 8 mutations in CBF-AML increases the relapse rate of patients with AML and might confer a poor prognosis.5 Our study provides the first evidence that KIT exon 8 mutations represent gain-of-function mutations that induce hyperactivation of KIT in response to its natural ligand stem cell factor (SCF).
Study design
Cell proliferation of Ba/F3 cells, apoptosis analysis, and application of specific inhibitors were performed as previously described.7
Results and discussion
In this study, we characterized the functional properties of mutations in exon 8 of the KIT gene that are associated with CBF-AML. Table 1 summarizes the amino acid (AA) sequences of all patients with exon 8 mutations that were published by Böll et al9 (patient nos. 1-4) and Gari et al4 (patient nos. 5-11). Because of the marked heterogeneity of these mutations, we first aimed to identify a common mutated region. The deletion at position 419 (Δ419) is present in about 93%5 of all exon 8 mutations and was found as a single abnormality in patient no. 1.9 We also created a mutation that only contained the deletion of 2 codons (Δ418_419) that is frequently observed in patients with an exon 8 mutation (4 of 11 patients, nos. 5, 7, 9, and 10 in Table 1). The mutation containing deletions of AA 417 to 419 and an additional insertion of isoleucine (T417I/Δ418_419) was described in 2 patients by Gari et al (patient nos. 5 and 9). Despite the heterogeneity of KIT exon 8 mutations, the combination of a point mutation at AA 417 with deletions of codons 418 and 419 was found in 4 of 11 patients and might define a common mutation pattern. Thus, the representative KIT-T417I/Δ418_419 mutant was selected for further detailed analyses of receptor physiology and signaling.
Published KIT exon 8 AA sequences in AML patients
. | 416 . | 417 . | 418 . | 419 . | 420 . | 421 . | 422 . |
|---|---|---|---|---|---|---|---|
| Sources . | Leu . | Thr . | Tyr . | Asp . | Arg . | Leu . | Val . |
| Patient no. | |||||||
| 1* | Leu | Thr | Tyr | — | Arg | Leu | Val |
| 2* | Leu | Thr | Tyr | — | Trp | Gly | Val |
| 3* | Leu | Thr | Arg | — | Trp | Leu | Val |
| 4* | Leu | Thr | Ala | — | Arg | Leu | Val |
| 5 | Leu | Ile | — | — | Arg | Leu | Val |
| 6 | Leu | Thr | Tyr | — | — | Phe | Val |
| 7 | Leu | Val | — | — | Arg | Leu | Val |
| 8 | Leu | Arg | Gly | — | Arg | Leu | Val |
| 9 | Leu | Ile | — | — | Arg | Leu | Val |
| 10 | Leu | Asn | — | — | Arg | Leu | Val |
| 11 | Leu | Thr | Tyr† | Asp | Gly | Leu | Val |
| Constructs | |||||||
| Δ419 | Leu | Thr | Tyr | — | Arg | Leu | Val |
| Δ418_419 | Leu | Thr | — | — | Arg | Leu | Val |
| T417I/Δ418_419 | Leu | Ile | — | — | Arg | Leu | Val |
. | 416 . | 417 . | 418 . | 419 . | 420 . | 421 . | 422 . |
|---|---|---|---|---|---|---|---|
| Sources . | Leu . | Thr . | Tyr . | Asp . | Arg . | Leu . | Val . |
| Patient no. | |||||||
| 1* | Leu | Thr | Tyr | — | Arg | Leu | Val |
| 2* | Leu | Thr | Tyr | — | Trp | Gly | Val |
| 3* | Leu | Thr | Arg | — | Trp | Leu | Val |
| 4* | Leu | Thr | Ala | — | Arg | Leu | Val |
| 5 | Leu | Ile | — | — | Arg | Leu | Val |
| 6 | Leu | Thr | Tyr | — | — | Phe | Val |
| 7 | Leu | Val | — | — | Arg | Leu | Val |
| 8 | Leu | Arg | Gly | — | Arg | Leu | Val |
| 9 | Leu | Ile | — | — | Arg | Leu | Val |
| 10 | Leu | Asn | — | — | Arg | Leu | Val |
| 11 | Leu | Thr | Tyr† | Asp | Gly | Leu | Val |
| Constructs | |||||||
| Δ419 | Leu | Thr | Tyr | — | Arg | Leu | Val |
| Δ418_419 | Leu | Thr | — | — | Arg | Leu | Val |
| T417I/Δ418_419 | Leu | Ile | — | — | Arg | Leu | Val |
Sequences of patients no. 5 to no. 11 were published by Gari et al.4 “Sources” indicate the AA sequence of KIT wild type. The lower part of the panel shows the AA sequence of mutations generated by in vitro site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The correct sequence of all constructs was confirmed by complete nucleotide sequencing. The shaded areas indicate differences of the AA sequence of patient samples with exon 8 mutations to KIT-WT.
The protein sequence of KIT in exon 8 was reconstructed in 4 patients with AML M4eo that carried exon 8 mutations (patient nos. 1-4).9
ins Phe Phe.
All 3 representative KIT exon 8 mutations (Table 1, constructs) and the transforming D816V mutant were generated by in vitro mutagenesis. After transient expression in 293 cells and immunoblotting with a phospho-specific KIT antibody, no autophosphorylation of exon 8 mutants could be detected, whereas the D816V mutant was strongly autophosphorylated (data not shown). In cross-linking experiments, KIT exon 8 mutant T417I/Δ418_419 showed spontaneous receptor dimerization. In addition, a significantly higher ratio of cross-linked to noncross-linked receptor compared to KIT-wild-type (WT) was observed after SCF stimulation (data not shown).
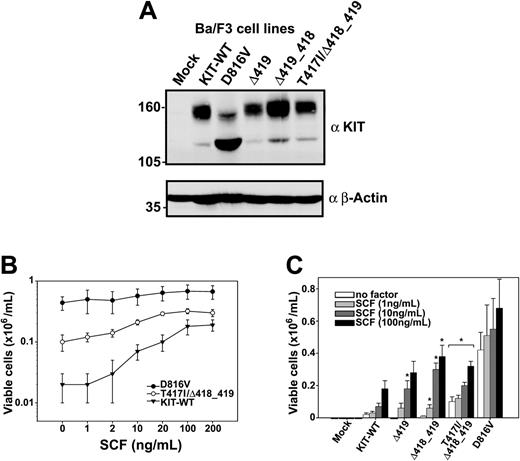
The constructs were then stably expressed in Ba/F3 cells and identical expression levels were confirmed by Western blotting (Figure 1A) and fluorescence-activated cell sorting (FACS) analyses (data not shown).
Expression of KIT receptor in transduced Ba/F3 cells, and hyperproliferative response of representative exon 8 mutants to SCF. (A) The expression of the indicated KIT constructs in Ba/F3 cells was confirmed by Western blotting using a polyclonal KIT antibody (α KIT) as described previously.18 The β-Actin reblot (αβ-Actin) served as a control for protein loading. (B-C) Ba/F3 cells transduced with the indicated constructs (B: •, D816V; ○, T417I/Δ418_419; ▾, KIT-WT) were grown in the presence or absence of SCF as indicated (C: open bars, no factor; light gray bars, 1 ng/mL; dark gray bars, 10 ng/mL; filled bars, 100 ng/mL) and counted after 72 hours in a standard hemacytometer after staining with trypan blue. All values obtained from D816V and T417I/Δ418_419 cell lines and all values of Δ419 and Δ418_419 labeled with an asterisk were significantly different from the corresponding wild-type value (P < .05). Figures show mean values and SDs from 3 independent experiments. Data were statistically tested using a 2-sided paired t test (Excel, Microsoft, Redmond, WA).
Expression of KIT receptor in transduced Ba/F3 cells, and hyperproliferative response of representative exon 8 mutants to SCF. (A) The expression of the indicated KIT constructs in Ba/F3 cells was confirmed by Western blotting using a polyclonal KIT antibody (α KIT) as described previously.18 The β-Actin reblot (αβ-Actin) served as a control for protein loading. (B-C) Ba/F3 cells transduced with the indicated constructs (B: •, D816V; ○, T417I/Δ418_419; ▾, KIT-WT) were grown in the presence or absence of SCF as indicated (C: open bars, no factor; light gray bars, 1 ng/mL; dark gray bars, 10 ng/mL; filled bars, 100 ng/mL) and counted after 72 hours in a standard hemacytometer after staining with trypan blue. All values obtained from D816V and T417I/Δ418_419 cell lines and all values of Δ419 and Δ418_419 labeled with an asterisk were significantly different from the corresponding wild-type value (P < .05). Figures show mean values and SDs from 3 independent experiments. Data were statistically tested using a 2-sided paired t test (Excel, Microsoft, Redmond, WA).
Ba/F3 cells expressing the T417I/Δ418_419 mutant proliferated spontaneously after cytokine deprivation, albeit at a lower rate than cells with KIT-D816V, whereas KIT-WT-transduced cells were unable to proliferate in the absence of interleukin 3 (IL-3; Figure 1B). Because exon 8 mutations are localized between immunoglobulin-like loops 4 (that is responsible for dimerization of the ligand-stimulated receptor10 ) and 5, we hypothesized that these mutants might exert their effects mainly on stimulation with SCF. At SCF concentrations ranging from 1 to 10 ng/mL, KIT-T417I/Δ418_419 mutant-expressing cells showed a significantly higher viability compared to KIT-WT-expressing cells (P < .01, for all values between 1 and 20 ng/mL SCF). Similar results were also observed for the Δ418_419 and Δ419 mutants, but they did not reach statistical significance at all data points (Figure 1C). Maximal stimulation of KIT-WT and exon 8 mutants in terms of proliferation could be fully reverted by the KIT inhibitor imatinib (Table 2) in contrast to the D816V mutant, which was resistant as described previously.11
Response of KIT transduced Ba/F3 cells to selective PTK inhibitors
. | IC50, μM . | . | . | ||
|---|---|---|---|---|---|
. | Imatinib . | PD98059 . | LY294002 . | ||
| KIT-WT | 0.07 | 6.0 | 1.5 | ||
| T417I/Δ418_419 | 0.05 | 8.5 | 2.5 | ||
| D816V | 3.0* | 10.0 | 2.5 | ||
| KIT-WT + IL-3 | 7.0* | > 25 | > 10 | ||
. | IC50, μM . | . | . | ||
|---|---|---|---|---|---|
. | Imatinib . | PD98059 . | LY294002 . | ||
| KIT-WT | 0.07 | 6.0 | 1.5 | ||
| T417I/Δ418_419 | 0.05 | 8.5 | 2.5 | ||
| D816V | 3.0* | 10.0 | 2.5 | ||
| KIT-WT + IL-3 | 7.0* | > 25 | > 10 | ||
Ba/F3 cells were grown in presence of 100 ng/mL SCF or 10 ng/mL IL-3 as indicated. Inhibitory concentration of 50% (IC50) values in the relevant cell lines of KIT inhibitor imatinib, MEK1 inhibitor PD98059, and Pl3-kinase inhibitor LY294002 were assessed by counting viable cells after 72 hours. Results represent means of 3 independent experiments. Final concentrations of dimethyl sulfoxide (DMSO) did not exceed 0.25%, which was nontoxic for Ba/F3 cells in control experiments. Values labeled with an asterisk were significantly different from the corresponding wild-type value.
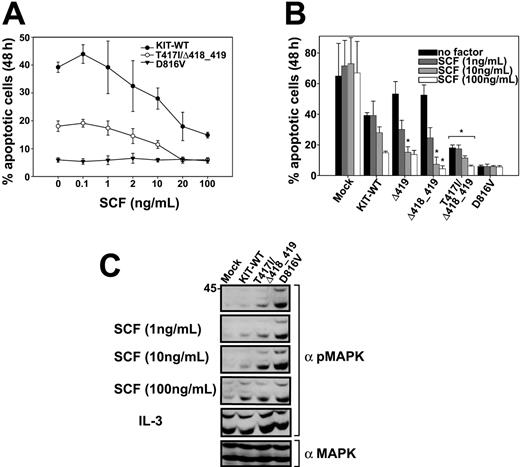
To support our findings, we analyzed the antiapoptotic activity of KIT exon 8 mutants. The KIT-D816V mutant showed a very low rate of apoptotic cell death that was independent from the concentration of SCF (Figure 2A-B). The T417I/Δ418_419 mutant conferred a significantly higher resistance to apoptotic cell death induced by IL-3 withdrawal to Ba/F3 cells compared to the KIT-WT construct (Figure 2A). A similar effect was seen in Δ418_419 mutant-expressing cells in the presence of SCF, but it was not statistically significant at all concentrations (Figure 2B).
Resistance of KIT mutants against apoptotic cell death and hyperphosphorylation of MAPK in response to SCF. (A-B) Ba/F3 cells transduced with the indicated constructs (A: •, KIT-WT; ○, T417I/Δ418_419; ▾, D816V) were grown for 48 hours in the presence or absence of SCF as indicated (B: ▪, no factor;  , 1 ng/mL;
, 1 ng/mL;  , 10 ng/mL; □, 100 ng/mL) and subsequently analyzed by flow cytometry after staining with annexin V-PE and 7-AAD. All values obtained from D816V and T417I/Δ418_419 cell lines and all values of Δ419 and Δ418_419 labeled with an asterisk were significantly different from the corresponding wild-type value (P < .05). Figures show mean values and SDs from 3 independent experiments. Data were statistically tested using a 2-sided paired t test (Excel, Microsoft). (C) Ba/F3 cells transduced with the indicated constructs were starved for 24 hours in the presence of 0.3% fetal bovine serum (FBS) and stimulated with the indicated concentrations of SCF or IL-3 (50 ng/mL) for 5 minutes at 37°C and 5% CO2. Crude lysates were analyzed by Western blotting using a polyclonal phospho-specific mitogen-activated protein kinase antibody (α pMAPK). Equal expression of MAPK in the same lysates was determined by immunoblotting with a polyclonal MAPK antibody (α MAPK); only the reblot for the 10 ng/mL dose of SCF is shown.
, 10 ng/mL; □, 100 ng/mL) and subsequently analyzed by flow cytometry after staining with annexin V-PE and 7-AAD. All values obtained from D816V and T417I/Δ418_419 cell lines and all values of Δ419 and Δ418_419 labeled with an asterisk were significantly different from the corresponding wild-type value (P < .05). Figures show mean values and SDs from 3 independent experiments. Data were statistically tested using a 2-sided paired t test (Excel, Microsoft). (C) Ba/F3 cells transduced with the indicated constructs were starved for 24 hours in the presence of 0.3% fetal bovine serum (FBS) and stimulated with the indicated concentrations of SCF or IL-3 (50 ng/mL) for 5 minutes at 37°C and 5% CO2. Crude lysates were analyzed by Western blotting using a polyclonal phospho-specific mitogen-activated protein kinase antibody (α pMAPK). Equal expression of MAPK in the same lysates was determined by immunoblotting with a polyclonal MAPK antibody (α MAPK); only the reblot for the 10 ng/mL dose of SCF is shown.
Resistance of KIT mutants against apoptotic cell death and hyperphosphorylation of MAPK in response to SCF. (A-B) Ba/F3 cells transduced with the indicated constructs (A: •, KIT-WT; ○, T417I/Δ418_419; ▾, D816V) were grown for 48 hours in the presence or absence of SCF as indicated (B: ▪, no factor;  , 1 ng/mL;
, 1 ng/mL;  , 10 ng/mL; □, 100 ng/mL) and subsequently analyzed by flow cytometry after staining with annexin V-PE and 7-AAD. All values obtained from D816V and T417I/Δ418_419 cell lines and all values of Δ419 and Δ418_419 labeled with an asterisk were significantly different from the corresponding wild-type value (P < .05). Figures show mean values and SDs from 3 independent experiments. Data were statistically tested using a 2-sided paired t test (Excel, Microsoft). (C) Ba/F3 cells transduced with the indicated constructs were starved for 24 hours in the presence of 0.3% fetal bovine serum (FBS) and stimulated with the indicated concentrations of SCF or IL-3 (50 ng/mL) for 5 minutes at 37°C and 5% CO2. Crude lysates were analyzed by Western blotting using a polyclonal phospho-specific mitogen-activated protein kinase antibody (α pMAPK). Equal expression of MAPK in the same lysates was determined by immunoblotting with a polyclonal MAPK antibody (α MAPK); only the reblot for the 10 ng/mL dose of SCF is shown.
, 10 ng/mL; □, 100 ng/mL) and subsequently analyzed by flow cytometry after staining with annexin V-PE and 7-AAD. All values obtained from D816V and T417I/Δ418_419 cell lines and all values of Δ419 and Δ418_419 labeled with an asterisk were significantly different from the corresponding wild-type value (P < .05). Figures show mean values and SDs from 3 independent experiments. Data were statistically tested using a 2-sided paired t test (Excel, Microsoft). (C) Ba/F3 cells transduced with the indicated constructs were starved for 24 hours in the presence of 0.3% fetal bovine serum (FBS) and stimulated with the indicated concentrations of SCF or IL-3 (50 ng/mL) for 5 minutes at 37°C and 5% CO2. Crude lysates were analyzed by Western blotting using a polyclonal phospho-specific mitogen-activated protein kinase antibody (α pMAPK). Equal expression of MAPK in the same lysates was determined by immunoblotting with a polyclonal MAPK antibody (α MAPK); only the reblot for the 10 ng/mL dose of SCF is shown.
To characterize the signaling properties of KIT exon 8 mutants, phosphorylation of the KIT downstream targets mitogen-activated protein kinase (MAPK), AKT, signal transducer and activator of transcription 3 (STAT3), and STAT5 were studied by immunoblotting with phospho-specific antibodies.
SCF induced a significantly stronger phosphorylation of MAPK in the T417I/Δ418_419 mutant compared to KIT-WT at physiologic concentrations of SCF (1-10 ng/mL),12 as shown in Figure 2C. The KIT-D816V-expressing cell line showed a strong constitutive MAPK phosphorylation that could not be further enhanced by exogenous SCF.
AKT was phosphorylated on Ser473 in response to SCF, but no significant difference of the T417I/Δ418_419 mutant compared to KIT-WT was observed (data not shown). In contrast to MAPK and AKT, SCF did not induce phosphorylation of STAT3 or STAT5 in KIT-WT and exon 8 mutant-expressing Ba/F3 cell lines (data not shown). To further support our findings, the mitogen-activated protein kinase kinase-1 (MEK1) inhibitor PD98059 and the phosphatidylinositol 3-kinase (PI3-kinase) inhibitor LY294002 were used in proliferation experiments. Both compounds significantly inhibited the growth of SCF-stimulated KIT-WT, exon 8 mutant, and D816V Ba/F3 cells at concentrations that did not significantly affect IL-3-induced proliferation (Table 2). PD98059 also inhibited the autonomous proliferation of Ba/F3 cells expressing the T417I/Δ418_419 mutant (data not shown).
In contrast to the previously described KIT D816 mutations in patients with AML,3 exon 8 mutations are localized in the extracellular portion of the receptor. This class of mutations has been described in several receptor tyrosine kinases including KIT, FMS, fibroblast growth factor receptor (FGFR), epidermal growth factor receptor (EGFR), and TRKA, although they differ significantly in their transforming potential.8,13-17 Interestingly, colony-stimulating factor-1 receptor (CSF-1R) point mutants found in v-FMS (AA 301 and 374) are also located in the immunoglobulin-like domain 48 and show a very similar phenotype compared to the KIT exon 8 mutations. These mutants have a weak transforming potential in Rat and FDCP-1 cells that can be enhanced by ligand stimulation. In addition, they neither induce constitutive FMS autophosphorylation nor affect surface expression or internalization of the receptor. The immunoglobulin-like domain 4 is reported to mediate receptor dimerization, but not ligand binding.10 Mutations in this area might therefore lower the threshold of ligand concentration that is needed for receptor activation. This fact could explain the different biologic effects of exon 8 mutations that cause hyperactivation in response to SCF in contrast to exon 17 mutations at Asp816 that lead to ligand-independent, direct activation of the kinase domain.
Taken together, our data show that KIT exon 8 mutations represent gain-of-function mutations that are sensitive to KIT-selective protein tyrosine kinase (PTK) inhibitors. These findings point to an important functional role for KIT exon 8 mutations in the pathogenesis of CBF-AML and provide the basis for a targeted therapy with KIT PTK inhibitors.
Prepublished online as Blood First Edition Paper, December 23, 2004; DOI 10.1182/blood-2004-06-2068.
Presented in part in oral form at the 46th annual meeting of the American Society of Hematology, San Diego, CA, December 4-7, 2004.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG Sp556/3-1) and the Deutsche Krebshilfe (10-1997-Sp2).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr Elisabeth Buchdunger and Novartis for providing imatinib and Leonie K. Ashman (Institute of Medical and Veterinary Science, Adelaide, Australia) for providing cDNA for KIT-WT. We also thank Ksenia Bagrintseva for helpful discussions; Susan King and Stefan Bohlander for critical reading of the manuscript; and Ruth Schwab, Sabine Eichenlaub, and Karin Nispel for excellent technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal